Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Culture Conditions and DNA Isolation
2.2. Whole-Genome Sequencing and Genome Annotation
2.3. Phylogenetic Analysis
2.4. Detection of Genetic Elements Associated with Probiotic Characteristics
2.5. Cell-Free Supernatant Preparation
2.6. Sulforhodamine B Colorimetric Assay
2.7. Colony Formation Assay
2.8. Wound Healing Assay
2.9. Statistical Analysis
3. Results
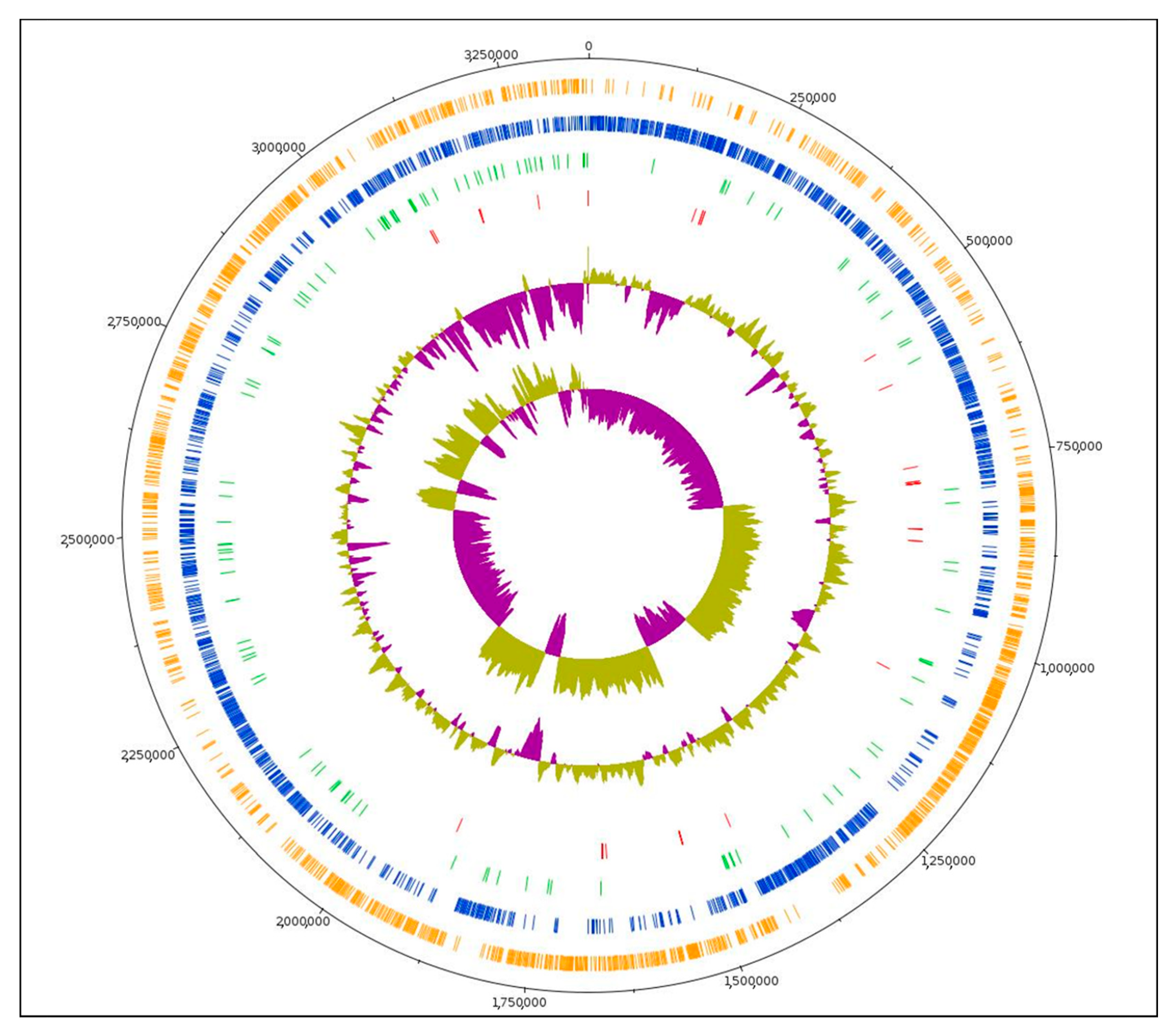
3.1. Genome Features
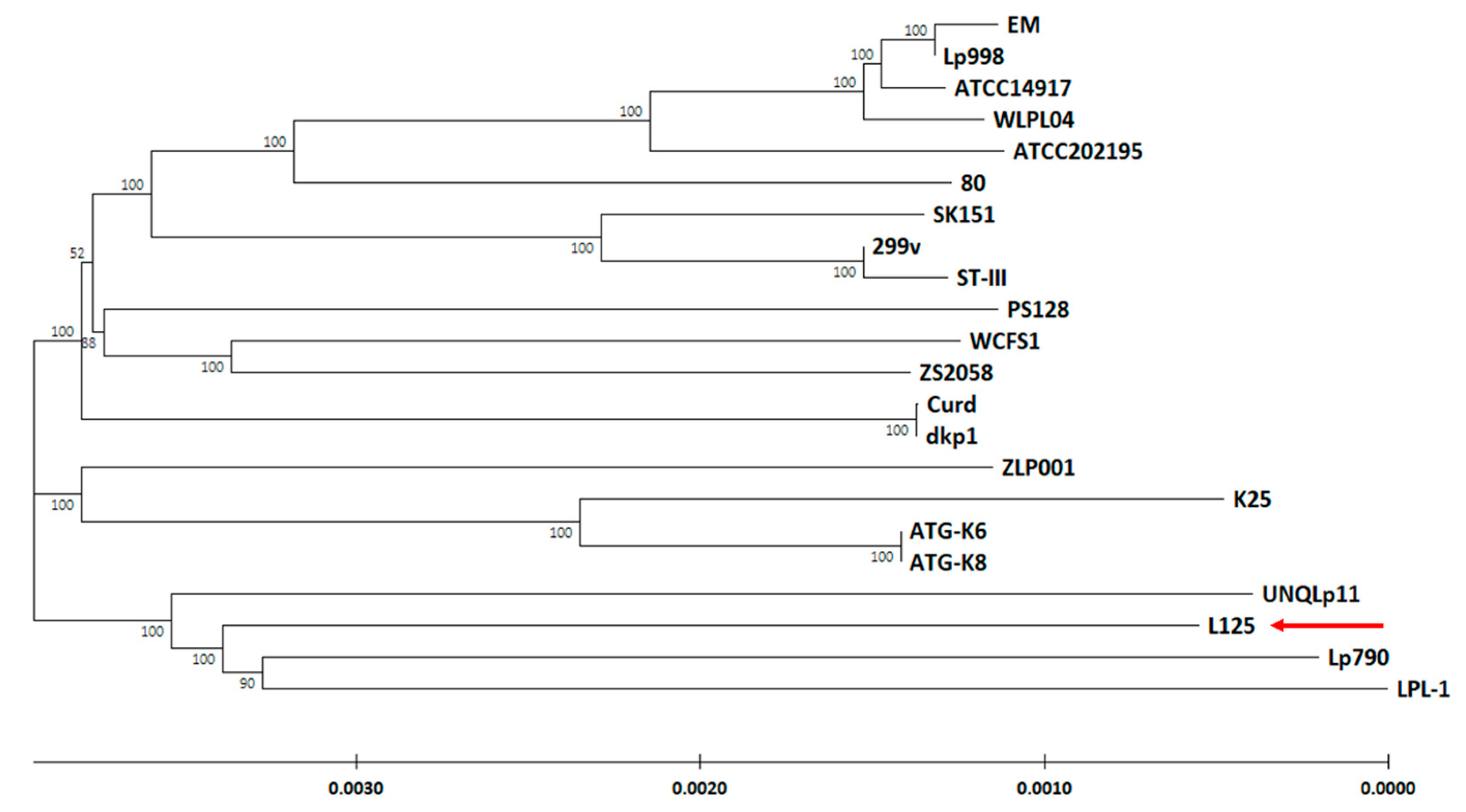
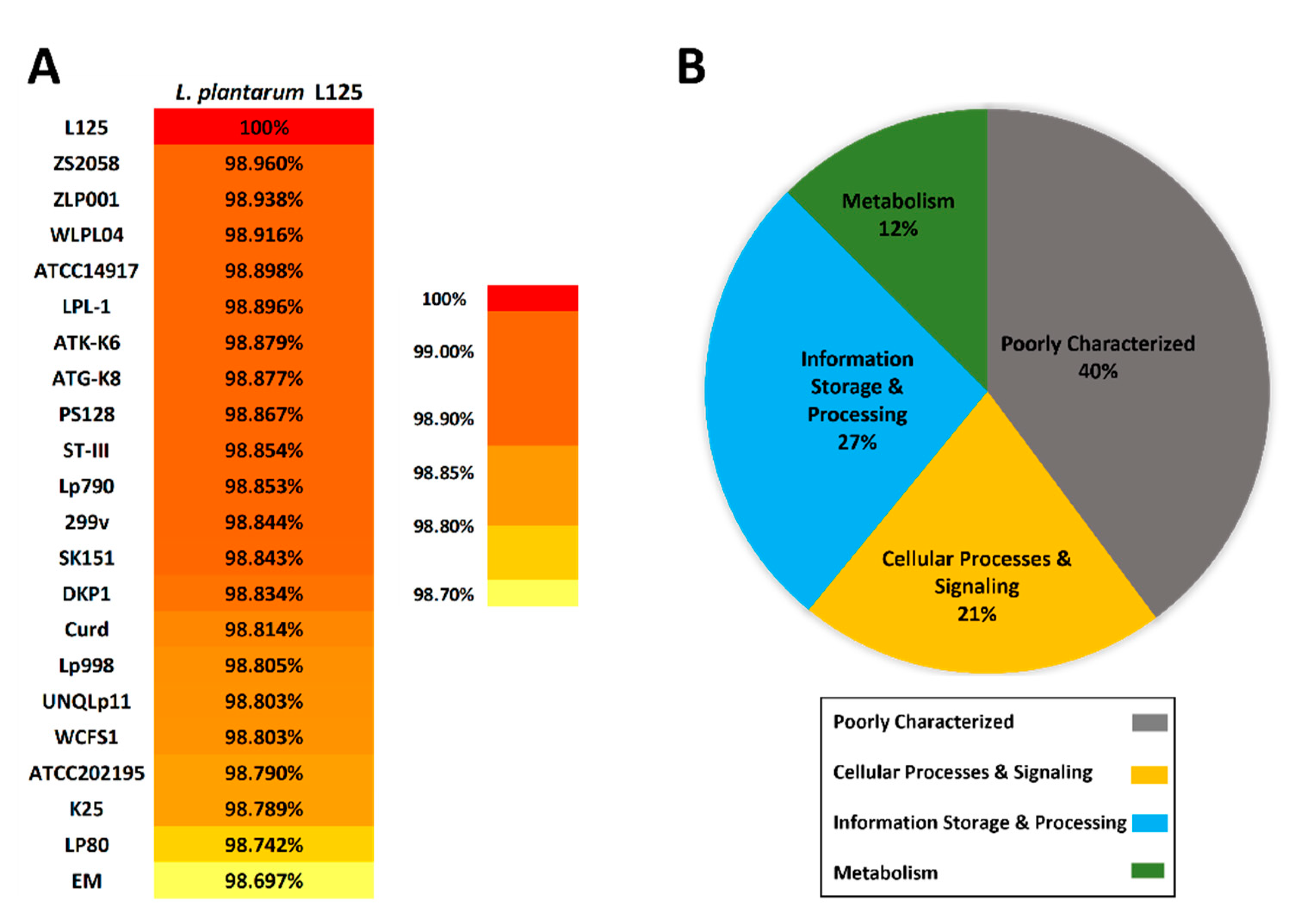
3.2. Phylogenetic Analysis and Unique Genome Characteristics of L. plantarum L125
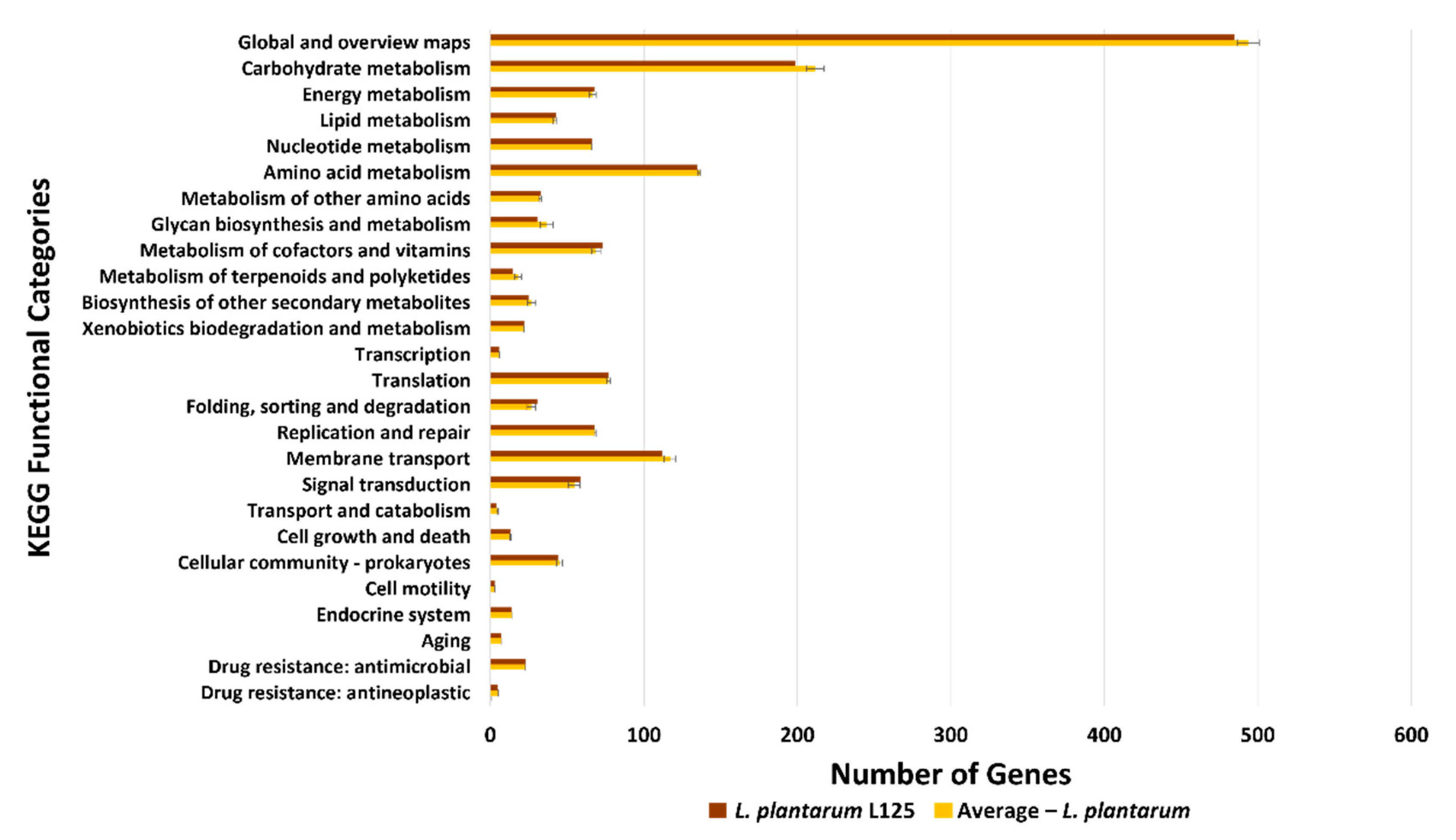
3.3. Functional Classification
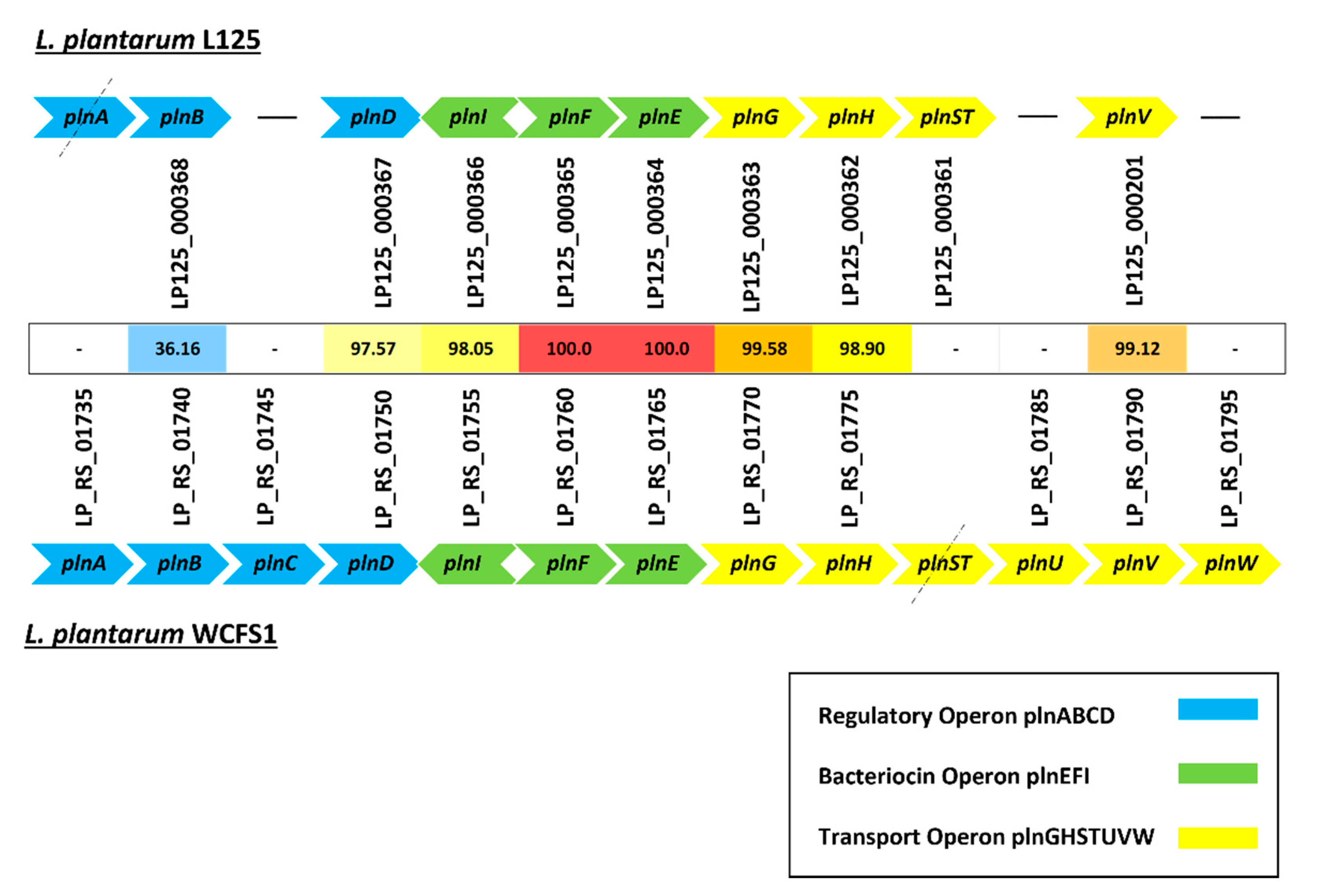
3.4. Identification of Genes Implicated in Stress Response, Microbe–Host Interactions and Bacteriocin Biosynthesis
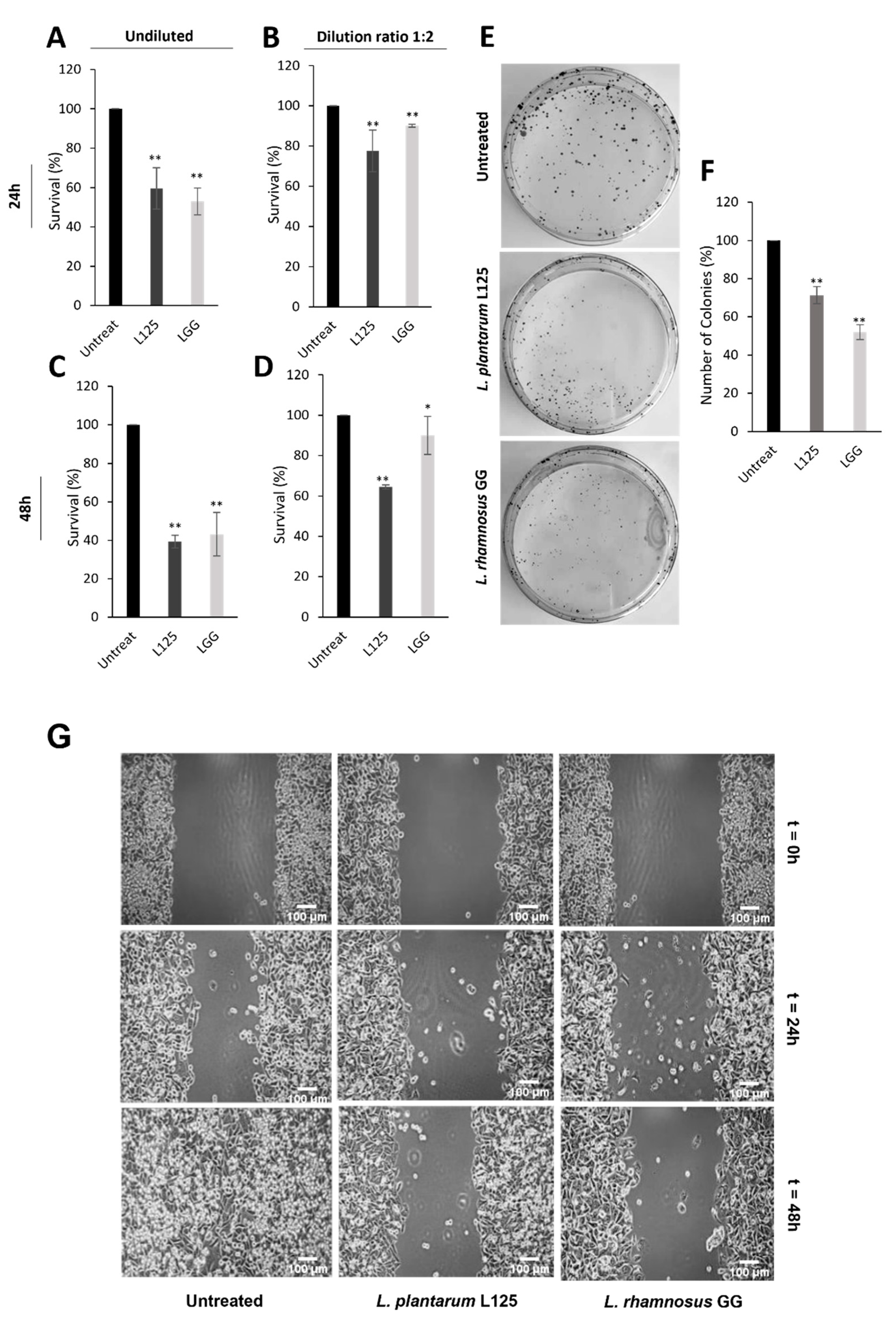
3.5. Investigation of Potential Health-Promoting Effects Induced by L. plantarum L125
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siezen, R.J.; Tzeneva, V.A.; Castioni, A.; Wels, M.; Phan, H.T.; Rademaker, J.L.; Starrenburg, M.J.; Kleerebezem, M.; Molenaar, D.; van Hylckama Vlieg, J.E. Phenotypic and genomic diversity of Lactobacillus plantarum strains isolated from various environmental niches. Environ. Microbiol. 2010, 12, 758–773. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.; Harris, H.M.B.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J.; et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- Martino, M.E.; Bayjanov, J.R.; Caffrey, B.E.; Wels, M.; Joncour, P.; Hughes, S.; Gillet, B.; Kleerebezem, M.; van Hijum, S.A.; Leulier, F. Nomadic lifestyle of Lactobacillus plantarum revealed by comparative genomics of 54 strains isolated from different habitats. Environ. Microbiol. 2016, 18, 4974–4989. [Google Scholar] [CrossRef]
- Benbara, T.; Lalouche, S.; Drider, D.; Bendali, F. Lactobacillus plantarum S27 from chicken faeces as a potential probiotic to replace antibiotics: In vivo evidence. Benef. Microbes. 2020, 11, 163–173. [Google Scholar] [CrossRef]
- Ruiz, M.J.; Zbrun, M.V.; Signorini, M.L.; Zimmermann, J.A.; Soto, L.P.; Rosmini, M.R.; Frizzo, L.S. In vitro screening and in vivo colonization pilot model of Lactobacillus plantarum LP5 and Campylobacter coli DSPV 458 in mice. Arch. Microbiol. 2021, 203, 4161–4171. [Google Scholar] [CrossRef] [PubMed]
