Different HCV Exposure Drives Specific miRNA Profile in PBMCs of HIV Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Groups
2.2. High Throughput Sequencing of Small RNA
2.3. Data Processing Pipeline
2.4. Statistical Analysis
2.4.1. Data Preprocessing
2.4.2. Data Exploratory Analysis
2.4.3. Differential Expression Analysis
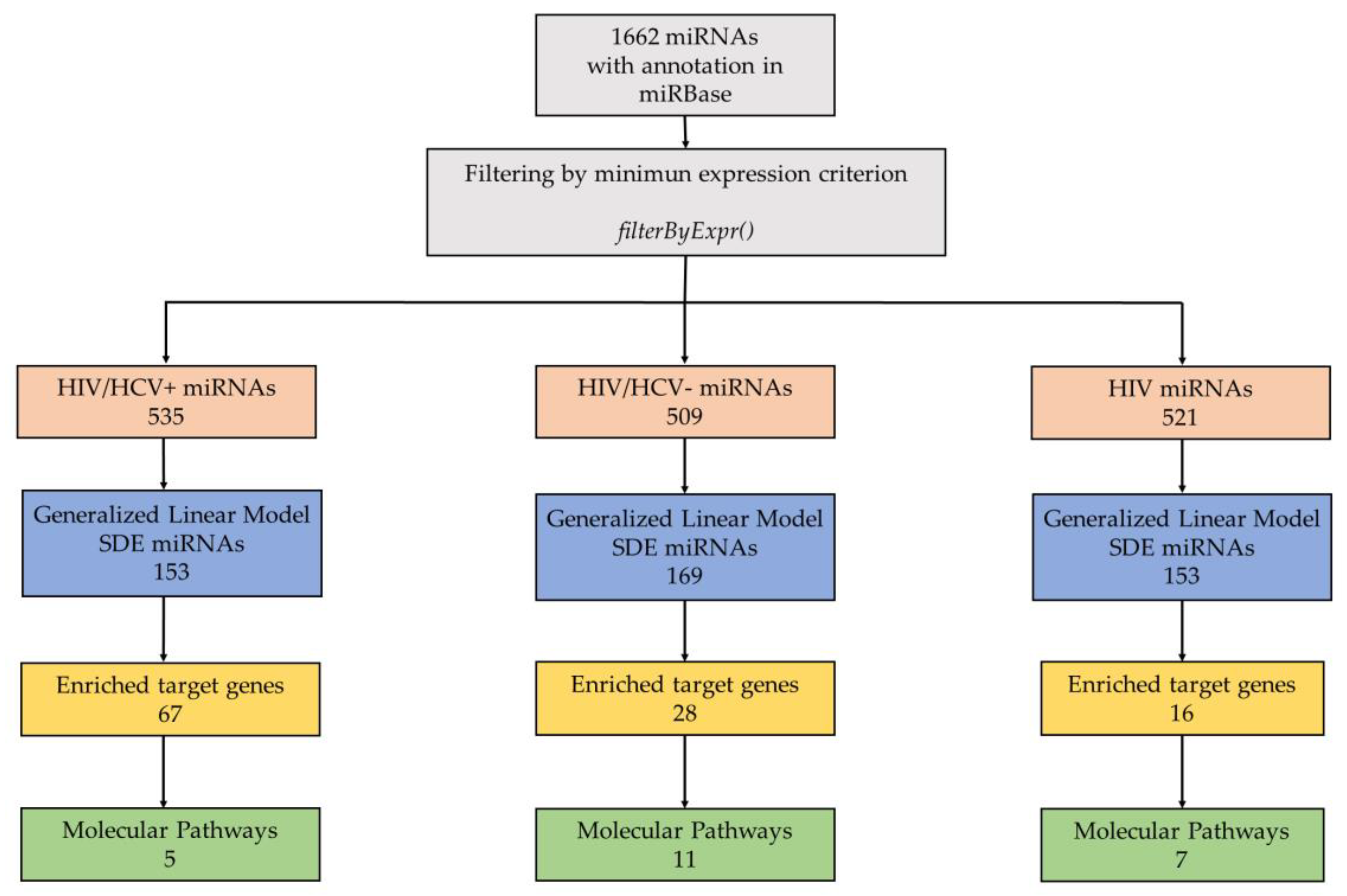
2.5. miRNA-Based Target Prediction and Pathway Enrichment Analysis of Target Genes
3. Results
3.1. Clinical Characteristics of Each Group of Patients
3.2. Differentially Expressed miRNAs between Study Groups
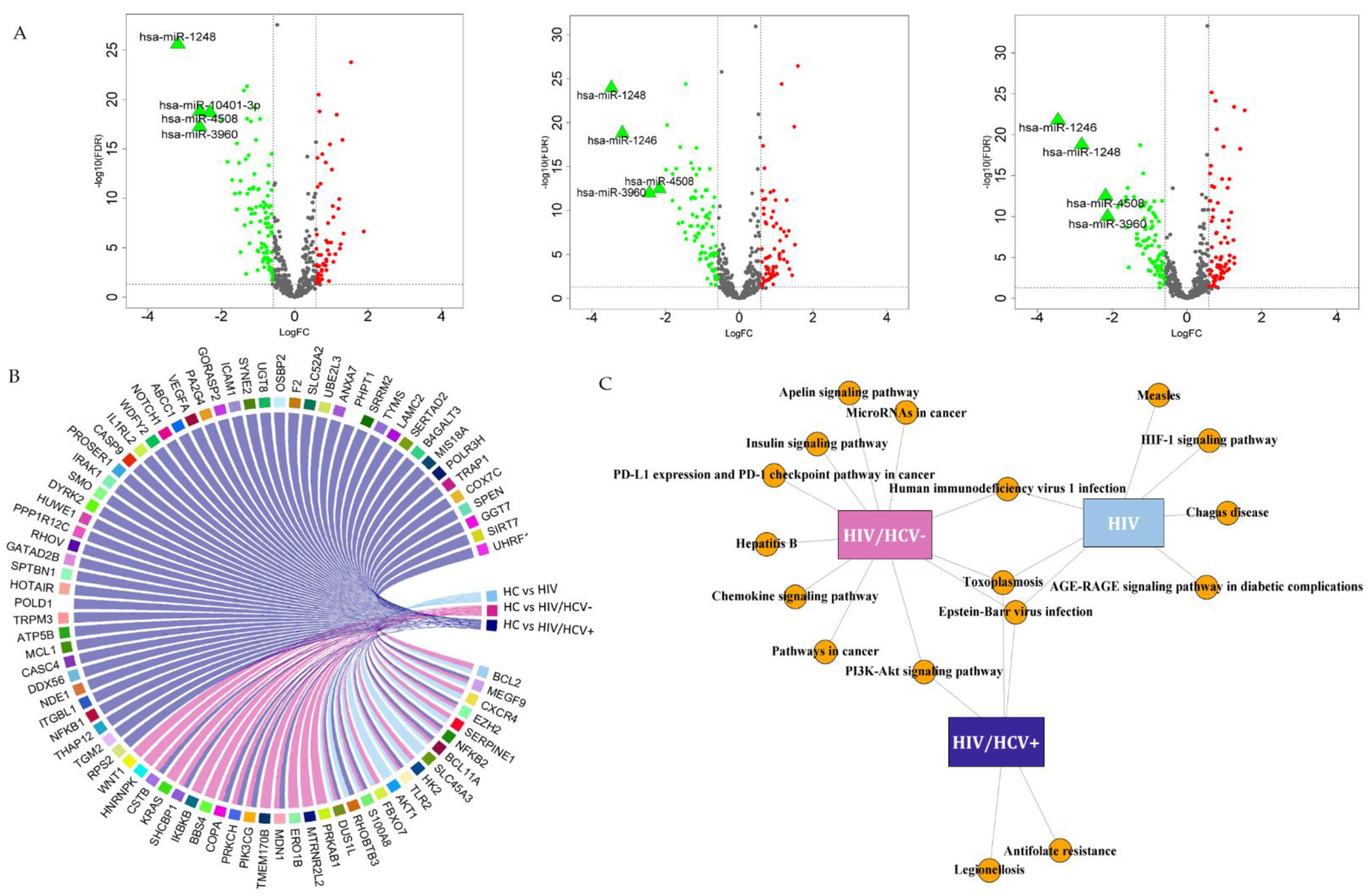
3.2.1. HIV/HCV+ Co-Infected Patients
3.2.2. HIV/HCV- Spontaneous Clearance
3.2.3. HIV+ Infected Group
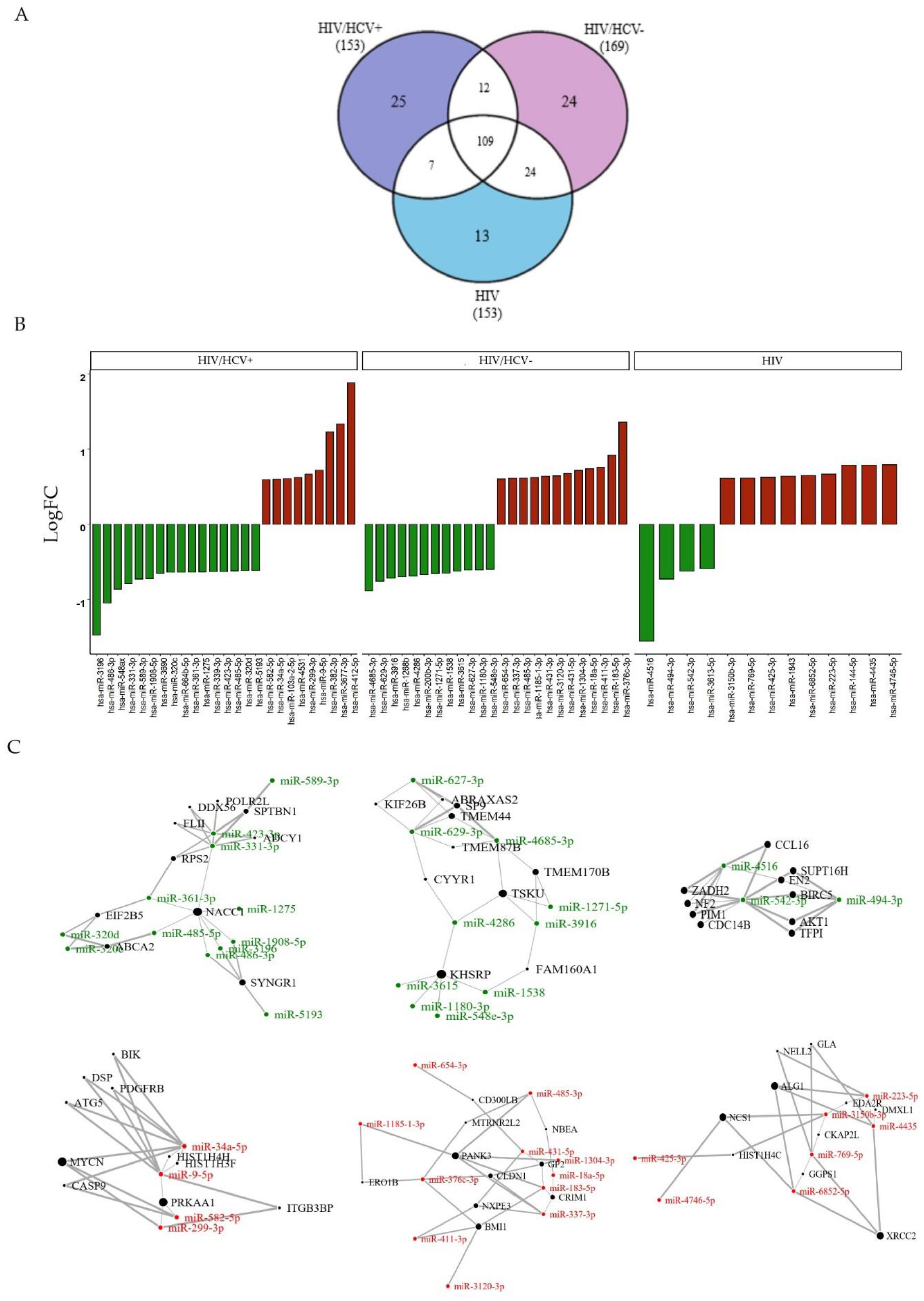
3.2.4. Specific Viral Infection Group Signatures Due to HIV+, HIV/HCV+ or HIV/HCV-
4. Discussion
4.1. HIV/HCV+ miRNA Profile
4.2. HIV/HCV- Spontaneous Clearance Profile
4.3. HIV miRNA Profile
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Arora, U.; Garg, P.; Agarwal, S.; Nischal, N.; Shalimar; Wig, N. Complexities in the treatment of coinfection with HIV, hepatitis B, hepatitis C, and tuberculosis. Lancet Infect. Dis. 2021. [Google Scholar] [CrossRef]
- Hernandez, M.D.; Sherman, K.E. HIV/hepatitis C coinfection natural history and disease progression. Curr. Opin. HIV AIDS 2011, 6, 478–482. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.A.; Feld, J.J. Acute hepatitis C: Management in the rapidly evolving world of HCV. Curr. Gastroenterol. Rep. 2014, 16, 371. [Google Scholar] [CrossRef]
- Grebely, J.; Page, K.; Sacks-Davis, R.; van der Loeff, M.S.; Rice, T.M.; Bruneau, J.; Morris, M.D.; Hajarizadeh, B.; Amin, J.; Cox, A.L.; et al. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology 2014, 59, 109–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerlach, J.T.; Diepolder, H.M.; Zachoval, R.; Gruener, N.H.; Jung, M.C.; Ulsenheimer, A.; Schraut, W.W.; Schirren, C.A.; Waechtler, M.; Backmund, M.; et al. Acute hepatitis C: High rate of both spontaneous and treatment-induced viral clearance. Gastroenterology 2003, 125, 80–88. [Google Scholar] [CrossRef]
- Villano, S.A.; Vlahov, D.; Nelson, K.E.; Cohn, S.; Thomas, D.L. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology 1999, 29, 908–914. [Google Scholar] [CrossRef]
- Liu, L.; Fisher, B.E.; Thomas, D.L.; Cox, A.L.; Ray, S.C. Spontaneous clearance of primary acute hepatitis C virus infection correlated with high initial viral RNA level and rapid HVR1 evolution. Hepatology 2012, 55, 1684–1691. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.L.; Thio, C.L.; Martin, M.P.; Qi, Y.; Ge, D.; O’Huigin, C.; Kidd, J.; Kidd, K.; Khakoo, S.I.; Alexander, G.; et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 2009, 461, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.E. The impact of chronic hepatitis C virus infection on mortality. J. Infect. Dis. 2012, 206, 461–463. [Google Scholar] [CrossRef] [Green Version]
