Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Site-Directed Mutagenesis
2.3. Cell Lines, Cell Culture, and Transient Transfections
2.4. Purification and Analysis of HBV Cytoplasmic Intermediates and cccDNA
2.5. cccDNA Chromatin Immunoprecipitation Assays (ChIP)
2.6. Epifluorescence
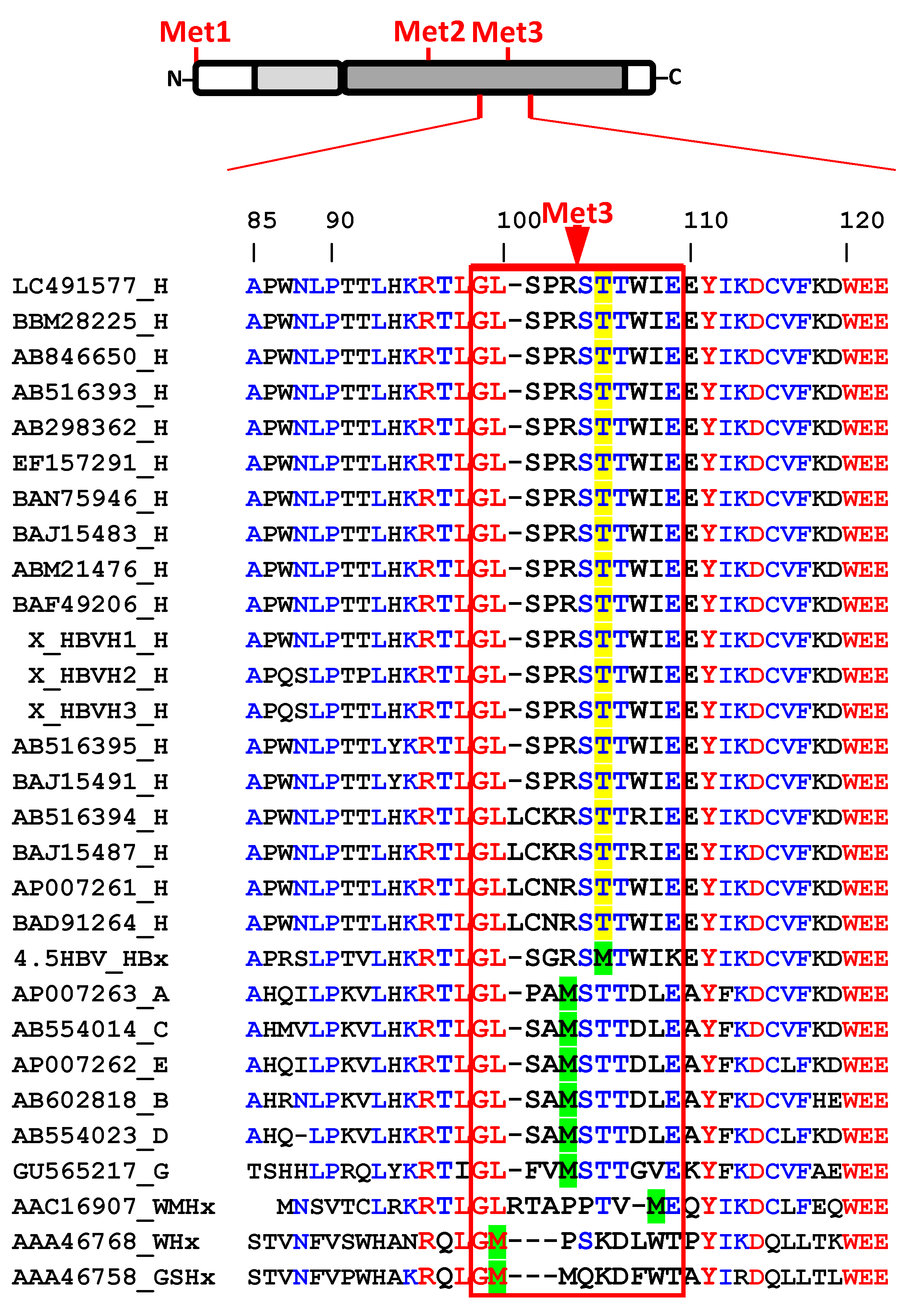
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Hepatitis Report, 2017. 2017. Available online: https://www.who.int/hepatitis/publications/global-hepatitis-report2017/en/ (accessed on 15 June 2021).
- Revill, P.; Tu, T.; Netter, H.; Yuen, L.; Locarnini, S.; Littlejohn, M. The evolution and clinical impact of hepatitis B virus genome diversity. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 618–634. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Sarica, N.; Neuveut, C. Early Steps of Hepatitis B Life Cycle: From Capsid Nuclear Import to cccDNA Formation. Viruses 2021, 13, 757. [Google Scholar] [CrossRef] [PubMed]
- Araujo, N. Hepatitis B virus intergenotypic recombinants worldwide: An overview. Infect. Genet. Evol. 2015, 36, 500–510. [Google Scholar] [CrossRef]
- Pujol, F.; Jaspe, R.; Loureiro, C.; Chemin, I. Hepatitis B virus American genotypes: Pathogenic variants? Clin. Res. Hepatol. Gastroenterol. 2020, 44, 825–835. [Google Scholar] [CrossRef]
- Kramvis, A. Genotypes and genetic variability of hepatitis B virus. Intervirology 2014, 57, 141–150. [Google Scholar] [CrossRef]
- Livingston, S.; Simonetti, J.; McMahon, B.; Bulkow, L.; Hurlburt, K.; Homan, C.; Snowball, M.; Cagle, H.; Williams, J.; Chulanov, V. Hepatitis B virus genotypes in Alaska Native people with hepatocellular carcinoma: Preponderance of genotype F. J. Infect. Dis. 2007, 195, 5–11. [Google Scholar] [CrossRef]
- Ching, L.; Gounder, P.; Bulkow, L.; Spradling, P.; Bruce, M.; Negus, S.; Snowball, M.; McMahon, B. Incidence of hepatocellular carcinoma according to hepatitis B virus genotype in Alaska Native people. Liver Int. 2016, 36, 1507–1515. [Google Scholar] [CrossRef]
- Gounder, P.; Bulkow, L.; Snowball, M.; Negus, S.; Spradling, P.; McMahon, B. Hepatocellular Carcinoma Risk in Alaska Native Children and Young Adults with Hepatitis B Virus: Retrospective Cohort Analysis. J. Pediatr. 2016, 178, 206–213. [Google Scholar] [CrossRef]
- Kowalec, K.; Minuk, G.; Børresen, M.; Koch, A.; McMahon, B.; Simons, B.; Osiowy, C. Genetic diversity of hepatitis B virus genotypes B6, D and F among circumpolar indigenous individuals. J. Viral Hepat. 2013, 20, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Pineau, P.; Ruiz, E.; Deharo, E.; Bertani, S. On hepatocellular carcinoma in South America and early-age onset of the disease. Clin. Res. Hepatol. Gastroenterol. 2019, 43, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Panduro, A.; Maldonado-Gonzalez, M.; Fierro, N.; Roman, S. Distribution of HBV genotypes F and H in Mexico and Central America. Antivir. Ther. 2013, 18, 475–484. [Google Scholar] [CrossRef]
- Roman, S.; Panduro, A. HBV endemicity in Mexico is associated with HBV genotypes H and G. World J. Gastroenterol. 2013, 19, 5446–5453. [Google Scholar] [CrossRef]
- Roman, S.; Tanaka, Y.; Khan, A.; Kurbanov, F.; Kato, H.; Mizokami, M.; Panduro, A. Occult hepatitis B in the genotype H-infected Nahuas and Huichol native Mexican population. J. Med. Virol. 2010, 82, 1527–1536. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Tachiquín, M.; Valdez-Salazar, H.; Juárez-Barreto, V.; Dehesa-Violante, M.; Torres, J.; Muñoz-Hernández, O.; Alvarez-Muñoz, M. Molecular analysis of hepatitis B virus “a” determinant in asymptomatic and symptomatic Mexican carriers. Virol. J. 2007, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Melegari, M.; Scaglioni, P.; Wands, J. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J. Virol. 1998, 72, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.; Wang, L.; Schneider, R. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 2001, 294, 2376–2378. [Google Scholar] [CrossRef]
- Leupin, O.; Bontron, S.; Schaeffer, C.; Strubin, M. Hepatitis B virus X protein stimulates viral genome replication via a DDB1-dependent pathway distinct from that leading to cell death. J. Virol. 2005, 79, 4238–4245. [Google Scholar] [CrossRef]
- Tang, H.; Delgermaa, L.; Huang, F.; Oishi, N.; Liu, L.; He, F.; Zhao, L.; Murakami, S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 2005, 79, 5548–5556. [Google Scholar] [CrossRef]
- Keasler, V.; Hodgson, A.; Madden, C.; Slagle, B. Enhancement of hepatitis B virus replication by the regulatory X protein in vitro and in vivo. J. Virol. 2007, 81, 2656–2662. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, J.; Arzberger, S.; Durantel, D.; Belloni, L.; Strubin, M.; Levrero, M.; Zoulim, F.; Hantz, O.; Protzer, U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 2011, 55, 996–1003. [Google Scholar] [CrossRef]