- Nordström, E.A.; Teixeira, C.; Montelius, C.; Jeppsson, B.; Larsson, N. Lactiplantibacillus plantarum 299v (LP299V®): Three decades of research. Benef. Microbes. 2021, 12, 441–465. [Google Scholar] [CrossRef] [PubMed]
- Farhangfar, A.; Gandomi, H.; Basti, A.A.; Misaghi, A.; Noori, N. Study of growth kinetic and gastrointestinal stability of acid-bile resistant Lactobacillus plantarum strains isolated from Siahmazgi traditional cheese. Vet. Res. Forum 2021, 12, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liu, Y.; Sun, M.; Zhang, H.; Mu, G.; Tuo, Y. Physiological function analysis of Lactobacillus plantarum Y44 based on genotypic and phenotypic characteristics. J. Dairy Sci. 2020, 103, 5916–5930. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Zhao, K.; Chen, S.; Ren, Z.; Wei, H.; Wan, C. Whole genome and acid stress comparative transcriptome analysis of Lactiplantibacillus plantarum ZDY2013. Arch. Microbiol. 2021, 203, 2795–2807. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document. The International Scientific Association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [Green Version]
- Juhász, J.; Ligeti, B.; Gajdács, M.; Makra, N.; Ostorházi, E.; Farkas, F.B.; Stercz, B.; Tóth, Á.; Domokos, J.; Pongor, S.; et al. Colonization dynamics of multidrug-resistant klebsiella pneumoniae are dictated by microbiota-Cluster group behavior over individual antibiotic susceptibility: A metataxonomic analysis. Antibiotics 2021, 10, 268. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.J.; Chen, Y.F.; Yang, H.J.; Yang, J.; Xue, J.G.; Li, C.K.; Kwok, L.Y.; Zhang, H.P.; Sun, T.S. Screening for Lactobacillus plantarum with potential inhibitory activity against enteric pathogens. Ann. Microbiol. 2014, 65, 1257–1265. [Google Scholar] [CrossRef]
- Bibalan, M.H.; Eshaghi, M.; Rohani, M.; Esghaei, M.; Darban-Sarokhalil, D.; Pourshafie, M.R.; Talebi, M. Isolates of Lactobacillus plantarum and L. reuteri display greater antiproliferative and antipathogenic activity than other Lactobacillus isolates. J. Med. Microbiol. 2017, 66, 1416–1420. [Google Scholar] [CrossRef]
- Kassayova, M.; Bobrov, N.; Strojny, L.; Orendas, P.; Demeckova, V.; Jendzelovsky, R.; Kubatka, P.; Kiskova, T.; Kruzliak, P.; Adamkov, M.; et al. Anticancer and immunomodulatory effects of Lactobacillus plantarum LS/07, inulin and melatonin in NMU-induced rat model of breast cancer. Anticancer Res. 2016, 36, 2719–2728. [Google Scholar]