- Brochado-Kith, Ó.; Gómez Sanz, A.; Real, L.M.; Crespo García, J.; Ryan Murúa, P.; Macías, J.; Cabezas González, J.; Troya, J.; Pineda, J.A.; Arias Loste, M.T.; et al. MicroRNA Profile of HCV Spontaneous Clarified Individuals, Denotes Previous HCV Infection. J. Clin. Med. 2019, 8, 849. [Google Scholar] [CrossRef] [Green Version]
- Funderburg, N.T.; Mehta, N.N. Lipid Abnormalities and Inflammation in HIV Inflection. Curr. HIV/AIDS Rep. 2016, 13, 218–225. [Google Scholar] [CrossRef] [Green Version]
- Ono, C.; Fukuhara, T.; Li, S.; Wang, J.; Sato, A.; Izumi, T.; Fauzyah, Y.; Yamamoto, T.; Morioka, Y.; Dokholyan, N.V.; et al. Various miRNAs compensate the role of miR-122 on HCV replication. PLoS Pathog. 2020, 16, e1008308. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, F.; Argyris, E.; Chen, K.; Liang, Z.; Tian, H.; Huang, W.; Squires, K.; Verlinghieri, G.; Zhang, H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 2007, 13, 1241–1247. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venables, W.; Ripley, B. Modern Applied Statistics; Fourth, S., Ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moghoofei, M.; Najafipour, S.; Mostafaei, S.; Tavakoli, A.; Bokharaei-Salim, F.; Ghorbani, S.; Javanmard, D.; Ghaffari, H.; Monavari, S.H. MicroRNAs Profiling in HIV, HCV, and HIV/HCV Co-Infected Patients. Curr. HIV Res. 2021, 19, 27–34. [Google Scholar] [CrossRef]
- Yousefpouran, S.; Mostafaei, S.; Manesh, P.V.; Iranifar, E.; Bokharaei-Salim, F.; Nahand, J.S.; Mirzaei, H.; Taran, M.; Babaei, F.; Sayad, B.; et al. The assessment of selected MiRNAs profile in HIV, HBV, HCV, HIV/HCV, HIV/HBV Co-infection and elite controllers for determination of biomarker. Microb Pathog 2020, 147, 104355. [Google Scholar] [CrossRef]
- Dobiásová, M.; Frohlich, J. The plasma parameter log (TG/HDL-C) as an atherogenic index: Correlation with lipoprotein particle size and esterification rate in apoB-lipoprotein-depleted plasma (FER(HDL)). Clin. Biochem. 2001, 34, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Hernando, C.; Suárez, Y.; Rayner, K.J.; Moore, K.J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 2011, 22, 86–92. [Google Scholar] [CrossRef]
- Yang, Z.; Cappello, T.; Wang, L. Emerging role of microRNAs in lipid metabolism. Acta Pharm. Sin. B 2015, 5, 145–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubé, M.P.; Cadden, J.J. Lipid metabolism in treated HIV Infection. Best Pract. Res. Clin. Endocrinol. Metab. 