In Vitro Replication Inhibitory Activity of Xanthorrhizol against Severe Acute Respiratory Syndrome Coronavirus 2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents, Plasmids, and Antibodies
2.2. Cell Culture
2.3. Viruses and Plaque Assay
2.4. Cell Viability Assay
2.5. Real-Time Reverse-Transcription Quantitative PCR (RT-qPCR)
2.6. SCoV2 Entry Assay
2.7. SCoV1 Subgenomic Replicon Replication Assay
2.8. Immunoblotting Analysis
2.9. Expression and Purification of Recombinant SCoV2 Enzymes
2.10. Nsp12 RdRp Assay
2.11. Nsp5 Protease Assay
2.12. Nsp13 Helicase Assay
2.13. Interferon β Reporter Assay
2.14. Data Analysis and Statistical Analysis
3. Results
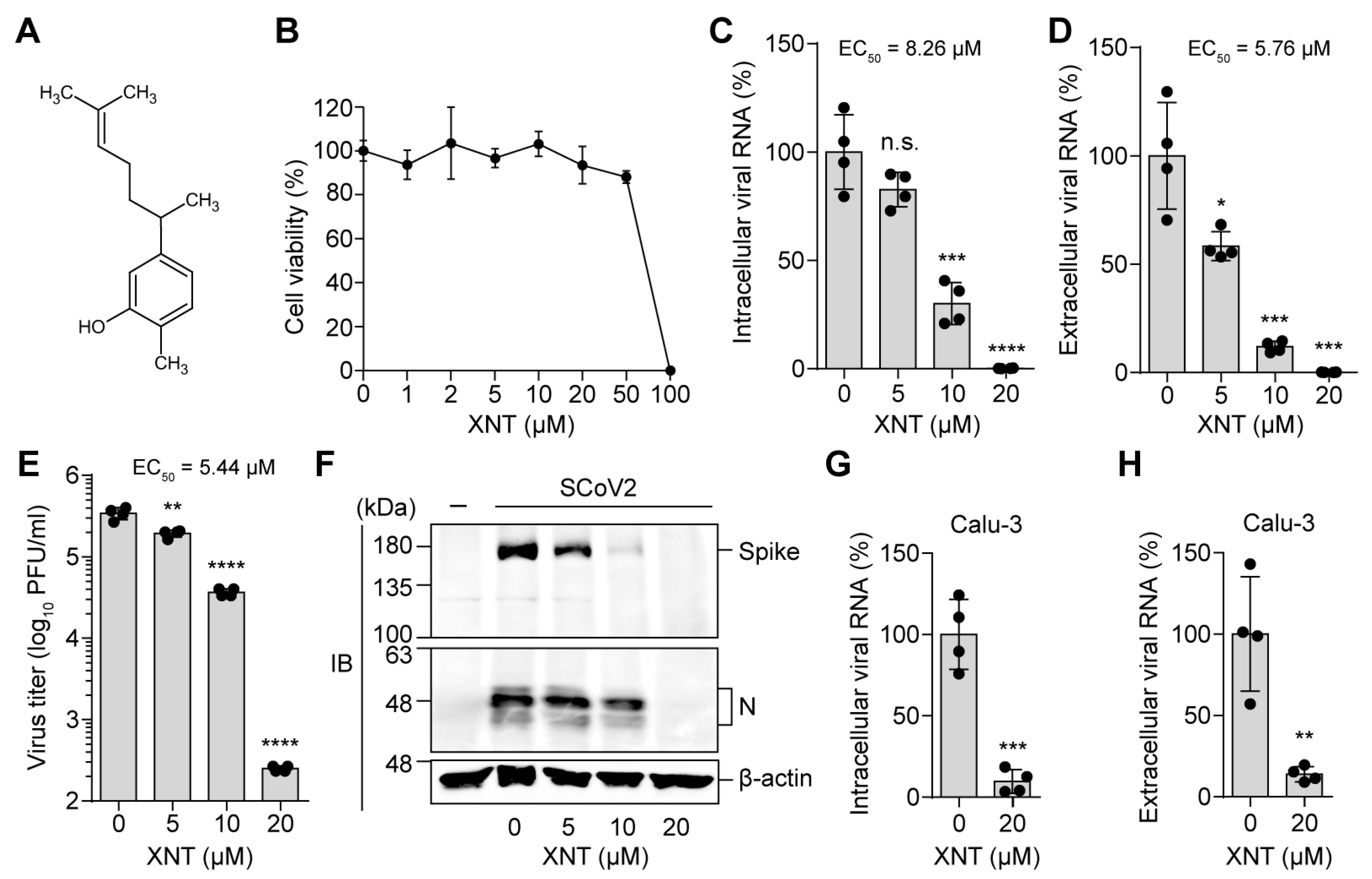
3.1. Identification of XNT as an Antiviral Agent against SCoV2
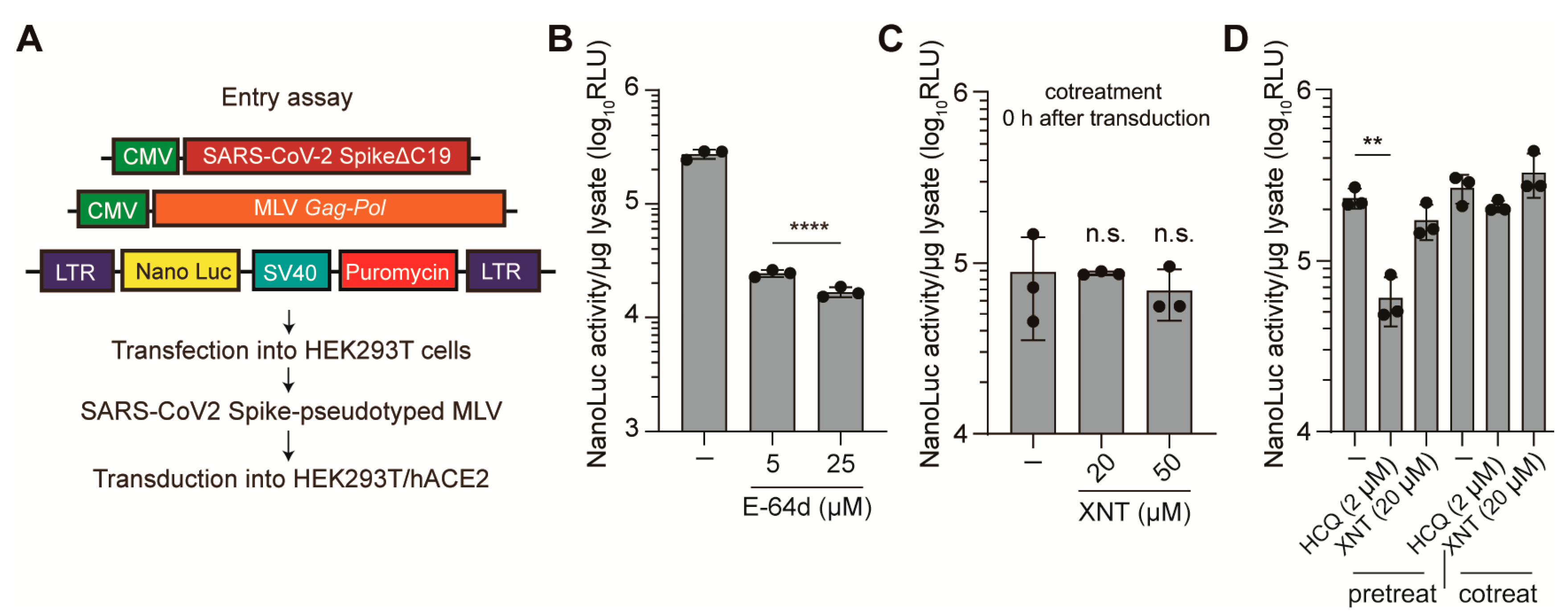
3.2. Effect of XNT on SCoV2 Cellular Entry
3.3. Inhibition of SCoV1 Subgenomic Replicon Replication by XNT
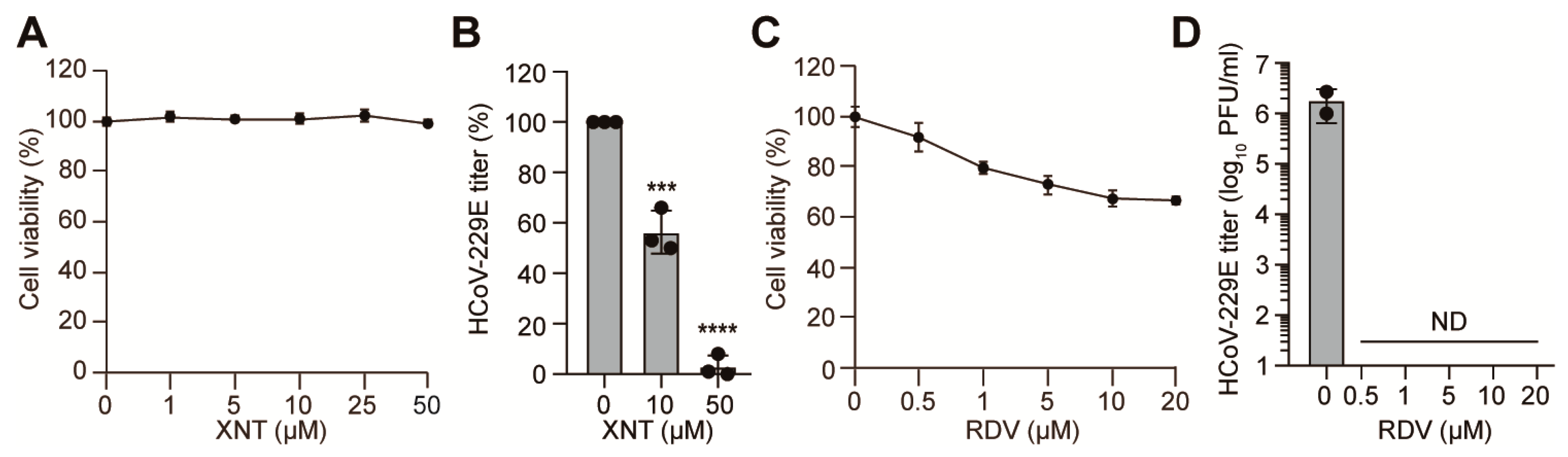
3.4. Inhibition of HCoV-229E by XNT
3.5. Antiviral Efficacy of XNT against SCoV2 Variants
3.6. XNT Lacks Inhibitory Activity against Nsp5, Nsp12, and Nsp13
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Doxey, A.C.; Mossman, K.; Irving, A.T. Unraveling the zoonotic origin and transmission of SARS-CoV-2. Trends Ecol. Evol. 2021, 36, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Wilson, N.; Kvalsvig, A.; Barnard, L.T.; Baker, M.G. Case-fatality risk estimates for COVID-19 calculated by using a lag time for fatality. Emerg. Infect. Dis. 2020, 26, 1339–1441. [Google Scholar] [CrossRef]
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 17 September 2021).
- V’kovski, P.; Kratzel, A.; Steiner, S.; Stalder, H.; Thiel, V. Coronavirus biology and replication: Implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021, 19, 155–170. [Google Scholar] [CrossRef]
- Kasuga, Y.; Zhu, B.; Jang, K.-J.; Yoo, J.-S. Innate immune sensing of coronavirus and viral evasion strategies. Exp. Mol. Med. 2021, 53, 723–736. [Google Scholar] [CrossRef]
- Christian, M.D.; Poutanen, S.M.; Loutfy, M.R.; Muller, M.P.; Low, D.E. Severe acute respiratory syndrome. Clin. Infect. Dis. 2004, 38, 1420–1427. [Google Scholar] [CrossRef]
- Tai, D.Y. Pharmacologic treatment of SARS: Current knowledge and recommendations. Ann. Acad. Med. Singap. 2007, 36, 438–443. [Google Scholar] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the treatment of Covid-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.M.; Zhang, X. The World Medicines Situation 2011, Traditional Medicines: Global Situation, Issues and Challenges; World Health Organization: Geneva, Switzerland, 2011. [Google Scholar]