- Konishi, H.; Fujiya, M.; Tanaka, H.; Ueno, N.; Moriichi, K.; Sasajima, J.; Ikuta, K.; Akutsu, H.; Tanabe, H.; Kohgo, Y. Probiotic-derived ferrichrome inhibits colon cancer progression via JNK-mediated apoptosis. Nat. Commun. 2016, 7, 12365. [Google Scholar] [CrossRef] [PubMed]
- Harahap, I.A.; Mariyatun, M.; Hasan, P.N.; Pamungkaningtyas, F.H.; Widada, J.; Utami, T.; Cahyanto, M.N.; Juffrie, M.; Dinoto, A.; Nurfiani, S.; et al. Recovery of indigenous probiotic Lactobacillus plantarum Mut-7 on healthy Indonesian adults after consumption of fermented milk containing these bacteria. J. Food Sci. Technol. 2021, 58, 3525–3532. [Google Scholar] [CrossRef]
- Frediansyah, A.; Romadhoni, F.; Suryani; Nurhayati, R.; Wibowo, A.T. Fermentation of Jamaican cherries juice using Lactobacillus plantarum elevates antioxidant potential and inhibitory activity against Type II diabetes-related enzymes. Molecules 2021, 26, 2868. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, J.; Zhao, X.; Zhao, W.; Yu, Z.; Chen, C.; Yang, Z. Complete genome sequencing of exopolysaccharide-producing Lactobacillus plantarum K25 provides genetic evidence for the probiotic functionality and cold endurance capacity of the strain. Biosci. Biotechnol. Biochem. 2018, 82, 1225–1233. [Google Scholar] [CrossRef] [Green Version]
- Pavli, F.G.; Argyri, A.A.; Papadopoulou, O.S.; Nychas, G.E.; Chroianopoulos, N.; Tassou, C.C. Probiotic potential of lactic acid bacteria from traditional fermented dairy and meat products: Assessment by in vitro tests and molecular characterization. J. Prob. Health 2016, 4, 1000157. [Google Scholar] [CrossRef]
- Pavli, F.G.; Argyri, A.A.; Chroianopoulos, N.; Nychas, G.E.; Tassou, C.C. Effect of Lactobacillus plantarum L125 strain with probiotic potential on physicochemical, microbiological and sensorial characteristics of dry-fermented sausages. LWT-Food Sci. Technol. 2020, 118, 108810. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 4 August 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernandez-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Ramulu, H.G.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [Green Version]