2011, 25, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Collins, L.F.; Adekunle, R.O.; Cartwright, E.J. Metabolic Syndrome in HIV/HCV Co-infected Patients. Curr. Treat. Options Infect. Dis. 2019, 11, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Noren Hooten, N.; Fitzpatrick, M.; Wood, W.H.; De, S.; Ejiogu, N.; Zhang, Y.; Mattison, J.A.; Becker, K.G.; Zonderman, A.B.; Evans, M.K. Age-related changes in microRNA levels in serum. Aging 2013, 5, 725–740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Q.; Hoffman, B.; Liu, Q. PI3K-Akt signaling pathway upregulates hepatitis C virus RNA translation through the activation of SREBPs. Virology 2016, 490, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Boštjančič, E.; Bandelj, E.; Luzar, B.; Poljak, M.; Glavač, D. Hepatic expression of miR-122, miR-126, miR-136 and miR-181a and their correlation to histopathological and clinical characteristics of patients with hepatitis C. J. Viral Hepat. 2015, 22, 146–157. [Google Scholar] [CrossRef]
- Li, X.; Zhang, W.; Xu, K.; Lu, J. miR-34a promotes liver fibrosis in patients with chronic hepatitis via mediating Sirt1/p53 signaling pathway. Pathol. Res. Pract. 2020, 216, 152876. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Chen, E.; Haleyurgirisetty, M.; Lee, S.; Hewlett, I.; Devadas, K. Comparison of miRNA Expression Profiles between HIV-1 and HIV-2 Infected Monocyte-Derived Macrophages (MDMs) and Peripheral Blood Mononuclear Cells (PBMCs). Int. J. Mol. Sci. 2020, 21, 6970. [Google Scholar] [CrossRef]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3-mediated maturation of miR-126-5p promotes ovarian cancer progression via PTEN-mediated PI3K/Akt/mTOR pathway. Cancer Gene Ther. 2021, 28, 335–349. [Google Scholar] [CrossRef]
- Gnanamony, M.; Demirkhanyan, L.; Ge, L.; Sojitra, P.; Bapana, S.; Norton, J.A.; Gondi, C.S. Circular dumbbell miR-34a-3p and -5p suppresses pancreatic tumor cell-induced angiogenesis and activates macrophages. Oncol. Lett. 2021, 21, 75. [Google Scholar] [CrossRef]
- Chugh, P.; Bradel-Tretheway, B.; Monteiro-Filho, C.M.; Planelles, V.; Maggirwar, S.B.; Dewhurst, S.; Kim, B. Akt inhibitors as an HIV-1 infected macrophage-specific anti-viral therapy. Retrovirology 2008, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Tian, Y.; Machida, K.; Lai, M.M.; Luo, G.; Foung, S.K.; Ou, J.H. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J. Biol. Chem. 2012, 287, 41922–41930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Zhu, L.; Qiu, C.; Xu, X.; Zhang, L.; Ding, X.; Liao, Q.; Xu, J.; Zhang, X. MicroRNA miR-126-5p Enhances the Inflammatory Responses of Monocytes to Lipopolysaccharide Stimulation by Suppressing Cylindromatosis in Chronic HIV-1 Infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Guo, J.T.; Jiang, D.; Guo, H.; Taylor, J.M.; Block, T.M. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J. Virol. 2008, 82, 8215–8223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherry, S.; Kunte, A.; Wang, H.; Coyne, C.; Rawson, R.B.; Perrimon, N. COPI activity coupled with fatty acid biosynthesis is required for viral replication. PLoS Pathog. 2006, 2, e102. [Google Scholar] [CrossRef] [Green Version]