- Nugraha, R.V.; Ridwansyah, H.; Ghozali, M.; Khairani, A.F.; Atik, N. Traditional herbal medicine candidates as complementary treatments for COVID-19: A review of their mechanisms, pros and cons. Evid.-Based Complementary Altern. Med. 2020, 2020, 2560645. [Google Scholar] [CrossRef]
- Rizzuti, B.; Grande, F.; Conforti, F.; Jimenez-Alesanco, A.; Ceballos-Laita, L.; Ortega-Alarcon, D.; Vega, S.; Reyburn, H.T.; Abian, O.; Velazquez-Campoy, A. Rutin is a low micromolar inhibitor of SARS-CoV-2 main protease 3CLpro: Implications for drug design of quercetin analogs. Biomedicines 2021, 9, 375. [Google Scholar] [CrossRef]
- Tagde, P.; Tagde, S.; Tagde, P.; Bhattacharya, T.; Monzur, S.M.; Rahman, M.H.; Otrisal, P.; Behl, T.; Ul Hassan, S.S.; Abdel-Daim, M.M.; et al. Nutraceuticals and herbs in reducing the risk and improving the treatment of COVID-19 by targeting SARS-CoV-2. Biomedicines 2021, 9, 1266. [Google Scholar] [CrossRef]
- Rahmat, E.; Lee, J.; Kang, Y. Javanese turmeric (Curcuma xanthorrhiza Roxb.): Ethnobotany, phytochemistry, biotechnology, and pharmacological activities. Evid.-Based Complementary Altern. Med. 2021, 2021, 9960813. [Google Scholar] [CrossRef]
- Oon, S.F.; Nallappan, M.; Tee, T.T.; Shohaimi, S.; Kassim, N.K.; Sa’ariwijaya, M.S.F.; Cheah, Y.H. Xanthorrhizol: A review of its pharmacological activities and anticancer properties. Cancer Cell Int. 2015, 15, 100. [Google Scholar] [CrossRef] [Green Version]
- Du, Z.T.; Zheng, S.; Chen, G.; Lv, D. A short synthesis of bisabolane sesquiterpenes. Molecules 2011, 16, 8053–8061. [Google Scholar] [CrossRef]
- Jantan, I.; Saputri, F.C.; Qaisar, M.N.; Buang, F. Correlation between chemical composition of Curcuma domestica and Curcuma xanthorrhiza and their antioxidant effect on human low-density lipoprotein oxidation. Evid.-Based Complementary Altern. Med. 2012, 2012, 438356. [Google Scholar] [CrossRef] [Green Version]
- Balasubramanian, A.; Pilankatta, R.; Teramoto, T.; Sajith, A.M.; Nwulia, E.; Kulkarni, A.; Padmanabhan, R. Inhibition of dengue virus by curcuminoids. Antivir. Res. 2019, 162, 71–78. [Google Scholar] [CrossRef]
- Mounce, B.C.; Cesaro, T.; Carrau, L.; Vallet, T.; Vignuzzi, M. Curcumin inhibits Zika and chikungunya virus infection by inhibiting cell binding. Antivir. Res. 2017, 142, 148–157. [Google Scholar] [CrossRef]