- Biswas, A.; Staals, R.H.; Morales, S.E.; Fineran, P.C.; Brown, C.M. CRISPRDetect: A flexible algorithm to define CRISPR arrays. BMC Genom. 2016, 17, 356. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [Green Version]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humprhis, S.; Eplhinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- de Jong, A.; van Hijum, S.A.; Bijlsma, J.J.; Kok, J.; Kuipers, O.P. BAGEL: A web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006, 34, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Plessas, S.; Kiousi, D.E.; Rathosi, M.; Alexopoulos, A.; Kourkoutas, Y.; Mantzourani, I.; Galanis, A.; Bezirtzoglou, E. Isolation of a Lactobacillus paracasei strain with probiotic attributes from kefir grains. Biomedicines 2020, 8, 594. [Google Scholar] [CrossRef]
- Saxami, G.; Karapetsas, A.; Lamprianidou, E.; Kotsianidis, I.; Chlichlia, A.; Tassou, C.; Zoumpourlis, V.; Galanis, A. Two potential probiotic Lactobacillus strains isolated from olive microbiota exhibit adhesion and anti-proliferative effects in cancer cell lines. J. Funct. Foods 2016, 24, 461–471. [Google Scholar] [CrossRef]
- Franken, N.A.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- van den Nieuwboer, M.; van Hemert, S.; Claassen, E.; de Vos, W.M. Lactobacillus plantarum WCFCS1 and its host interaction: A dozen years after the genome. Microb. Biotechnol. 2016, 9, 452–465. [Google Scholar] [CrossRef] [Green Version]
- Zago, M.; Fornasari, M.E.; Carminati, D.; Burns, P.; Suarez, V.; Vinderola, G.; Reinheimer, J.; Giraffa, G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011, 28, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ji, H.; Zhang, D.; Liu, H.; Wang, S.; Wang, J.; Wang, Y. Complete genome sequencing of Lactobacillus plantarum ZLP001, a potential probiotic That enhances intestinal epithelial barrier function and defense against pathogens in pigs. Front. Physiol. 2018, 9, 1689. [Google Scholar] [CrossRef]
- Goel, A.; Halami, P.M.; Tamang, J.P. Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front. Microbiol. 2020, 11, 40. [Google Scholar] [CrossRef]
- Guo, H.; Suzuki, T.; Rubinstein, J.L. Structure of a bacterial ATP synthase. Elife 2019, 8, e43128. [Google Scholar] [CrossRef]
- Todorov, S.D. Bacteriocins from Lactobacillus plantarum—Production, genetic organization and mode of action: Producao, organizacao genetica e modo de acao. Braz. J. Microbiol. 2009, 40, 209–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diep, D.B.; Straume, D.; Kjos, M.; Torres, C.; Nes, I.F. An overview of the mosaic bacteriocin pln loci from Lactobacillus plantarum. Peptides 2009, 30, 1562–1574. [Google Scholar] [CrossRef]