- Murakami, S.; Cheong, J.; Kaneko, S. Human hepatitis B virus X gene encodes a regulatory domain which represses transactivation of X protein. J. Biol. Chem. 1994, 269, 15118–15123. [Google Scholar] [CrossRef]
- Tang, H.; Oishi, N.; Kaneko, S.; Murakami, S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006, 97, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sarkar, D. Hepatitis B Virus X Protein: Structure–Function Relationships and Role in Viral Pathogenesis. In Transcription Factors; (Handbook of Experimental Pharmacology book series); Springer: Berlin/Heidelberg, Germany, 2004; Volume 166, pp. 377–407. [Google Scholar]
- Gong, D.; Chen, E.; Huang, F.; Leng, X.; Cheng, X.; Tang, H. Role and functional domain of hepatitis B virus X protein in regulating HBV transcription and replication in vitro and in vivo. Viruses 2013, 5, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Slagle, B.; Bouchard, M. Hepatitis B Virus X and Regulation of Viral Gene Expression. Cold Spring Harb. Perspect. Med. 2016, 6, a021402. [Google Scholar] [CrossRef]
- Hernández, S.; Jiménez, G.; Alarcón, V.; Prieto, C.; Muñoz, F.; Riquelme, C.; Venegas, M.; Brahm, J.; Loyola, A.; Villanueva, R. Replication of a chronic hepatitis B virus genotype F1b construct. Arch. Virol. 2016, 161, 583–594. [Google Scholar] [CrossRef]
- Hernandez, S.; Venegas, M.; Brahm, J.; Villanueva, R. Full-genome sequence of a hepatitis B virus genotype F1b clone from a chronically infected Chilean patient. Genome Announc. 2014, 2, e01075-14. [Google Scholar] [CrossRef] [PubMed]
- Misra, K.; Mukherji, A.; Kumar, V. The conserved amino-terminal region (amino acids 1-20) of the hepatitis B virus X protein shows a transrepression function. Virus Res. 2004, 105, 157–165. [Google Scholar] [CrossRef]
- Hodgson, A.; Hyser, J.; Keasler, V.; Cang, Y.; Slagle, B. Hepatitis B virus regulatory HBx protein binding to DDB1 is required but is not sufficient for maximal HBV replication. Virology 2012, 426, 73–82. [Google Scholar] [CrossRef]
- Kumar, V.; Jayasuryan, N.; Kumar, R. A truncated mutant (residues 58–140) of the hepatitis B virus X protein retains transactivation function. Proc. Natl. Acad. Sci. USA 1996, 93, 5647–5652. [Google Scholar] [CrossRef]
- Arii, M.; Takada, S.; Koike, K. Identification of three essential regions of hepatitis B virus X protein for trans-activation function. Oncogene 1992, 7, 397–403. [Google Scholar]
- Nijhara, R.; Jana, S.; Goswami, S.; Kumar, V.; Sarkar, D. An internal segment (residues 58-119) of the hepatitis B virus X protein is sufficient to activate MAP kinase pathways in mouse liver. FEBS Lett. 2001, 504, 59–64. [Google Scholar] [CrossRef]
- Gottlob, K.; Pagano, S.; Levrero, M.; Graessmann, A. Hepatitis B virus X protein transcription activation domains are neither required nor sufficient for cell transformation. Cancer Res. 1998, 58, 3566–3570. [Google Scholar]
- Reddi, H.; Kumar, R.; Jain, S.; Kumar, V. A carboxy-terminal region of the hepatitis B virus X protein promotes DNA interaction of CREB and mimics the native protein for transactivation function. Virus Genes 2003, 26, 227–238. [Google Scholar] [CrossRef]
- Huh, K.; Siddiqui, A. Characterization of the mitochondrial association of hepatitis B virus X protein, HBx. Mitochondrion 2002, 1, 349–359. [Google Scholar] [CrossRef]
- Takada, S.; Shirakata, Y.; Kaneniwa, N.; Koike, K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 1999, 18, 6965–6973. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Cha, E.; Lim, J.; Kwon, S.; Kim, D.; Cho, H.; Han, K. Structural characterization of an intrinsically unfolded mini-HBX protein from hepatitis B virus. Mol. Cells 2012, 34, 165–169. [Google Scholar] [CrossRef]
- De Moura, P.R.; Rui, E.; Gonçalves, K.d.A.; Kobarg, J. The cysteine residues of the hepatitis B virus onco-protein HBx are not required for its interaction with RNA or with human p53. Virus Res. 2005, 108, 121–131. [Google Scholar] [CrossRef]
- Ramakrishnan, D.; Xing, W.; Beran, R.; Chemuru, S.; Rohrs, H.; Niedziela-Majka, A.; Marchand, B.; Mehra, U.; Zábranský, A.; Doležal, M.; et al. Hepatitis B Virus X Protein Function Requires Zinc Binding. J. Virol. 2019, 93, e00250-19. [Google Scholar] [CrossRef]