- Aranda, J.F.; Madrigal-Matute, J.; Rotllan, N.; Fernández-Hernando, C. MicroRNA modulation of lipid metabolism and oxidative stress in cardiometabolic diseases. Free Radic. Biol. Med. 2013, 64, 31–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, X.; Xi, Q.Y.; Wei, S.; Wu, D.; Ye, R.S.; Chen, T.; Qi, Q.E.; Jiang, Q.Y.; Wang, S.B.; Wang, L.N.; et al. Critical role of miR-125b in lipogenesis by targeting stearoyl-CoA desaturase-1 (SCD-1). J. Anim. Sci. 2016, 94, 65–76. [Google Scholar] [CrossRef] [Green Version]
- Hong, G.; Zhang, W.; Li, H.; Shen, X.; Guo, Z. Separate enrichment analysis of pathways for up- and downregulated genes. J. R. Soc. Interface 2014, 11, 20130950. [Google Scholar] [CrossRef] [Green Version]
- Guévin, C.; Manna, D.; Bélanger, C.; Konan, K.V.; Mak, P.; Labonté, P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology 2010, 405, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.Y.; Suzuki, H.; Honda, M.; Okada, H.; Kaneko, S.; Inoue, I.; Ebisui, E.; Hashimoto, K.; Carninci, P.; Kanki, K.; et al. Prevention of hepatocellular carcinoma by targeting MYCN-positive liver cancer stem cells with acyclic retinoid. Proc. Natl. Acad. Sci. USA 2018, 115, 4969–4974. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.; Song, X.; Yu, C. Identification of genes in hepatocellular carcinoma induced by non-alcoholic fatty liver disease. Cancer Biomark. 2020, 29, 69–78. [Google Scholar] [CrossRef]
- Kocabayoglu, P.; Lade, A.; Lee, Y.A.; Dragomir, A.C.; Sun, X.; Fiel, M.I.; Thung, S.; Aloman, C.; Soriano, P.; Hoshida, Y.; et al. β-PDGF receptor expressed by hepatic stellate cells regulates fibrosis in murine liver injury, but not carcinogenesis. J. Hepatol. 2015, 63, 141–147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhi, X.; Lin, L.; Yang, S.; Bhuvaneshwar, K.; Wang, H.; Gusev, Y.; Lee, M.H.; Kallakury, B.; Shivapurkar, N.; Cahn, K.; et al. βII-Spectrin (SPTBN1) suppresses progression of hepatocellular carcinoma and Wnt signaling by regulation of Wnt inhibitor kallistatin. Hepatology 2015, 61, 598–612. [Google Scholar] [CrossRef]
- Chen, S.; Li, J.; Zhou, P.; Zhi, X. SPTBN1 and cancer, which links? J. Cell Physiol. 2020, 235, 17–25. [Google Scholar] [CrossRef]
- Zhang, W.; Jiang, X.; Bao, J.; Wang, Y.; Liu, H.; Tang, L. Exosomes in Pathogen Infections: A Bridge to Deliver Molecules and Link Functions. Front. Immunol. 2018, 9, 90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, E.K.; Fahmy, S.A.; Effat, H.; Wahab, A.H.A. Circulating MiR-210 and MiR-1246 as Potential Biomarkers for Differentiating Hepatocellular Carcinoma from Metastatic Tumors in the Liver. J. Med. Biochem. 2019, 38, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Chettimada, S.; Lorenz, D.R.; Misra, V.; Wolinsky, S.M.; Gabuzda, D. Small RNA sequencing of extracellular vesicles identifies circulating miRNAs related to inflammation and oxidative stress in HIV patients. BMC Immunol. 2020, 21, 57. [Google Scholar] [CrossRef]