- Tai, H.F.; Foo, H.L.; Rahim, R.A.; Loh, T.C.; Abdullah, M.P.; Yoshinobu, K. Molecular characterisation of new organisation of plnEF and plw loci of bacteriocin genes harbour concomitantly in Lactobacillus plantarum I-UL4. Microb. Cell Fact. 2015, 14, 89. [Google Scholar] [CrossRef] [Green Version]
- Saxami, G.; Karapetsas, A.; Chondrou, P.; Vasiliadis, S.; Lamprianidou, E.; Kotsianidis, I.; Ypsilantis, P.; Botaitis, S.; Simopoulos, C.; Galanis, A. Potentially probiotic Lactobacillus strains with anti-proliferative activity induce cytokine/chemokine production and neutrophil recruitment in mice. Benef. Microb. 2017, 8, 615–623. [Google Scholar] [CrossRef] [PubMed]
- Torriani, S.; Clementi, F.; Vancanneyt, M.; Hoste, B.; Dellaglio, F.; Kersters, K. Differentiation of Lactobacillus plantarum, L. pentosus and L. paraplantarum species by RAPD-PCR and AFLP. Syst. Appl. Microbiol. 2001, 24, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Kleerebezem, M.; Boekhorst, J.; van Kranenburg, R.; Molenaar, D.; Kuipers, O.P.; Leer, R.; Tarchini, R.; Peters, S.A.; Sandbrink, H.M.; Fiers, M.W.; et al. Complete genome sequence of Lactobacillus plantarum WCFCS1. Proc. Natl. Acad. Sci. USA 2003, 100, 1990–1995. [Google Scholar] [CrossRef] [Green Version]
- Molenaar, D.; Bringel, F.; Schuren, F.H.; de Vos, W.M.; Siezen, R.J.; Kleerebezem, M. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 2005, 187, 6119–6127. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Ruan, L.; Sun, M.; Ganzle, M. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl. Environ. Microbiol. 2015, 81, 7233–7243. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Chang, H.C.; Kim, H.Y. Complete genome sequence of Lactobacillus plantarum EM, a putative probiotic strain with the cholesterol-lowering effect and antimicrobial activity. Curr. Microbiol. 2020, 77, 1871–1882. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Zhao, J.; Xia, Y.; Lai, P.F.; Ai, L. A surface protein from Lactobacillus plantarum increases the adhesion of lactobacillus strains to human epithelial cells. Front. Microbiol. 2018, 9, 2858. [Google Scholar] [CrossRef] [Green Version]
- Yadav, A.K.; Tyagi, A.; Kumar, A.; Panwar, S.; Grover, S.; Saklani, A.C.; Hemalatha, R.; Batish, V.K. Adhesion of lactobacilli and their anti-infectivity potential. Crit. Rev. Food Sci. Nutr. 2017, 57, 2042–2056. [Google Scholar] [CrossRef]
- Siciliano, R.A.; Lippolis, R.; Mazzeo, M.F. Proteomics for the investigation of surface-exposed proteins in probiotics. Front. Nutr. 2019, 6, 52. [Google Scholar] [CrossRef]