- Chen, T.Y.; Chen, D.Y.; Wen, H.W.; Ou, J.L.; Chiou, S.S.; Chen, J.M.; Wong, M.L.; Hsu, W.L. Inhibition of enveloped viruses infectivity by curcumin. PLoS ONE 2013, 8, e62482. [Google Scholar] [CrossRef] [Green Version]
- Bormann, M.; Alt, M.; Schipper, L.; van de Sand, L.; Le-Trilling, V.T.K.; Rink, L.; Heinen, N.; Madel, R.J.; Otte, M.; Wuensch, K.; et al. Turmeric root and its bioactive ingredient curcumin effectively neutralize SARS-CoV-2 in vitro. Viruses 2021, 13, 1914. [Google Scholar] [CrossRef]
- Wahyuni, T.S.; Permatasari, A.A.; Widiandani, T.; Fuad, A.; Widyawaruyanti, A.; Aoki-Utsubo, C.; Hotta, H. Antiviral activities of Curcuma genus against hepatitis C virus. Nat. Prod. Commun. 2018, 13, 1579–1582. [Google Scholar] [CrossRef] [Green Version]
- Ahn, D.G.; Lee, W.; Choi, J.K.; Kim, S.J.; Plant, E.P.; Almazan, F.; Taylor, D.R.; Enjuanes, L.; Oh, J.W. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 2011, 91, 1–10. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, J.H.; Kim, Y.G.; Lim, H.S.; Oh, J.W. Protein kinase C-related kinase 2 regulates hepatitis C virus RNA polymerase function by phosphorylation. J. Biol. Chem. 2004, 279, 50031–50041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, K.-O.; Sosnovtsev, S.V.; Belliot, G.; King, A.D.; Green, K.Y. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology 2006, 353, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Lee, Y.J.; Yoon, J.S.; Ahn, J.Y.; Kim, J.H.; Choi, J.Y.; Oh, J.W. Genome sequences of two GH clade SARS-CoV-2 strains isolated from patients with COVID-19 in South Korea. Microbiol. Resour. Announc. 2021, 10, e01384-20. [Google Scholar] [CrossRef]
- Kim, M.; Cho, H.; Lee, S.H.; Park, W.J.; Kim, J.M.; Moon, J.S.; Kim, G.W.; Lee, W.; Jung, H.G.; Yang, J.S.; et al. An infectious cDNA clone of a growth attenuated Korean isolate of MERS coronavirus KNIH002 in clade B. Emerg. Microbes Infect. 2020, 9, 2714–2726. [Google Scholar] [CrossRef]
- Zust, R.; Cervantes-Barragan, L.; Habjan, M.; Maier, R.; Neuman, B.W.; Ziebuhr, J.; Szretter, K.J.; Baker, S.C.; Barchet, W.; Diamond, M.S.; et al. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 2011, 12, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.; Lee, J.E.; Cho, H.; Jung, H.G.; Lee, W.; Seo, H.Y.; Lee, S.H.; Ahn, D.G.; Kim, S.J.; Yu, J.W.; et al. Antiviral efficacy of orally delivered neoagarohexaose, a nonconventional TLR4 agonist, against norovirus infection in mice. Biomaterials 2020, 263, 120391. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, J.; Martin, E.T.; Heugel, J.; Wright, N.; Morrow, R.; Englund, J.A. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics 2007, 119, e70–e76. [Google Scholar] [CrossRef]