- Li, T.; Robert, E.; van Breugel, P.; Strubin, M.; Zheng, N. A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat. Struct. Mol. Biol. 2010, 17, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liu, M.; Wu, J.; Shi, Y. Structural and biochemical analysis of Bcl-2 interaction with the hepatitis B virus protein HBx. Proc. Natl. Acad. Sci. USA 2016, 113, 2074–2079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Chen, H.; Cao, J.; Xiong, H.; Mo, X.; Li, T.; Kang, X.; Zhao, J.; Yin, B.; Zhao, X.; et al. Structural and functional analyses of hepatitis B virus X protein BH3-like domain and Bcl-xL interaction. Nat. Commun. 2019, 10, 3192. [Google Scholar] [CrossRef]
- Dandri, M.; Schirmache, P.; Rogler, C. Woodchuck hepatitis virus X protein is present in chronically infected woodchuck liver and woodchuck hepatocellular carcinomas which are permissive for viral replication. J. Virol. 1996, 70, 5246–5254. [Google Scholar] [CrossRef]
- Sirma, H.; Giannini, C.; Poussin, K.; Paterlini, P.; Kremsdorf, D.; Bréchot, C. Hepatitis B virus X mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of HBx. Oncogene 1999, 18, 4848–4859. [Google Scholar] [CrossRef]
- Su, Q.; Schröder, C.; Hofmann, W.; Otto, G.; Pichlmayr, R.; Bannasch, P. Expression of hepatitis B virus X protein in HBV-infected human livers and hepatocellular carcinomas. Hepatology 1998, 27, 1109–1120. [Google Scholar] [CrossRef]
- Hoare, J.; Henkler, F.; Dowling, J.; Errington, W.; Goldin, R.; Fish, D.; McGarvey, M. Subcellular localisation of the X protein in HBV infected hepatocytes. J. Med. Virol. 2001, 64, 419–426. [Google Scholar] [CrossRef]
- Weil, R.; Sirma, H.; Giannini, C.; Kremsdorf, D.; Bessia, C.; Dargemont, C.; Bréchot, C.; Israël, A. Direct association and nuclear import of the hepatitis B virus X protein with the NF-kappaB inhibitor IkappaBalpha. Mol. Cell. Biol. 1999, 19, 6345–6354. [Google Scholar] [CrossRef][Green Version]
- Kornyeyev, D.; Ramakrishnan, D.; Voitenleitner, C.; Livingston, C.; Xing, W.; Hung, M.; Kwon, H.; Fletcher, S.; Beran, R. Spatiotemporal Analysis of Hepatitis B Virus X Protein in Primary Human Hepatocytes. J. Virol. 2019, 93, e00248-19. [Google Scholar] [CrossRef] [PubMed]
- Henkler, F.; Hoare, J.; Waseem, N.; Goldin, R.; McGarvey, M.; Koshy, R.; King, I. Intracellular localization of the hepatitis B virus HBx protein. J. Gen. Virol. 2001, 82, 871–882. [Google Scholar] [CrossRef]
- Cha, M.; Ryu, D.; Jung, H.; Chang, H.; Ryu, W. Stimulation of hepatitis B virus genome replication by HBx is linked to both nuclear and cytoplasmic HBx expression. J. Gen. Virol. 2009, 90, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Sun, T.; Park, S.; Shen, G.; Liu, J. The role of hepatitis B virus X protein is related to its differential intracellular localization. Acta Biochim. Biophys. Sin. 2011, 43, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Prieto, C.; Montecinos, J.; Jiménez, G.; Riquelme, C.; Garrido, D.; Hernández, S.; Loyola, A.; Villanueva, R. Phosphorylation of Phylogenetically Conserved Amino Acid Residues Confines HBx within Different Cell Compartments of Human Hepatocarcinoma Cells. Molecules 2021, 26, 1254. [Google Scholar] [CrossRef] [PubMed]
- Benn, J.; Su, F.; Doria, M.; Schneider, R. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 1996, 70, 4978–4985. [Google Scholar] [CrossRef]
- Lee, Y.; Yun, Y. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J. Biol. Chem. 1998, 273, 25510–25515. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.; Wang, L.; Schneider, R. Activation of focal adhesion kinase by hepatitis B virus HBx protein: Multiple functions in viral replication. J. Virol. 2006, 80, 4406–4414. [Google Scholar] [CrossRef]
- Lee, Y.; Kang-Park, S.; Do, S.; Lee, Y. The hepatitis B virus-X protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 2001, 276, 16969–16977. [Google Scholar] [CrossRef]
- Cha, M.; Kim, C.; Park, Y.; Ryu, W. Hepatitis B virus X protein is essential for the activation of Wnt/beta-catenin signaling in hepatoma cells. Hepatology 2004, 39, 1683–1693. [Google Scholar] [CrossRef] [PubMed]
- Oropeza, C.; Tarnow, G.; Sridhar, A.; Taha, T.; Shalaby, R.; McLachlan, A. The Regulation of HBV Transcription and Replication. Adv. Exp. Med. Biol. 2020, 1179, 39–69. [Google Scholar] [PubMed]
- Keasler, V.; Hodgson, A.; Madden, C.; Slagle, B. Hepatitis B virus HBx protein localized to the nucleus restores HBx-deficient virus replication in HepG2 cells and in vivo in hydrodynamically-injected mice. Virology 2009, 390, 122–129. [Google Scholar] [CrossRef]
- Rossner, M. Review: Hepatitis B virus X-gene product: A promiscuous transcriptional activator. J. Med. Virol. 1992, 36, 101–117. [Google Scholar] [CrossRef]
- Lin, Y.; Nomura, T.; Cheong, J.; Dorjsuren, D.; Iida, K.; Murakami, S. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 1997, 272, 7132–7139. [Google Scholar] [CrossRef]
