Bone Marrow-Derived Mesenchymal Stem Cells Migrate toward Hormone-Insensitive Prostate Tumor Cells Expressing TGF-β via N-Cadherin
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Conditioned Medium
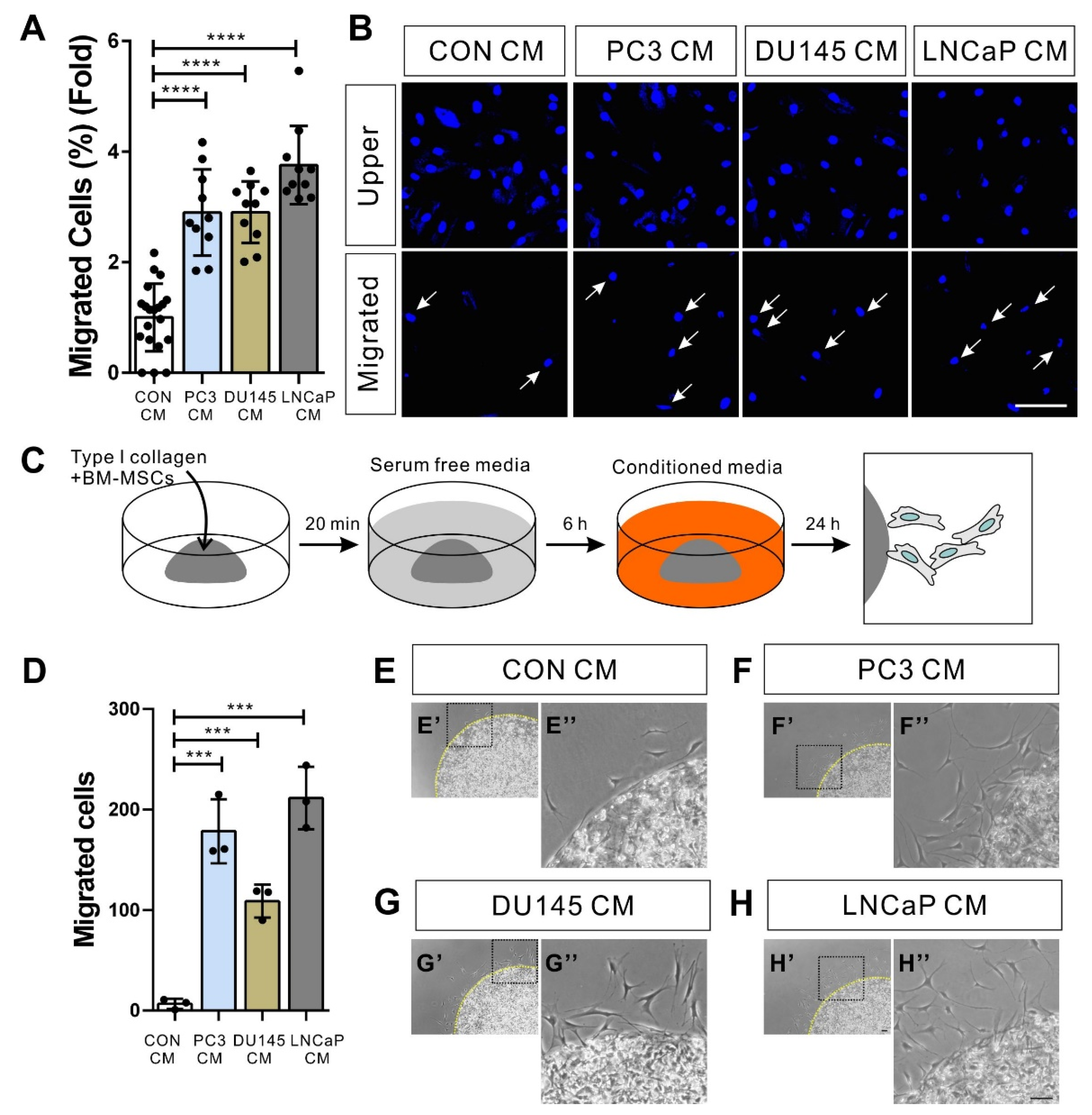
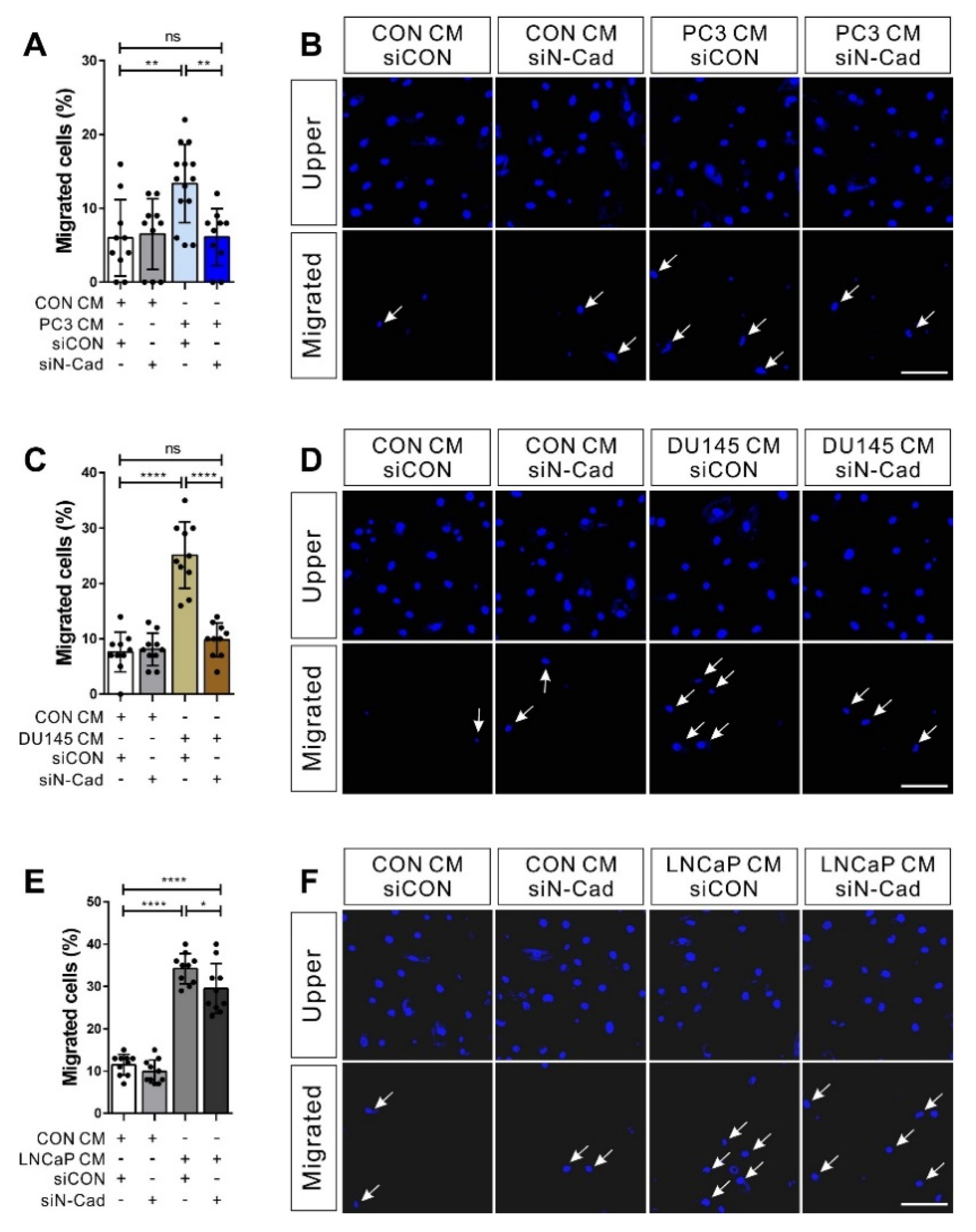