- China National Institute for Viral Disease Control and Prevention. Specific Primers and Probes for Detection 2019 Novel Coronavirus. Available online: http://ivdc.chinacdc.cn/kyjz/202001/t20200121_211337.html (accessed on 10 October 2021).
- Millet, J.K.; Tang, T.; Nathan, L.; Jaimes, J.A.; Hsu, H.L.; Daniel, S.; Whittaker, G.R. Production of pseudotyped particles to study highly pathogenic coronaviruses in a biosafety level 2 setting. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [Green Version]
- Ujike, M.; Huang, C.; Shirato, K.; Makino, S.; Taguchi, F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016, 97, 1853–1864. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, D.G.; Choi, J.K.; Taylor, D.R.; Oh, J.W. Biochemical characterization of a recombinant SARS coronavirus nsp12 RNA-dependent RNA polymerase capable of copying viral RNA templates. Arch. Virol. 2012, 157, 2095–2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Sacco, M.D.; Hurst, B.; Townsend, J.A.; Hu, Y.; Szeto, T.; Zhang, X.; Tarbet, B.; Marty, M.T.; Chen, Y.; et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020, 30, 678–692. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Tanner, J.A.; Zheng, B.J.; Watt, R.M.; He, M.L.; Lu, L.Y.; Jiang, J.Q.; Shum, K.T.; Lin, Y.P.; Wong, K.L.; et al. Bismuth complexes inhibit the SARS coronavirus. Angew. Chem. Int. Ed. 2007, 46, 6464–6468. [Google Scholar] [CrossRef]
- Emeny, J.M.; Morgan, M.J. Regulation of the interferon system: Evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 1979, 43, 247–252. [Google Scholar] [CrossRef]
- Chu, H.; Chan, J.F.-W.; Yuen, T.T.-T.; Shuai, H.; Yuan, S.; Wang, Y.; Hu, B.; Yip, C.C.-Y.; Tsang, J.O.-L.; Huang, X.; et al. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 2020, 1, e14–e23. [Google Scholar] [CrossRef]
- Bravo, J.P.K.; Dangerfield, T.L.; Taylor, D.W.; Johnson, K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 2021, 81, 1548–1552. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell 2020, 182, 685–712. [Google Scholar] [CrossRef]
- Kaur, U.; Chakrabarti, S.S.; Ojha, B.; Pathak, B.K.; Singh, A.; Saso, L.; Chakrabarti, S. Targeting host cell proteases to prevent SARS-CoV-2 invasion. Curr. Drug Targets 2021, 22, 192–201. [Google Scholar] [CrossRef]