- Lin, Y.; Nomura, T.; Yamashita, T.; Dorjsuren, D.; Tang, H.; Murakami, S. The transactivation and p53-interacting functions of hepatitis B virus X protein are mutually interfering but distinct. Cancer Res. 1997, 57, 5137–5142. [Google Scholar]
- Qadri, I.; Conaway, J.; Conaway, R.; Schaack, J.; Siddiqui, A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc. Natl. Acad. Sci. USA 1996, 93, 10578–10583. [Google Scholar] [CrossRef]
- Qadri, I.; Maguire, H.; Siddiqui, A. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc. Natl. Acad. Sci. USA 1995, 92, 1003–1007. [Google Scholar] [CrossRef]
- Cougot, D.; Wu, Y.; Cairo, S.; Caramel, J.; Renard, C.; Lévy, L.; Buendia, M.; Neuveut, C. The hepatitis B virus X protein functionally interacts with CREB-binding protein/p300 in the regulation of CREB-mediated transcription. J. Biol. Chem. 2007, 282, 4277–4287. [Google Scholar] [CrossRef]
- Slagle, B.; Bouchard, M. Role of HBx in hepatitis B virus persistence and its therapeutic implications. Curr. Opin. Virol. 2018, 30, 32–38. [Google Scholar] [CrossRef]
- Bogaert, A.; Fernandez, E.; Gevaert, K. N-Terminal Proteoforms in Human Disease. Trends Biochem. Sci. 2020, 45, 308–320. [Google Scholar] [CrossRef] [PubMed]
- Kochetov, A. Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. Bioessays 2008, 30, 683–691. [Google Scholar] [CrossRef] [PubMed]
- James, C.; Smyth, J. Alternative mechanisms of translation initiation: An emerging dynamic regulator of the proteome in health and disease. Life Sci. 2018, 212, 138–144. [Google Scholar] [CrossRef]
- Zheng, Y.; Riegler, J.; Wu, J.; Yen, T. Novel short transcripts of hepatitis B virus X gene derived from intragenic promoter. J. Biol. Chem. 1994, 269, 22593–22598. [Google Scholar] [CrossRef]
- Zhang, P.; Raney, A.; McLachlan, A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology 1992, 191, 31–41. [Google Scholar] [CrossRef]
- Treinin, M.; Laub, O. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol. Cell. Biol. 1987, 7, 545–548. [Google Scholar]
- Nakatake, H.; Chisaka, O.; Yamamoto, S.; Matsubara, K.; Koshy, R. Effect of X protein on transactivation of hepatitis B virus promoters and on viral replication. Virology 1993, 195, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Kwee, L.; Lucito, R.; Aufiero, B.; Schneider, R. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivate class II and III promoters. J. Virol. 1992, 66, 4382–4389. [Google Scholar] [CrossRef]
- Leach, J.; Qiao, L.; Fang, Y.; Han, S.; Gilfor, D.; Fisher, P.; Grant, S.; Hylemon, P.; Peterson, D.; Dent, P. Regulation of p21 and p27 expression by the hepatitis B virus X protein and the alternate initiation site X proteins, AUG2 and AUG3. J. Gastroenterol. Hepatol. 2003, 18, 376–385. [Google Scholar] [CrossRef]
- Yuan, S.; Liao, G.; Zhang, M.; Zhu, Y.; Wang, K.; Xiao, W.; Jia, C.; Dong, M.; Sun, N.; Walch, A.; et al. Translatomic profiling reveals novel self-restricting virus-host interactions during HBV infection. J. Hepatol. 2021, 75, 74–85. [Google Scholar] [CrossRef]
- Tuttleman, J.; Pourcel, C.; Summers, J. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 1986, 47, 451–460. [Google Scholar] [CrossRef]
- Bock, C.; Schranz, I.; Schroder, C.; Zentgraf, H. Hepatitis B Virus Genome Is Organized into Nucleosomes in the Nucleus of the Infected Cell. Virus Genes 1994, 8, 215–229. [Google Scholar] [CrossRef]
- Newbold, J.; Xin, H.; Tencza, M.; Sherman, G.; Dean, J. The Covalently Closed Duplex Form of the Hepadnavirus Genome Exists In Situ as a Heterogeneous Population of Viral Minichromosomes. J. Virol. 1995, 69, 3350–3357. [Google Scholar] [CrossRef]
- Bock, C.; Schwinn, S.; Locarnini, S.; Fyfe, J.; Manns, M.; Trautwein, C.; Zentgraf, H. Structural organization of the hepatitis B virus minichromosome. J. Mol. Biol. 2001, 307, 183–196. [Google Scholar] [CrossRef]
- Belloni, L.; Pollicino, T.; Nicola, F.D.; Guerrieri, F.; Raffa, G.; Fanciulli, M.; Raimondo, G.; Levrero, M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA 2009, 106, 19975–19979. [Google Scholar] [CrossRef] [PubMed]
- Belloni, L.; Allweiss, L.; Guerrieri, F.; Pediconi, N.; Volz, T.; Pollicino, T.; Petersen, J.; Raimondo, G.; Dandri, M.; Levrero, M. IFN-α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Investig. 2012, 122, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Belloni, L.; Raffa, G.; Pediconi, N.; Squadrito, G.; Raimondo, G.; Levrero, M. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 2006, 130, 823–837. [Google Scholar] [CrossRef] [PubMed]