- Mumdzhiev, N.H.; Radicheva, D.V.; Radicheva, M.P.; Tenev, R.V.; Vasileva, Z.D. Spontaneous Clearance of Chronic HCV: The Key Ending Left in the Dark. Open Access Maced. J. Med. Sci. 2019, 7, 1657–1659. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.J.; von Hahn, T.; Tscherne, D.M.; Syder, A.J.; Panis, M.; Wölk, B.; Hatziioannou, T.; McKeating, J.A.; Bieniasz, P.D.; Rice, C.M. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 2007, 446, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Gupta, V.; Patterson-Fortin, J.; Bhattacharya, S.; Katlinski, K.; Wu, J.; Varghese, B.; Carbone, C.J.; Aressy, B.; Fuchs, S.Y.; et al. A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Rep. 2013, 5, 180–193. [Google Scholar] [CrossRef] [Green Version]
- Sung, P.S.; Shin, E.C.; Yoon, S.K. Interferon Response in Hepatitis C Virus (HCV) Infection: Lessons from Cell Culture Systems of HCV Infection. Int. J. Mol. Sci. 2015, 16, 23683–23694. [Google Scholar] [CrossRef] [Green Version]
- Huang, M.; Jiang, J.D.; Peng, Z. Recent advances in the anti-HCV mechanisms of interferon. Acta Pharm. Sin. B 2014, 4, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.Q.; Zhao, M.; Kong, W.H.; Peng, J.S.; Wang, F.; Qiu, H.Y.; Zhu, Z.R.; Tang, L.; Sang, M.; Wu, J.G.; et al. Antiretroviral Therapy Fails to Restore Levels of HIV-1 Restriction miRNAs in PBMCs of HIV-1-infected MSM. Medicine 2015, 94, e2116. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Ding, C.; Jiang, X.; Gao, Y. Advances in Developing CAR T-Cell Therapy for HIV Cure. Front. Immunol. 2020, 11, 361. [Google Scholar] [CrossRef] [PubMed]
- Whisnant, A.W.; Bogerd, H.P.; Flores, O.; Ho, P.; Powers, J.G.; Sharova, N.; Stevenson, M.; Chen, C.H.; Cullen, B.R. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 2013, 4, e000193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palazon, A.; Goldrath, A.W.; Nizet, V.; Johnson, R.S. HIF transcription factors, inflammation, and immunity. Immunity 2014, 41, 518–528. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, X.; Pedroza-Pacheco, I.; Nawroth, I.; Kliszczak, A.E.; Magri, A.; Paes, W.; Rubio, C.O.; Yang, H.; Ashcroft, M.; Mole, D.; et al. Hypoxic microenvironment shapes HIV-1 replication and latency. Commun. Biol. 2020, 3, 376. [Google Scholar] [CrossRef]
- Huang, H.; Santoso, N.; Power, D.; Simpson, S.; Dieringer, M.; Miao, H.; Gurova, K.; Giam, C.Z.; Elledge, S.J.; Zhu, J. FACT Proteins, SUPT16H and SSRP1, Are Transcriptional Suppressors of HIV-1 and HTLV-1 That Facilitate Viral Latency. J. Biol. Chem. 2015, 290, 27297–27310. [Google Scholar] [CrossRef] [Green Version]
- Duverger, A.; Wolschendorf, F.; Anderson, J.C.; Wagner, F.; Bosque, A.; Shishido, T.; Jones, J.; Planelles, V.; Willey, C.; Cron, R.Q.; et al. Kinase control of latent HIV-1 infection: PIM-1 kinase as a major contributor to HIV-1 reactivation. J. Virol. 2014, 88, 364–376. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, D.; Roy, D.; Cassol, E. Examining Relationships between Metabolism and Persistent Inflammation in HIV Patients on Antiretroviral Therapy. Mediators Inflamm. 2018, 2018, 6238978. [Google Scholar] [CrossRef]