- Choudhary, J.; Dubey, R.C.; Sengar, G.; Dheeman, S. Evaluation of probiotic potential and safety assessment of Lactobacillus pentosus MMP4 isolated from mare’s lactation. Probiotics Antimicrob. Proteins 2019, 11, 403–412. [Google Scholar] [CrossRef]
- da Silva Sabo, S.; Vitolo, M.; Gonzalez, J.M.D.; Oliveira, R.P.S. Overview of Lactobacillus plantarum as a promising bacteriocin producer among lactic acid bacteria. Food Res. Int. 2014, 64, 527–536. [Google Scholar] [CrossRef]
- Chen, C.C.; Lai, C.C.; Huang, H.L.; Huang, W.Y.; Toh, H.S.; Weng, T.C.; Chuang, Y.C.; Lu, Y.C.; Tang, H.J. Antimicrobial activity of Lactobacillus species against carbapenem-resistant enterobacteriaceae. Front. Microbiol. 2019, 10, 789. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, R.; Altermann, E.; Anderson, R.C.; McNabb, W.C.; Moughan, P.J.; Roy, N.C. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediat. Inflamm. 2013, 2013, 237921. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Liu, Y.M. Antibacterial effect of bacteriocin isolated from Lactobacillus plantarum ATCC 8014 on postoperative infection of mandibular fracture in vivo. J. Craniofac. Surg. 2017, 28, 679–682. [Google Scholar] [CrossRef]
- Nowak, A.; Paliwoda, A.; Blasiak, J. Anti-proliferative, pro-apoptotic and anti-oxidative activity of lactobacillus and bifidobacterium strains: A review of mechanisms and therapeutic perspectives. Crit. Rev. Food Sci. Nutr. 2019, 59, 3456–3467. [Google Scholar] [CrossRef]
- Chondrou, P.; Karapetsas, A.; Kiousi, D.E.; Tsela, D.; Tiptiri-Kourpeti, A.; Anestopoulos, I.; Kotsianidis, I.; Bezirtzoglou, E.; Pappa, A.; Galanis, A. Lactobacillus paracasei K5 displays adhesion, anti-proliferative activity and apoptotic effects in human colon cancer cells. Benef. Microbes. 2018, 9, 975–983. [Google Scholar] [CrossRef]
- Tiptiri-Kourpeti, A.; Spyridopoulou, K.; Santarmaki, V.; Aindelis, G.; Tompoulidou, E.; Lamprianidou, E.E.; Saxami, G.; Ypsilantis, P.; Lampri, E.S.; Simopoulos, C.; et al. Lactobacillus casei exerts anti-proliferative effects accompanied by apoptotic cell death and up-regulation of TRAIL in colon carcinoma cells. PLoS ONE 2016, 11, e0147960. [Google Scholar] [CrossRef]
- Aindelis, G.; Chlichlia, K. Modulation of anti-tumour immune responses by probiotic bacteria. Vaccines 2020, 8, 329. [Google Scholar] [CrossRef] [PubMed]
- Shin, R.; Itoh, Y.; Kataoka, M.; Iino-Miura, S.; Miura, R.; Mizutani, T.; Fujisawa, T. Anti-tumor activity of heat-killed Lactobacillus plantarum BF-LP284 on Meth-A tumor cells in BALB/c mice. Int. J. Food Sci. Nutr. 2016, 67, 641–649. [Google Scholar] [CrossRef]