- Rivière, L.; Gerossier, L.; Ducroux, A.; Dion, S.; Deng, Q.; Michel, M.-L.; Buendia, M.-A.; Hantz, O.; Neuveut, C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J. Hepatol. 2015, 63, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, V.; Hernández, S.; Rubio, L.; Alvarez, F.; Flores, Y.; Varas-Godoy, M.; Ferrari, G.D.; Kann, M.; Villanueva, R.; Loyola, A. The enzymes LSD1 and Set1A cooperate with the viral protein HBx to establish an active hepatitis B viral chromatin state. Sci. Rep. 2016, 6, 25901. [Google Scholar] [CrossRef] [PubMed]
- Benhenda, S.; Ducroux, A.; Riviere, L.; Sobhian, B.; Ward, M.D.; Dion, S.; Hantz, O.; Protzer, U.; Michel, M.L.; Benkirane, M.; et al. Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. J. Virol. 2013, 87, 4360–4371. [Google Scholar] [CrossRef]
- Tropberger, P.; Mercier, A.; Robinson, M.; Zhong, W.; Ganem, D.; Holdorf, M. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc. Natl. Acad. Sci. USA 2015, 112, E5715–E5724. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.H.; Hu, J.L.; Cheng, S.T.; Yu, H.B.; Wong, V.K.W.; Law, B.Y.K.; Yang, Y.F.; Huang, Y.; Liu, Y.; Chen, W.X.; et al. SIRT3 restricts hepatitis B virus transcription and replication through epigenetic regulation of covalently closed circular DNA involving suppressor of variegation 3-9 homolog 1 and SET domain containing 1A histone methyltransferases. Hepatology 2018, 68, 1260–1276. [Google Scholar] [CrossRef]
- Alvarez-Astudillo, F.; Garrido, D.; Varas-Godoy, M.; Gutiérrez, J.; Villanueva, R.; Loyola, A. The histone variant H3.3 regulates the transcription of the hepatitis B virus. Ann. Hepatol. 2021, 21, 100261. [Google Scholar] [CrossRef]
- Salerno, D.; Chiodo, L.; Alfano, V.; Floriot, O.; Cottone, G.; Paturel, A.; Pallocca, M.; Plissonnier, M.; Jeddari, S.; Belloni, S.; et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut 2020, 69, 2016–2024. [Google Scholar] [CrossRef]
- Chong, C.; Cheng, C.; Tsoi, S.; Huang, F.; Liu, F.; Fung, J.; Seto, W.; Lai, K.; Lai, C.; Yuen, M.; et al. HBV X protein mutations affect HBV transcription and association of histone-modifying enzymes with covalently closed circular DNA. Sci. Rep. 2020, 10, 802. [Google Scholar] [CrossRef]
- Saeed, U.; Kim, J.; Piracha, Z.; Kwon, H.; Jung, J.; Chwae, Y.; Park, S.; Shin, H.; Kim, K. Parvulin 14 and Parvulin 17 Bind to HBx and cccDNA and Upregulate Hepatitis B Virus Replication from cccDNA to Virion in an HBx-Dependent Manner. J. Virol. 2019, 93, e01840-18. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, W.; Song, J.; Wu, Y.; Ni, B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 2013, 33, 2810–2816. [Google Scholar] [CrossRef]
- Tang, D.; Zhao, H.; Wu, Y.; Peng, B.; Gao, Z.; Sun, Y.; Duan, J.; Qi, Y.; Li, Y.; Zhou, Z.; et al. Transcriptionally inactive hepatitis B virus episome DNA preferentially resides in the vicinity of chromosome 19 in 3D host genome upon infection. Cell Rep. 2021, 35, 109288. [Google Scholar] [CrossRef] [PubMed]
- Blum, H.E.; Zhang, Z.; Galun, E.; von Weizsäcker, F.; Garner, B.; Liang, T.; Wands, J. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 1992, 66, 1223–1227. [Google Scholar] [CrossRef] [PubMed]
- Zoulim, F.; Saputelli, J.; Seeger, C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 1994, 68, 2026–2030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Torii, N.; Hu, Z.; Jacob, J.; Liang, T. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 2001, 108, 1523–1531. [Google Scholar] [CrossRef] [PubMed]
- Slagle, B.; Andrisani, O.; Bouchard, M.; Lee, C.; Ou, J.; Siddiqui, A. Technical standards for hepatitis B virus X protein (HBx) research. Hepatology 2015, 61, 1416–1424. [Google Scholar] [CrossRef]
- Altinel, K.; Hashimoto, K.; Wei, Y.; Neuveut, C.; Gupta, I.; Suzuki, A.; Santos, A.D.; Moreau, P.; Xia, T.; Kojima, S.; et al. Single-Nucleotide Resolution Mapping of Hepatitis B Virus Promoters in Infected Human Livers and Hepatocellular Carcinoma. J. Virol. 2016, 90, 10811–10822. [Google Scholar] [CrossRef]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Kozak, M. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. Cell 1987, 196, 947–950. [Google Scholar] [CrossRef]
- Kozak, M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Genes 2005, 361, 13–37. [Google Scholar] [CrossRef]
- Hinnebusch, A. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 2011, 75, 434–467. [Google Scholar] [CrossRef] [PubMed]
- Nakai, K.; Horton, P. PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999, 24, 34–36. [Google Scholar] [CrossRef]
- Babu, M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem. Soc. Trans. 2016, 44, 1185–1200. [Google Scholar] [CrossRef]