- Lee, W.; Lee, S.H.; Ahn, D.G.; Cho, H.; Sung, M.H.; Han, S.H.; Oh, J.W. The antiviral activity of poly-γ-glutamic acid, a polypeptide secreted by Bacillus sp., through induction of CD14-dependent type I interferon responses. Biomaterials 2013, 34, 9700–9708. [Google Scholar] [CrossRef]
- Ziebuhr, J.; Snijder, E.J.; Gorbalenya, A.E. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 2000, 81, 853–879. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, T.; Huang, M.; Wu, D.; Ren, Y.; Zhang, X.; Han, Y.; Mu, J.; Wang, R.; Qiu, Y.; Zhang, D.Y.; et al. SARS-coronavirus-2 Nsp13 possesses NTPase and RNA helicase activities that can be inhibited by bismuth salts. Virol. Sin. 2020, 35, 321–329. [Google Scholar] [CrossRef]
- Rohaim, M.A.; El Naggar, R.F.; Clayton, E.; Munir, M. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 2021, 150, 104641. [Google Scholar] [CrossRef]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Ward, A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019, 10, 2342. [Google Scholar] [CrossRef] [Green Version]
- Guendel, I.; Agbottah, E.T.; Kehn-Hall, K.; Kashanchi, F. Inhibition of human immunodeficiency virus type-1 by cdk inhibitors. AIDS Res. Ther. 2010, 7, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holcakova, J.; Tomasec, P.; Bugert, J.J.; Wang, E.C.; Wilkinson, G.W.; Hrstka, R.; Krystof, V.; Strnad, M.; Vojtesek, B. The inhibitor of cyclin-dependent kinases, olomoucine II, exhibits potent antiviral properties. Antivir. Chem. Chemother. 2010, 20, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Perwitasari, O.; Yan, X.; O’Donnell, J.; Johnson, S.; Tripp, R.A. Repurposing kinase inhibitors as antiviral agents to control influenza A virus replication. Assay Drug Dev. Technol. 2015, 13, 638–649. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.; Lee, E.M.; Wen, Z.; Cheng, Y.; Huang, W.-K.; Qian, X.; Tcw, J.; Kouznetsova, J.; Ogden, S.C.; Hammack, C.; et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016, 22, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-J.; Park, K.-K.; Chung, W.-Y.; Hwang, J.-K.; Lee, S.K. Xanthorrhizol, a natural sesquiterpenoid, induces apoptosis and growth arrest in HCT116 human colon cancer cells. J. Pharmacol. Sci. 2009, 111, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Du, J.; Wei, N.; Guan, T.; Xu, H.; An, J.; Pritchard, K.A., Jr.; Shi, Y. Inhibition of CDKS by roscovitine suppressed LPS-induced NO production through inhibiting NFκB activation and BH4 biosynthesis in macrophages. Am. J. Physiol. Cell Physiol. 