- Kemp, M.Q.; Jeffy, B.D.; Romagnolo, D.F. Conjugated linoleic acid inhibits cell proliferation through a p53-dependent mechanism: Effects on the expression of G1-restriction points in breast and colon cancer cells. J. Nutr. 2003, 133, 3670–3677. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Zhang, Y.; Ye, L.; Wang, C. The anti-cancer effects and mechanisms of lactic acid bacteria exopolysaccharides in vitro: A review. Carbohydr. Polym. 2021, 253, 117308. [Google Scholar] [CrossRef]
- Yang, B.; Qi, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, H.; Chen, Y.Q. Characterization of the triple-component linoleic acid isomerase in Lactobacillus plantarum ZS2058 by genetic manipulation. J. Appl. Microbiol. 2017, 123, 1263–1273. [Google Scholar] [CrossRef]

| Attribute | Values |
|---|---|
| Genome Size (bp) | 3,354,135 |
| GC content (%) | 44.34 |
| Total Genes | 3220 |
| CDS (protein) | 3024 |
| Pseudogenes | 126 |
| tRNA genes | 62 |
| rRNA genes | 4 |
| ncRNA genes | 4 |
| Locus Tag | Description | Role |
|---|---|---|
| LP125_003204 | cation:proton antiporter | GI tract survival |
| LP125_001869 | PBP1A family penicillin-binding protein | GI tract survival |
| LP125_002196 | D-alanine--poly(phosphoribitol) ligase subunit DltA | Acid tolerance |
| LP125_002199 | D-alanyl-lipoteichoic acid biosynthesis protein DltD | Acid tolerance |
| LP125_001705 | glutamate decarboxylase | Acid tolerance |
| LP125_000817 | F0F1 ATP synthase subunit epsilon | Acid tolerance |
| LP125_000818 | F0F1 ATP synthase subunit beta | Acid tolerance |
| LP125_000819 | F0F1 ATP synthase subunit gamma | Acid tolerance |
| LP125_000820 | F0F1 ATP synthase subunit alpha | Acid tolerance |
| LP125_000821 | F0F1 ATP synthase subunit delta | Acid tolerance |
| LP125_000822 | F0F1 ATP synthase subunit B | Acid tolerance |
| LP125_000823 | F0F1 ATP synthase subunit C | Acid tolerance |
| LP125_000824 | F0F1 ATP synthase subunit A | Acid tolerance |
| LP125_003090 | choloylglycine hydrolase family protein | Bile Resistance |
| LP125_000497 | choloylglycine hydrolase family protein | Bile Resistance |
| LP125_000993 | linear amide C-N hydrolase | Bile Resistance |