- Buljan, M.; Chalancon, G.; Dunker, A.K.; Bateman, A.; Balaji, S.; Fuxreiter, M.; Babu, M.M. Alternative splicing of intrinsically disordered regions and rewiring of protein interactions. Curr. Opin. Struct. Biol. 2013, 23, 443–450. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, S.; Dunker, A.K. Intrinsically Disordered Proteins Link Alternative Splicing and Post-translational Modifications to Complex Cell Signaling and Regulation. J. Mol. Biol. 2018, 430, 2342–2359. [Google Scholar] [CrossRef]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int. J. Mol. Sci. 2016, 17, 1874. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Romero, P.; Rani, M.; Dunker, A.K.; Obradovic, Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform. 1999, 10, 30–40. [Google Scholar]
- Romero, P.; Obradovic, Z.; Dunker, K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform. 1997, 8, 110–124. [Google Scholar]
- Linding, R.; Jensen, L.J.; Diella, F.; Bork, P.; Gibson, T.J.; Russell, R.B. Protein disorder prediction: Implications for structural proteomics. Structure 2003, 11, 1453–1459. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
- Barik, A.; Katuwawala, A.; Hanson, J.; Paliwal, K.; Zhou, Y.; Kurgan, L. DEPICTER: Intrinsic Disorder and Disorder Function Prediction Server. J. Mol. Biol. 2020, 432, 3379–3387. [Google Scholar] [CrossRef] [PubMed]
- Decorsière, A.; Mueller, H.; van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.; Livingston, C.; Niu, C.; Fletcher, S.; Hantz, O.; et al. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 2016, 531, 386–389. [Google Scholar] [CrossRef]
- Murphy, C.; Xu, Y.; Li, F.; Nio, K.; Reszka-Blanco, N.; Li, X.; Wu, Y.; Yu, Y.; Xiong, Y.; Su, L. Hepatitis B Virus X Protein Promotes Degradation of SMC5/6 to Enhance HBV Replication. Cell Rep. 2016, 16, 2846–2854. [Google Scholar] [CrossRef]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef]
- Moolla, N.; Kew, M.; Arbuthnot, P. Regulatory elements of hepatitis B virus transcription. J. Viral Hepat. 2002, 9, 323–331. [Google Scholar] [CrossRef]
- Chang, H.K.; Chou, C.K.; Chang, C.; Su, T.S.; Hu, C.; Yoshida, M.; Ting, L.P. The enhancer sequence of human hepatitis B virus can enhance the activity of its surface gene promoter. Nucleic Acids Res. 1987, 15, 2261–2268. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Quarleri, J. Core promoter: A critical region where the hepatitis B virus makes decisions. World J. Gastroenterol. 2014, 20, 425–435. [Google Scholar] [CrossRef]
- Günther, S.; Li, B.C.; Miska, S.; Krüger, D.; Meisel, H.; Willl, H. A Novel Method for Efficient Amplification of Whole Hepatitis B Virus Genomes Permits Rapid Functional Analysis and Reveals Deletion Mutants in Immunosuppressed Patients. J. Virol. 1995, 69, 5437–5444. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, A.; Karayiannis, P. HBeAg negative variants and their role in the natural history of chronic hepatitis B virus infection. World J. Gastroenterol. 2014, 20, 7644–7652. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.P.; Yao, C.Y. Rapid and quantitative detection of hepatitis B virus. World J. Gastroenterol. 2015, 21, 11954–11963. [Google Scholar] [CrossRef] [PubMed]
- Escobar, T.M.; Loyola, A.; Reinberg, D. Parental nucleosome segregation and the inheritance of cellular identity. Nat. Rev. Genet. 2021, 22, 379–392. [Google Scholar] [CrossRef]
- Burstein, E.; Hoberg, J.E.; Wilkinson, A.S.; Rumble, J.M.; Csomos, R.A.; Komarck, C.M.; Maine, G.N.; Wilkinson, J.C.; Mayo, M.W.; Duckett, C.S. COMMD proteins, a novel family of structural and functional homologs of MURR1. J. Biol. Chem. 2005, 280, 22222–22232. [Google Scholar] [CrossRef]
- Van De Sluis, B.; Rothuizen, J.; Pearson, P.L.; van Oost, B.A.; Wijmenga, C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum. Mol. Genet. 2002, 11, 165–173. [Google Scholar] [CrossRef] [PubMed]
- De Bie, P.; van de Sluis, B.; Burstein, E.; Duran, K.J.; Berger, R.; Duckett, C.S.; Wijmenga, C.; Klomp, L.W. Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem. J. 2006, 398, 63–71. [Google Scholar] [CrossRef]
- Geng, H.; Wittwer, T.; Dittrich-Breiholz, O.; Kracht, M.; Schmitz, M.L. Phosphorylation of NF-kappaB p65 at Ser468 controls its COMMD1-dependent ubiquitination and target gene-specific proteasomal elimination. EMBO Rep. 2009, 10, 381–386. [Google Scholar] [CrossRef]
- Mao, X.; Gluck, N.; Chen, B.; Starokadomskyy, P.; Li, H.; Maine, G.N.; Burstein, E. COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding. J. Biol. Chem. 2011, 286, 32355–32365. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Chen, H.; Kaneko, S.; Girones, R.; Anderson, R.; Hornbuckle, W.; Tennant, B.; Cote, P.; Gerin, J.; Purcell, R.; Miller, R. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 1993, 67, 1218–1226. [Google Scholar] [CrossRef]