2009, 297, C742–C749. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.K.; Hong, C.H.; Huh, S.K.; Kim, S.S.; Oh, O.J.; Min, H.Y.; Park, K.K.; Chung, W.Y.; Hwang, J.K. Suppressive effect of natural sesquiterpenoids on inducible cyclooxygenase (COX-2) and nitric oxide synthase (iNOS) activity in mouse macrophage cells. J. Environ. Pathol. Toxicol. Oncol. 2002, 21, 141–148. [Google Scholar] [CrossRef]
- Kim, S.H.; Hong, K.O.; Chung, W.-Y.; Hwang, J.K.; Park, K.-K. Abrogation of cisplatin-induced hepatotoxicity in mice by xanthorrhizol is related to its effect on the regulation of gene transcription. Toxicol. Appl. Pharmacol. 2004, 196, 346–355. [Google Scholar]
- Chung, W.Y.; Park, J.H.; Kim, M.J.; Kim, H.O.; Hwang, J.K.; Lee, S.K.; Park, K.K. Xanthorrhizol inhibits 12-O-tetradecanoylphorbol-13-acetate-induced acute inflammation and two-stage mouse skin carcinogenesis by blocking the expression of ornithine decarboxylase, cyclooxygenase-2 and inducible nitric oxide synthase through mitogen-activated protein kinases and/or the nuclear factor-κB. Carcinogenesis 2007, 28, 1224–1231. [Google Scholar]
- Lim, C.S.; Jin, D.-Q.; Mok, H.; Oh, S.J.; Lee, J.U.; Hwang, J.K.; Ha, I.; Han, J.-S. Antioxidant and antiinflammatory activities of xanthorrhizol in hippocampal neurons and primary cultured microglia. J. Neurosci. Res. 2005, 82, 831–838. [Google Scholar] [CrossRef]
- King, C.; Sprent, J. Dual nature of type I interferons in SARS-CoV-2-induced inflammation. Trends Immunol. 2021, 42, 312–322. [Google Scholar] [CrossRef]
- Mazewski, C.; Perez, R.E.; Fish, E.N.; Platanias, L.C. Type I interferon (IFN)-regulated activation of canonical and non-canonical signaling pathways. Front. Immunol. 2020, 11, 606456. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.; Azfaralariff, A.; Law, D.; Najm, A.A.; Sanusi, S.A.; Lim, S.J.; Cheah, Y.H.; Fazry, S. Comprehensive computational target fishing approach to identify xanthorrhizol putative targets. Sci. Rep. 2021, 11, 1594. [Google Scholar] [CrossRef] [PubMed]
- Nusinzon, I.; Horvath, C.M. Positive and negative regulation of the innate antiviral response and beta interferon gene expression by deacetylation. Mol. Cell. Biol. 2006, 26, 3106–3113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shapira, L.; Ralph, M.; Tomer, E.; Cohen, S.; Kobiler, O. Histone deacetylase inhibitors reduce the number of herpes simplex virus-1 genomes initiating expression in individual cells. Front. Microbiol. 