| HC | HIV/HCV+ | HIV/HCV- | HIV+ | p-Values | ||||
|---|---|---|---|---|---|---|---|---|
| n | 32 | 45 | 36 | 36 | A | B | C | D |
| Sex (%) | 0.523 | 0.830 | 1.000 | 0.667 | ||||
| 16/32 (50.00%) | 27/45 (60.00%) | 20/36 (55.56%) | 18/36 (50.00%) | ||||
| Age (years) | 49.00 (43.75–55.00) | 50.00 (45.00–54.00) | 52.00 (48.00–56.00) | 49.50 (42.75–56.25) | 0.592 | 0.376 | 0.619 | 0.250 |
| Time of HIV Infection (Months) | - | 256.10 (115.12–324.76) | 275.70 (142.72–335.41) | 219.83 (88.64–265.33) | - | - | - | 0.187 |
| Risk (%) | - | - | - | <0.001 | ||||
| 0/0 (0%) | 26/40 (65.00%) | 21/31 (67.74%) | 0/30 (0.00%) | ||||
| 0/0 (0%) | 14/40 (35.00%) | 10/31 (32.26%) | 30/30 (100.00%) | ||||
| HIV clinical status (%) | - | - | - | 0.169 | ||||
| 0/0 (0%) | 22/43 (51.16%) | 13/32 (40.62%) | 23/33 (69.70%) | ||||
| 0/0 (0%) | 7/43 (16.28%) | 8/32 (25.00%) | 5/33 (15.15%) | ||||
| 0/0 (0%) | 14/43 (32.56%) | 11/32 (34.38%) | 5/33 (15.15%) | ||||
| HCV genotype (%) | - | - | - | - | ||||
| 0/0 (0%) | 16/39 (41.03%) | 0/2 (0.00%) | 0/0 (0%) | ||||
| 0/0 (0%) | 8/39 (20.51%) | 1/2 (50.00%) | 0/0 (0%) | ||||
| 0/0 (0%) | 1/39 (2.56%) | 0/2 (0.00%) | 0/0 (0%) | ||||
| 0/0 (0%) | 2/39 (5.13%) | 0/2 (0.00%) | 0/0 (0%) | ||||
| 0/0 (0%) | 12/39 (30.77%) | 1/2 (50.00%) | 0/0 (0%) | ||||
| IL28b (%) | 0.192 | 0.085 | 0.730 | 0.001 | ||||
| 15/29 (51.72%) | 14/45 (31.11%) | 27/36 (75.00%) | 19/36 (52.78%) | ||||
| 12/29 (41.38%) | 25/45 (55.56%) | 6/36 (16.67%) | 16/36 (44.44%) | ||||
| 2/29 (6.90%) | 6/45 (13.33%) | 3/36 (8.33%) | 1/36 (2.78%) | ||||
| CD4+ T cells (cells/mm3) | - | 712.00 (530.00–1047.60) | 732.70 (560.70–916.65) | 833.00 (697.42–1062.00) | - | - | - | 0.101 |
| CD4+ T cells (%) | - | 33.00 (27.00–41.50) | 35.00 (31.00–40.00) | 38.28 (31.50–44.75) | - | - | - | 0.460 |
| CD8+ T cells (cells/mm3) | - | 790.00 (634.00–1048.00) | 870.50 (636.50–1104.75) | 905.50 (795.42–1244.50) | - | - | - | 0.750 |
| CD8+ T cells (%) | - | 43.68 (12.35) | 37.50 (9.40) | 38.69 (8.44) | - | - | - | 0.158 |
| CD4/CD8 (%) | - | 0.81 (0.56–1.00) | 0.99 (0.75–1.16) | 1.06 (0.69–1.27) | - | - | - | 0.269 |
| HC | HIV/HCV+ | HIV/HCV- | HIV+ | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| n | 32 | 45 | 36 | 36 | A | B | C | D |
| Weight (kg) | 75.80 (64.60–88.00) | 64.00 (56.25–73.40) | 74.75 (62.62–80.75) | 67.00 (61.10–79.00) | 0.001 | 0.347 | 0.173 | 0.015 |
| BMI | 25.22 (22.78–28.58) | 22.48 (20.81–25.72) | 24.72 (22.07–27.89) | 24.75 (22.44–27.12) | 0.005 | 0.500 | 0.737 | 0.026 |
| Lipid Profile | ||||||||
| Glucose | 90.50 (80.75–93.75) | 88.00 (86.00–95.00) | 96.50 (89.50–102.25) | 92.50 (86.25–96.75) | 0.368 | 0.051 | 0.282 | 0.403 |