- Sozzi, V.; Shen, F.; Chen, J.; Colledge, D.; Jackson, K.; Locarnini, S.; Yuan, Z.; Revill, P. In vitro studies identify a low replication phenotype for hepatitis B virus genotype H generally associated with occult HBV and less severe liver disease. Virology 2018, 519, 190–196. [Google Scholar] [CrossRef] [PubMed]
- Klein, N.; Bouchard, M.; Wang, L.; Kobarg, C.; Schneider, R. Src kinases involved in hepatitis B virus replication. EMBO J. 1999, 18, 5019–5027. [Google Scholar] [CrossRef] [PubMed]
- Klein, N.; Schneider, R. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol. 1997, 17, 6427–6436. [Google Scholar] [CrossRef][Green Version]
- Kusunoki, H.; Tanaka, T.; Kohno, T.; Kimura, H.; Hosoda, K.; Wakamatsu, K.; Hamaguchi, I. Expression, purification and characterization of hepatitis B virus X protein BH3-like motif-linker-Bcl-x(L) fusion protein for structural studies. Biochem. Biophys. Rep. 2016, 9, 159–165. [Google Scholar] [CrossRef]
- Kusunoki, H.; Tanaka, T.; Kohno, T.; Kimura, H.; Hosoda, K.; Wakamatsu, K.; Hamaguchi, I. NMR characterization of the interaction between Bcl-x(L) and the BH3-like motif of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 2019, 518, 445–450. [Google Scholar] [CrossRef]
- Kusunoki, H.; Tanaka, T.; Kohno, T.; Wakamatsu, K.; Hamaguchi, I. Structural characterization of the BH3-like motif of hepatitis B virus X protein. Biochem. Biophys. Res. Commun. 2014, 450, 741–745. [Google Scholar] [CrossRef]
- Geng, X.; Harry, B.; Zhou, Q.; Skeen-Gaar, R.; Ge, X.; Lee, E.; Mitani, S.; Xue, D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2012, 109, 18465–18470. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Huang, C.; Qin, Y.; McCombs, J.E.; Yuan, Q.; Harry, B.L.; Palmer, A.E.; Xia, N.S.; Xue, D. Hepatitis B virus X protein targets Bcl-2 proteins to increase intracellular calcium, required for virus replication and cell death induction. Proc. Natl. Acad. Sci. USA 2012, 109, 18471–18476. [Google Scholar] [CrossRef] [PubMed]
- Reddi, H.; Kumar, V. Self-association of the hepatitis B virus X protein in the yeast two-hybrid system. Biochem. Biophys. Res. Commun. 2004, 317, 1017–1022. [Google Scholar] [CrossRef]
- De Klerk, E.; AC‘t Hoen, P. Alternative mRNA transcription, processing, and translation: Insights from RNA sequencing. Trends Genet. 2015, 31, 128–139. [Google Scholar] [CrossRef]
- Landry, J.; Mager, D.; Wilhelm, B. Complex controls: The role of alternative promoters in mammalian genomes. Trends Genet. 2003, 19, 640–648. [Google Scholar] [CrossRef]
- Stadelmayer, B.; Diederichs, A.; Chapus, F.; Rivoire, M.; Neveu, G.; Alam, A.; Fraisse, L.; Carter, K.; Testoni, B.; Zoulim, F. Full-length 5′RACE identifies all major HBV transcripts in HBV-infected hepatocytes and patient serum. J. Hepatol. 2020, 73, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Starokadomskyy, P.; Gluck, N.; Li, H.; Chen, B.; Wallis, M.; Maine, G.N.; Mao, X.; Zaidi, I.W.; Hein, M.Y.; McDonald, F.J.; et al. CCDC22 deficiency in humans blunts activation of proinflammatory NF-kappaB signaling. J. Clin. Investig. 2013, 123, 2244–2256. [Google Scholar] [CrossRef]
- Wang, X.; He, S.; Zheng, X.; Huang, S.; Chen, H.; Chen, H.; Luo, W.; Guo, Z.; He, X.; Zhao, Q. Transcriptional analysis of the expression, prognostic value and immune infiltration activities of the COMMD protein family in hepatocellular carcinoma. BMC Cancer 2021, 21, 1001. [Google Scholar] [CrossRef]
- Sitterlin, D.; Lee, T.; Prigent, S.; Tiollais, P.; Butel, J.; Transy, C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 1997, 71, 6194–6199. [Google Scholar] [CrossRef] [PubMed]
- Angers, S.; Li, T.; Yi, X.; MacCoss, M.J.; Moon, R.T.; Zheng, N. Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 2006, 443, 590–593. [Google Scholar] [CrossRef]
- Precious, B.; Childs, K.; Fitzpatrick-Swallow, V.; Goodbourn, S.; Randall, R.E. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 2005, 79, 13434–13441. [Google Scholar] [CrossRef] [PubMed]
- Hrecka, K.; Gierszewska, M.; Srivastava, S.; Kozaczkiewicz, L.; Swanson, S.K.; Florens, L.; Washburn, M.P.; Skowronski, J. Lentiviral Vpr usurps Cul4-DDB1[VprBP] E3 ubiquitin ligase to modulate cell cycle. Proc. Natl. Acad. Sci. USA 2007, 104, 11778–11783. [Google Scholar] [CrossRef]