2016, 7, 1970. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, Y.; Boukari, H.; Banerjee, I.; Sbalzarini, I.F.; Horvath, P.; Helenius, A. Histone deacetylase 8 is required for centrome cohesion and influenza A virus entry. PLoS Pathog. 2011, 7, e1002316. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.C.; Lin, C.C.; Lin, Y.H.; Supriyatna, S.; Teng, C.W. Protective and therapeutic effects of Curcuma xanthorrhiza on hepatotoxin-induced liver damage. Am. J. Chin. Med. 1995, 23, 243–254. [Google Scholar]
- Yamazaki, M.; Maebayashi, Y.; Iwase, N.; Kaneko, T. Studies on pharmacologically active principles from Indonesian crude drugs. I. Principle prolonging pentobarbital-induced sleeping time from Curcuma xanthorrhiza Roxb. Chem. Pharm. Bull. 1988, 36, 2070–2074. [Google Scholar] [CrossRef] [Green Version]
- Devaraj, S.; Esfahani, A.S.; Ismail, S.; Ramanathan, S.; Yam, M.F. Evaluation of the antinociceptive activity and acute oral toxicity of standardized ethanolic extract of the rhizome of Curcuma xanthorrhiza Roxb. Molecules 2010, 15, 2925–2934. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, M.; Cho, H.; Ahn, D.-G.; Jung, H.-G.; Seo, H.Y.; Kim, J.-S.; Lee, Y.-J.; Choi, J.Y.; Park, I.H.; Shin, J.-S.; et al. In Vitro Replication Inhibitory Activity of Xanthorrhizol against Severe Acute Respiratory Syndrome Coronavirus 2. Biomedicines 2021, 9, 1725. https://doi.org/10.3390/biomedicines9111725
Kim M, Cho H, Ahn D-G, Jung H-G, Seo HY, Kim J-S, Lee Y-J, Choi JY, Park IH, Shin J-S, et al. In Vitro Replication Inhibitory Activity of Xanthorrhizol against Severe Acute Respiratory Syndrome Coronavirus 2. Biomedicines. 2021; 9(11):1725. https://doi.org/10.3390/biomedicines9111725
Chicago/Turabian StyleKim, Minwoo, Hee Cho, Dae-Gyun Ahn, Hae-Gwang Jung, Han Young Seo, Ji-Su Kim, Youn-Jung Lee, Jun Yong Choi, In Ho Park, Jeon-Soo Shin, and et al. 2021. "In Vitro Replication Inhibitory Activity of Xanthorrhizol against Severe Acute Respiratory Syndrome Coronavirus 2" Biomedicines 9, no. 11: 1725. https://doi.org/10.3390/biomedicines9111725
APA StyleKim, M., Cho, H., Ahn, D.-G., Jung, H.-G., Seo, H. Y., Kim, J.-S., Lee, Y.-J., Choi, J. Y., Park, I. H., Shin, J.-S., Kim, S.-J., & Oh, J.-W. (2021). In Vitro Replication Inhibitory Activity of Xanthorrhizol against Severe Acute Respiratory Syndrome Coronavirus 2. Biomedicines, 9(11), 1725. https://doi.org/10.3390/biomedicines9111725