| Gluc > 110 | 2/26 (7.69%) | 3/37 (8.11%) | 5/28 (17.86%) | 2/26 (7.69%) | 1.000 | 0.480 | 1.000 | 0.377 |
| Total cholesterol | 207.50 (186.50–225.25) | 186.00 (166.00–203.00) | 192.00 (181.00–221.50) | 185.00 (172.50–204.75) | 0.010 | 0.178 | 0.278 | 0.343 |
| Chol > 200 | 15/26 (57.69%) | 10/35 (28.57%) | 11/27 (40.74%) | 10/26 (38.46%) | 0.043 | 0.337 | 0.267 | 0.560 |
| LDL | - | 108.00 (87.00–130.00) | 115.00 (100.00–133.00) | 115.50 (100.50–144.25) | - | - | - | 0.293 |
| LDL > 130 | - | 8/23 (34.78%) | 6/15 (40.00%) | 9/18 (50.00%) | - | - | - | 0.614 |
| HDL | 67.50 (50.75–78.75) | 50.00 (40.00–60.00) | 51.00 (45.00–57.00) | 46.00 (37.50–54.50) | <0.001 | 0.002 | 0.001 | 0.878 |
| TG | 85.00 (70.00–134.50) | 126.00 (80.00–167.00) | 131.00 (118.00–182.00) | 116.00 (97.25–178.75) | 0.328 | 0.038 | 0.157 | 0.535 |
| TG high | 2/26 (7.69%) | 4/35 (11.43%) | 6/25 (24.00%) | 5/24 (20.83%) | 0.960 | 0.224 | 0.352 | 0.412 |
| LDL/HDL | - | 2.23 (1.66–2.85) | 2.42 (1.91–2.95) | 2.43 (2.12–2.94) | - | - | - | 0.451 |
| AI | 3.00 (2.58–3.82) | 3.83 (3.07–4.42) | 4.00 (3.36–4.60) | 3.95 (3.67–5.07) | 0.085 | 0.011 | 0.005 | 0.447 |
| AI low risk | 23/26 (88.46%) | 29/35 (82.86%) | 17/21 (80.95%) | 17/24 (70.83%) | 0.806 | 0.759 | 0.229 | 0.519 |
| AI moderate risk | 3/26 (11.54%) | 5/35 (14.29%) | 4/21 (19.05%) | 6/24 (25.00%) | 1.000 | 0.759 | 0.385 | 0.584 |
| AIP | 0.18 (0.12–0.87) | 0.97 (0.32–1.23) | 0.92 (0.74–1.44) | 0.89 (0.69–1.43) | 0.019 | 0.004 | 0.005 | 0.568 |
| AIP high risk | 12/26 (46.15%) | 29/35 (82.86%) | 19/21 (90.48%) | 23/24 (95.83%) | 0.006 | 0.004 | <0.001 | 0.289 |
| LCI (×103) | - | 43.33 (27.17–75.66) | 70.21 (44.18–81.30) | 60.00 (35.84–122.80) | - | - | - | 0.731 |
| Liver Biochemical Parameters | ||||||||
| GOT (AST) | 18.50 (15.00–20.00) | 37.00 (29.00–46.00) | 23.00 (19.00–26.00) | 23.50 (20.50–26.75) | <0.001 | 0.055 | 0.007 | <0.001 |
| GOT > 40 | 0/32 (0.00%) | 12/36 (33.33%) | 0/28 (0.00%) | 2/25 (8.00%) | 0.001 | - | 0.366 | 0.001 |
| GPT (ALT) | 15.00 (12.25–20.75) | 45.00 (32.00–55.00) | 21.50 (17.00–25.00) | 27.50 (22.25–32.25) | <0.001 | 0.675 | 0.012 | <0.001 |
| GPT > 40 | 2/26 (7.69%) | 22/37 (59.46%) | 0/28 (0.00%) | 4/26 (15.38%) | <0.001 | 0.439 | 0.664 | <0.001 |
| GGT | 19.00 (12.00–28.00) | 48.00 (32.25–86.75) | 32.00 (26.75–39.50) | 26.50 (21.00–41.50) | 0.001 | <0.001 | 0.014 | 0.003 |
| GGT > 50 | 0/25 (0.00%) | 17/34 (50.00%) | 2/26 (7.69%) | 4/24 (16.67%) | <0.001 | 0.488 | 0.108 | <0.001 |
| APRI | - | 0.62 (0.50–0.67) | 0.35 (0.30–0.44) | - | - | - | - | - |
| FIB-4 | - | 1.56 (1.39–1.73) | 1.22 (1.18–1.31) | - | - | - | - | – |
| ALP | – | 85.00 (65.00–96.00) | 94.50 (70.25–113.00) | 77.00 (66.00–93.75) | - | - | - | 0.038 |
| Albumin | - | 4.40 (4.03–4.40) | 4.50 (4.43–4.68) | 4.15 (4.00–4.60) | - | - | - | 0.133 |
| Hemogram | ||||||||
| Vitamin D | - | 22.00 (14.00–27.00) | 15.50 (11.50–35.00) | 24.00 (21.75–37.00) | - | - | - | 0.491 |