- Vasilenko, N.L.; Snider, M.; Labiuk, S.L.; Lobanov, V.A.; Babiuk, L.A.; van Drunen Littel-van den Hurk, S. Bovine herpesvirus-1 VP8 interacts with DNA damage binding protein-1 (DDB1) and is monoubiquitinated during infection. Virus Res. 2012, 167, 56–66. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, R.; Zhou, Z.; Wen, S.; Lin, L.; Chen, S.; Shan, Y.; Cong, Y.; Wang, S. Macrophage migration inhibitory factor interacts with HBx and inhibits its apoptotic activity. Biochem. Biophys. Res. Commun. 2006, 342, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Shokri, S.; Mahmoudvand, S.; Taherkhani, R.; Farshadpour, F.; Jalalian, F.A. Complexity on modulation of NF-kappaB pathways by hepatitis B and C: A double-edged sword in hepatocarcinogenesis. J. Cell. Physiol. 2019, 234, 14734–14742. [Google Scholar] [CrossRef]
- Bohrer, A.; Yoshimoto, N.; Sekiguchi, A.; Rykulski, N.; Saito, K.; Takahashi, H. Alternative translational initiation of ATP sulfurylase underlying dual localization of sulfate assimilation pathways in plastids and cytosol in Arabidopsis thaliana. Front. Plant. Sci 2015, 5, 750. [Google Scholar] [CrossRef] [PubMed]
- Outten, C.; Culotta, V. Alternative start sites in the Saccharomyces cerevisiae GLR1 gene are responsible for mitochondrial and cytosolic isoforms of glutathione reductase. J. Biol. Chem. 2004, 279, 7785–7791. [Google Scholar] [CrossRef] [PubMed]
- Khoury, M.; Bourdon, J. The isoforms of the p53 protein. Cold Spring Harb. Perspect. Biol. 2010, 2, a000927. [Google Scholar] [CrossRef]
- Logette, E.; Wotawa, A.; Solier, S.; Desoche, L.; Solary, E.; Corcos, L. The human caspase-2 gene: Alternative promoters, pre-mRNA splicing and AUG usage direct isoform-specific expression. Oncogene 2003, 22, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Kadmiel, M.; Cidlowski, J. Glucocorticoid receptor signaling in health and disease. Trends Pharm. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Brubaker, S.; Gauthier, A.; Mills, E.; Ingolia, N.; Kagan, J. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell 2014, 156, 800–811. [Google Scholar] [CrossRef]
- Ivanov, P.; Anderson, P. Alternative translation initiation in immunity: MAVS learns new tricks. Trends Immunol. 2014, 35, 188–189. [Google Scholar] [CrossRef][Green Version]
- Goyama, S.; Schibler, J.; Mulloy, J. Alternative translation initiation generates the N-terminal truncated form of RUNX1 that retains hematopoietic activity. Exp. Hematol. 2019, 72, 27–35. [Google Scholar] [CrossRef]
- Liang, H.; Chen, X.; Yin, Q.; Ruan, D.; Zhao, X.; Zhang, C.; McNutt, M.; Yin, Y. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat. Commun. 2017, 8, 14771. [Google Scholar] [CrossRef]
- Trulley, P.; Snieckute, G.; Bekker-Jensen, D.; Menon, M.; Freund, R.; Kotlyarov, A.; Olsen, J.; Diaz-Muñoz, M.; Turner, M.; Bekker-Jensen, S.; et al. Alternative Translation Initiation Generates a Functionally Distinct Isoform of the Stress-Activated Protein Kinase MK2. Cell Rep. 2019, 27, 2859–2870. [Google Scholar] [CrossRef] [PubMed]
- Benassayag, C.; Montero, L.; Colombié, N.; Gallant, P.; Cribbs, D.; Morello, D. Human c-Myc isoforms differentially regulate cell growth and apoptosis in Drosophila melanogaster. Mol. Cell. Biol. 2005, 25, 9897–9909. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.; Diani, E.; Lievens, P. New insights into functional roles of the polypyrimidine tract-binding protein. Int. J. Mol. Sci. 2013, 14, 22906–22932. [Google Scholar] [CrossRef] [PubMed]
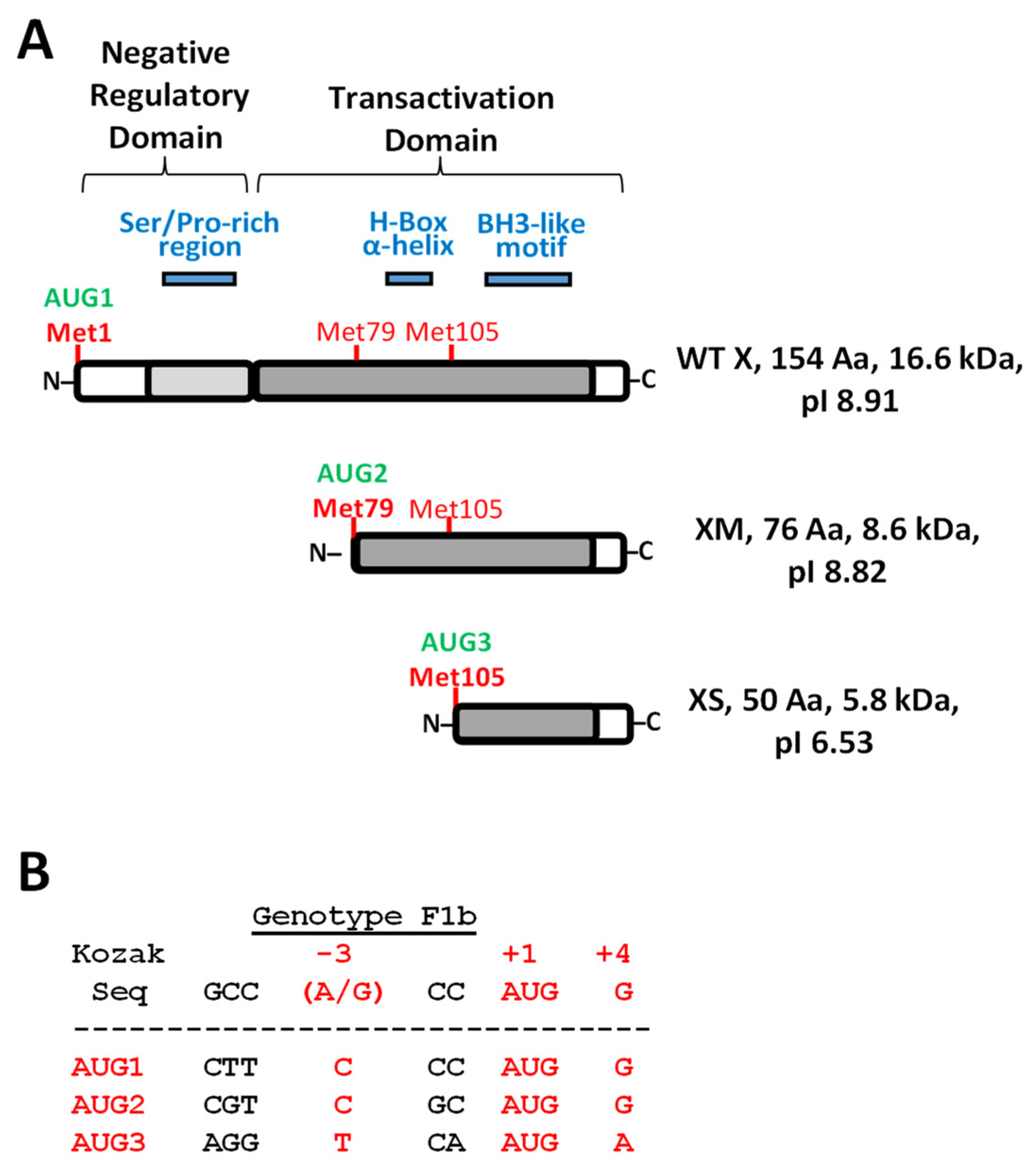
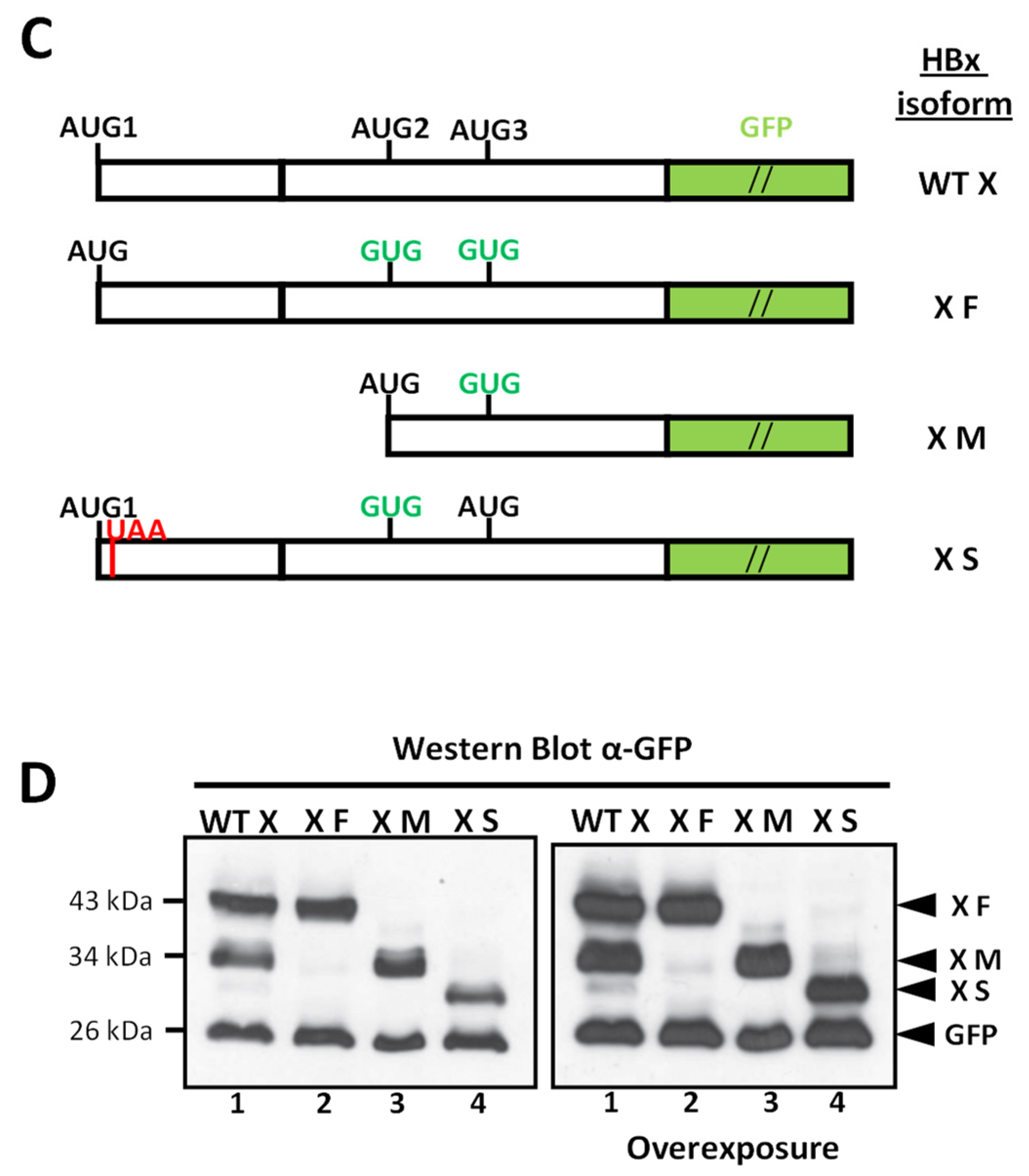
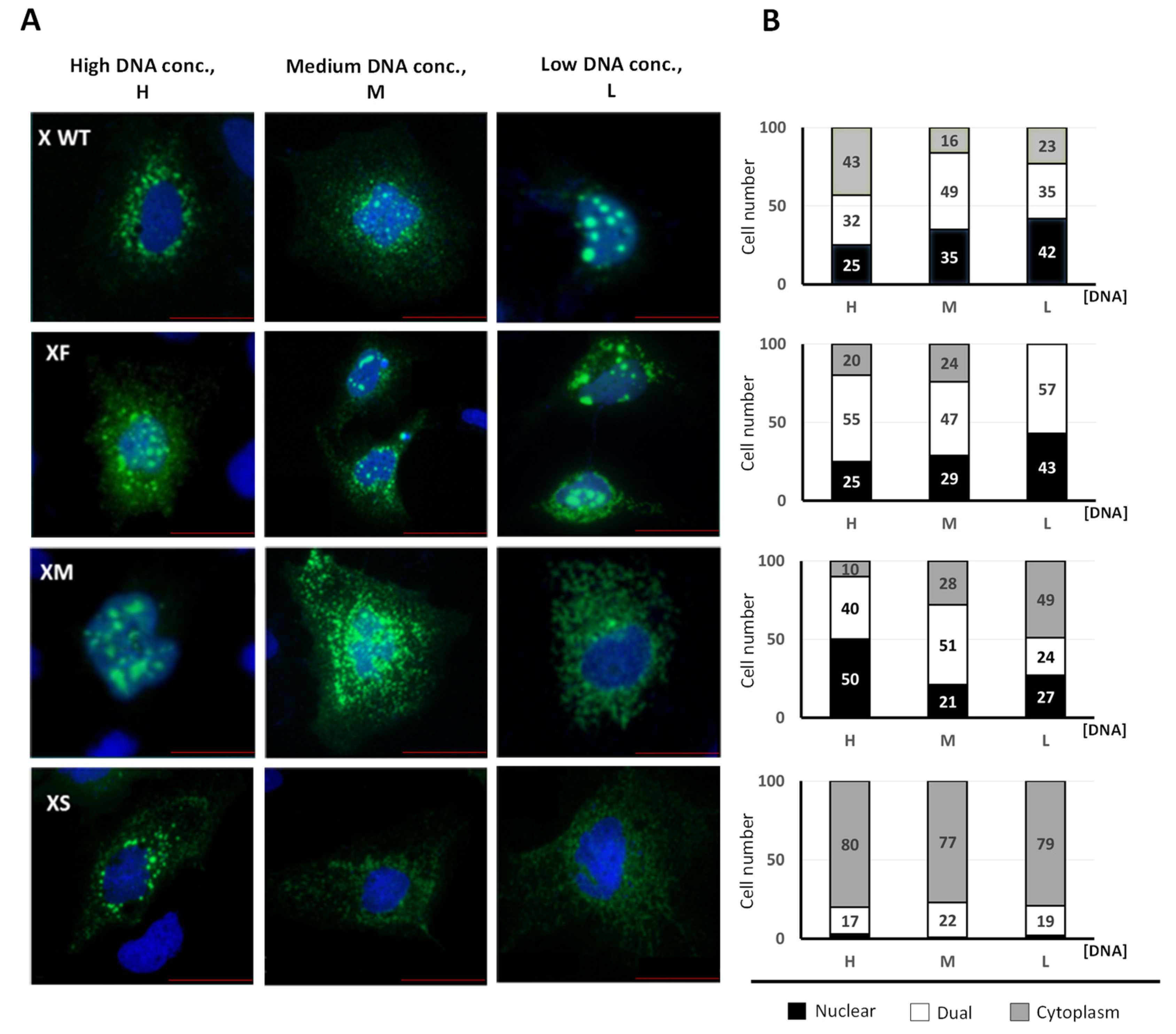

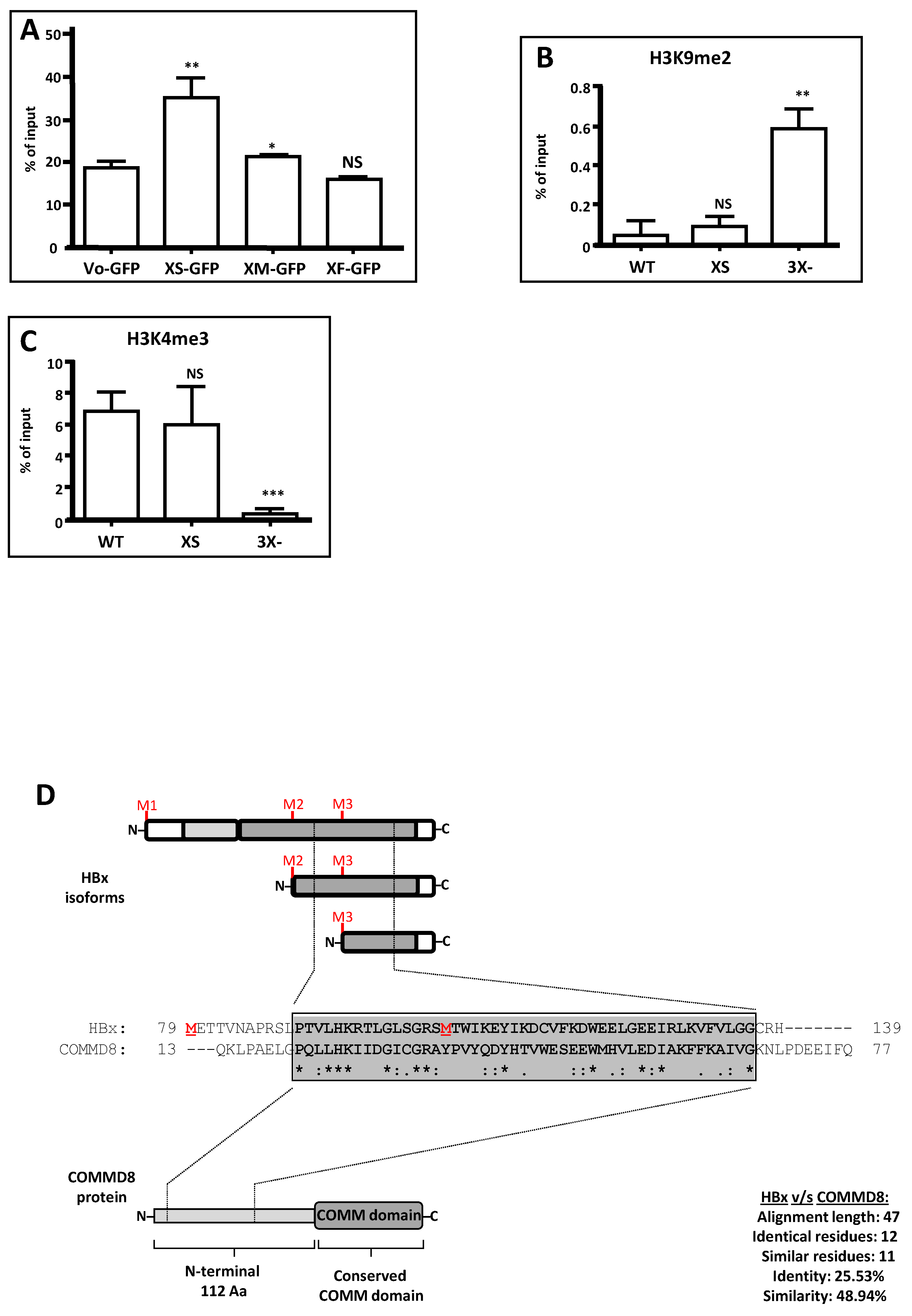
| WEB TOOL 2 | Predicted Disordered HBx Regions 1 | |||
|---|---|---|---|---|
| Region 1–78 | Region 79–104 | Region 105–154 | ||
| PONDR VL-XT 3 | 26–52 | 85–96 | – | |
| DisEMBL 1.5 4 | 24–50 | – | – | |
| Phyre 2 5 | 1–7 | 25–52 | 98–105 | 147–154 |
| PrDOS 6 | 1–3 | 26–44 | – | 150–154 |
| Depicter 7 | 1–5 | 15–78 | – | – |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hernández, S.; Álvarez-Astudillo, F.; Garrido, D.; Prieto, C.; Loyola, A.; Villanueva, R.A. Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication. Biomedicines 2021, 9, 1701. https://doi.org/10.3390/biomedicines9111701
Hernández S, Álvarez-Astudillo F, Garrido D, Prieto C, Loyola A, Villanueva RA. Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication. Biomedicines. 2021; 9(11):1701. https://doi.org/10.3390/biomedicines9111701
Chicago/Turabian StyleHernández, Sergio, Francisca Álvarez-Astudillo, Daniel Garrido, Cristian Prieto, Alejandra Loyola, and Rodrigo A. Villanueva. 2021. "Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication" Biomedicines 9, no. 11: 1701. https://doi.org/10.3390/biomedicines9111701
APA StyleHernández, S., Álvarez-Astudillo, F., Garrido, D., Prieto, C., Loyola, A., & Villanueva, R. A. (2021). Canonical and Divergent N-Terminal HBx Isoform Proteins Unveiled: Characteristics and Roles during HBV Replication. Biomedicines, 9(11), 1701. https://doi.org/10.3390/biomedicines9111701