| LP125_001391 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_001882 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_001897 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_003116 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_000218 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_001232 | LPXTG cell wall anchor domain-containing protein | Cell surface protein |
| LP125_000997 | collagen binding protein | Adhesion |
| LP125_002620 | MucBP domain-containing protein | Adhesion |
| LP125_000275 | MucBP domain-containing protein | Adhesion |
| LP125_000616 | MucBP domain-containing protein | Adhesion |
| LP125_002390 | NFACT family protein | Adhesion |
| LP125_000010 | NFACT family protein | Adhesion |
| LP125_002930 | elongation factor tu | Adhesion |
| LP125_002193 | molecular chaperone DnaJ | Heat Stress |
| LP125_002192 | molecular chaperone DnaK | Heat Stress |
| LP125_002191 | nucleotide exchange factor GrpE | Heat Stress |
| LP125_001567 | chaperonin GroEL | Heat Stress |
| LP125_001568 | co-chaperone GroES | Heat Stress |
| LP125_002661 | cold-shock protein | Cold Stress |
| LP125_002795 | cold-shock protein | Cold Stress |
| LP125_003063 | cold-shock protein | Cold Stress |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tegopoulos, K.; Stergiou, O.S.; Kiousi, D.E.; Tsifintaris, M.; Koletsou, E.; Papageorgiou, A.C.; Argyri, A.A.; Chorianopoulos, N.; Galanis, A.; Kolovos, P. Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells. Biomedicines 2021, 9, 1718. https://doi.org/10.3390/biomedicines9111718
Tegopoulos K, Stergiou OS, Kiousi DE, Tsifintaris M, Koletsou E, Papageorgiou AC, Argyri AA, Chorianopoulos N, Galanis A, Kolovos P. Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells. Biomedicines. 2021; 9(11):1718. https://doi.org/10.3390/biomedicines9111718
Chicago/Turabian StyleTegopoulos, Konstantinos, Odysseas Sotirios Stergiou, Despoina Eugenia Kiousi, Margaritis Tsifintaris, Ellie Koletsou, Aristotelis C. Papageorgiou, Anthoula A. Argyri, Nikos Chorianopoulos, Alex Galanis, and Petros Kolovos. 2021. "Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells" Biomedicines 9, no. 11: 1718. https://doi.org/10.3390/biomedicines9111718
APA StyleTegopoulos, K., Stergiou, O. S., Kiousi, D. E., Tsifintaris, M., Koletsou, E., Papageorgiou, A. C., Argyri, A. A., Chorianopoulos, N., Galanis, A., & Kolovos, P. (2021). Genomic and Phylogenetic Analysis of Lactiplantibacillus plantarum L125, and Evaluation of Its Anti-Proliferative and Cytotoxic Activity in Cancer Cells. Biomedicines, 9(11), 1718. https://doi.org/10.3390/biomedicines9111718