| Calcium (Ca2+) | 9.33 (9.21–9.65) | 9.40 (9.00–9.70) | 9.50 (9.25–9.70) | 9.35 (9.10–9.80) | 0.297 | 0.988 | 0.611 | 0.414 |
| Phosphorous (P) | 3.15 (2.60–3.55) | 3.30 (2.90–3.60) | 3.40 (3.15–3.80) | 3.40 (3.15–3.90) | 0.351 | 0.037 | 0.016 | 0.490 |
| Iron (Fe2+) | 86.50 (63.50–119.50) | 79.00 (78.50–79.50) | 83.00 (65.00–106.00) | - | 0.665 | 0.713 | - | - |
| Hemoglobin A1c | - | 5.30 (5.30–5.30) | 5.60 (5.50–5.80) | - | - | - | - | - |
| Leukocytes (×103) | 6.02 (5.28–7.21) | 6.91 (5.61–7.65) | 7.31 (6.07–8.32) | 6.90 (5.71–8.40) | 0.313 | 0.023 | 0.137 | 0.297 |
| Red blood cells | 4.90 (4.49–5.12) | 4.75 (4.24–5.09) | 4.98 (4.90–5.16) | 4.83 (4.77–4.96) | 0.289 | 0.506 | 0.780 | 0.283 |
| Hb | 14.15 (12.75–15.45) | 15.00 (14.50–15.90) | 15.45 (13.67–16.12) | 14.90 (13.80–15.67) | 0.008 | 0.018 | 0.146 | 0.376 |
| Hematocrit (%) | 42.20 (38.62–45.77) | 44.70 (43.00–47.50) | 46.75 (45.90–47.73) | 46.35 (45.63–49.38) | 0.049 | 0.013 | 0.004 | 0.363 |
| Platelets (×103) | 247.00 (206.50–267.50) | 219.00 (192.00–249.00) | 224.50 (202.00–280.75) | 234.50 (220.00–270.50) | 0.069 | 0.962 | 0.702 | 0.090 |
| LHD | - | 175.00 (167.00–195.50) | 167.50 (143.00–187.00) | 172.00 (162.50–203.50) | - | - | - | 0.466 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valle-Millares, D.; Brochado-Kith, Ó.; Martín-Carbonero, L.; Domínguez-Domínguez, L.; Ryan, P.; De los Santos, I.; De la Fuente, S.; Castro, J.M.; Lagarde, M.; Cuevas, G.; et al. Different HCV Exposure Drives Specific miRNA Profile in PBMCs of HIV Patients. Biomedicines 2021, 9, 1627. https://doi.org/10.3390/biomedicines9111627
Valle-Millares D, Brochado-Kith Ó, Martín-Carbonero L, Domínguez-Domínguez L, Ryan P, De los Santos I, De la Fuente S, Castro JM, Lagarde M, Cuevas G, et al. Different HCV Exposure Drives Specific miRNA Profile in PBMCs of HIV Patients. Biomedicines. 2021; 9(11):1627. https://doi.org/10.3390/biomedicines9111627
Chicago/Turabian StyleValle-Millares, Daniel, Óscar Brochado-Kith, Luz Martín-Carbonero, Lourdes Domínguez-Domínguez, Pablo Ryan, Ignacio De los Santos, Sara De la Fuente, Juan M. Castro, María Lagarde, Guillermo Cuevas, and et al. 2021. "Different HCV Exposure Drives Specific miRNA Profile in PBMCs of HIV Patients" Biomedicines 9, no. 11: 1627. https://doi.org/10.3390/biomedicines9111627
APA StyleValle-Millares, D., Brochado-Kith, Ó., Martín-Carbonero, L., Domínguez-Domínguez, L., Ryan, P., De los Santos, I., De la Fuente, S., Castro, J. M., Lagarde, M., Cuevas, G., Mayoral-Muñoz, M., Matarranz, M., Díez, V., Gómez-Sanz, A., Martínez-Román, P., Crespo-Bermejo, C., Palladino, C., Muñoz-Muñoz, M., Jiménez-Sousa, M. A., ... on Behalf of Multidisciplinary Group of Viral Coinfection HIV/Hepatitis (COVIHEP). (2021). Different HCV Exposure Drives Specific miRNA Profile in PBMCs of HIV Patients. Biomedicines, 9(11), 1627. https://doi.org/10.3390/biomedicines9111627