2.3. Transwell Migration Assay
2.4. Three-Dimensional (3D) Cell Migration Assay
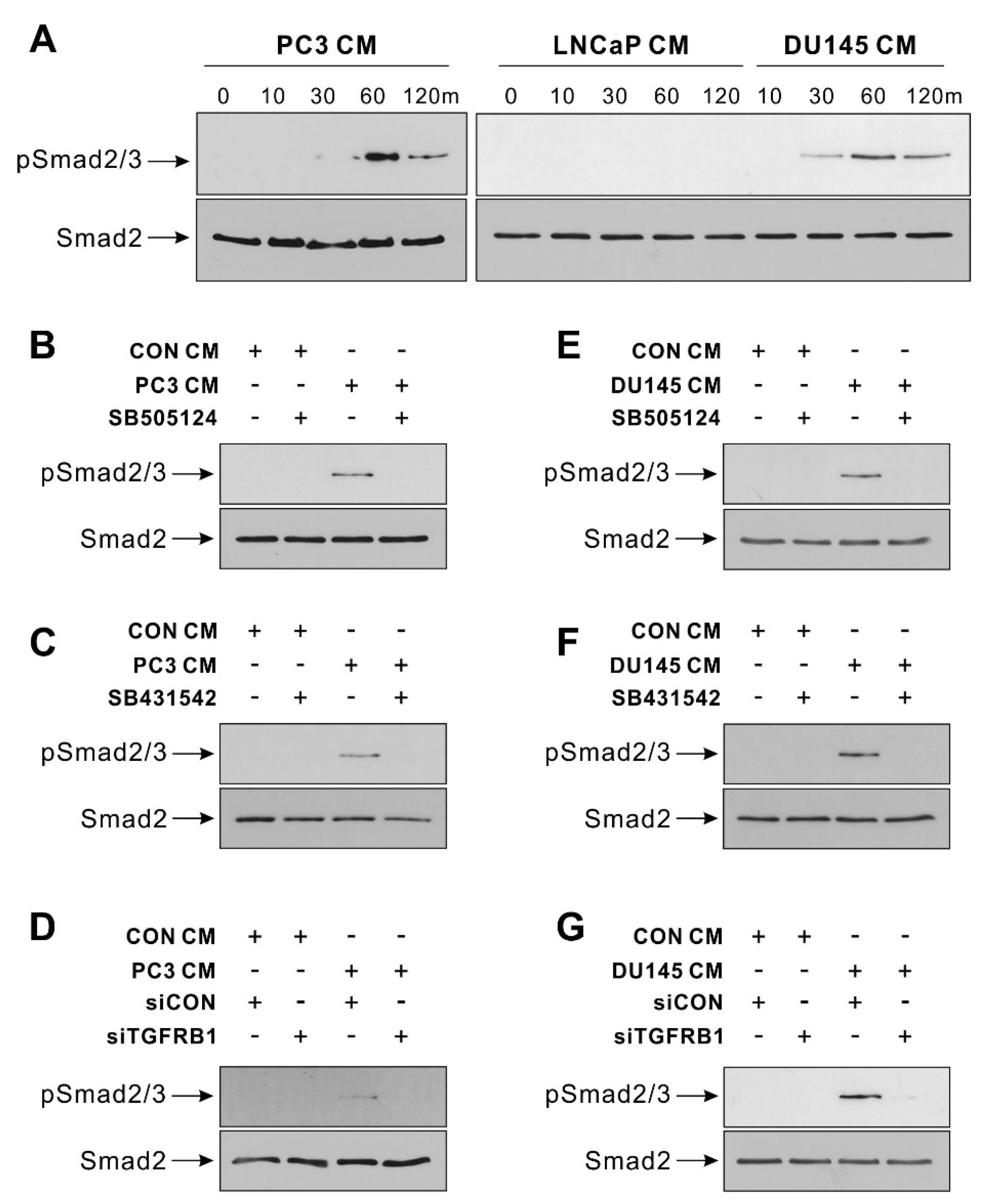
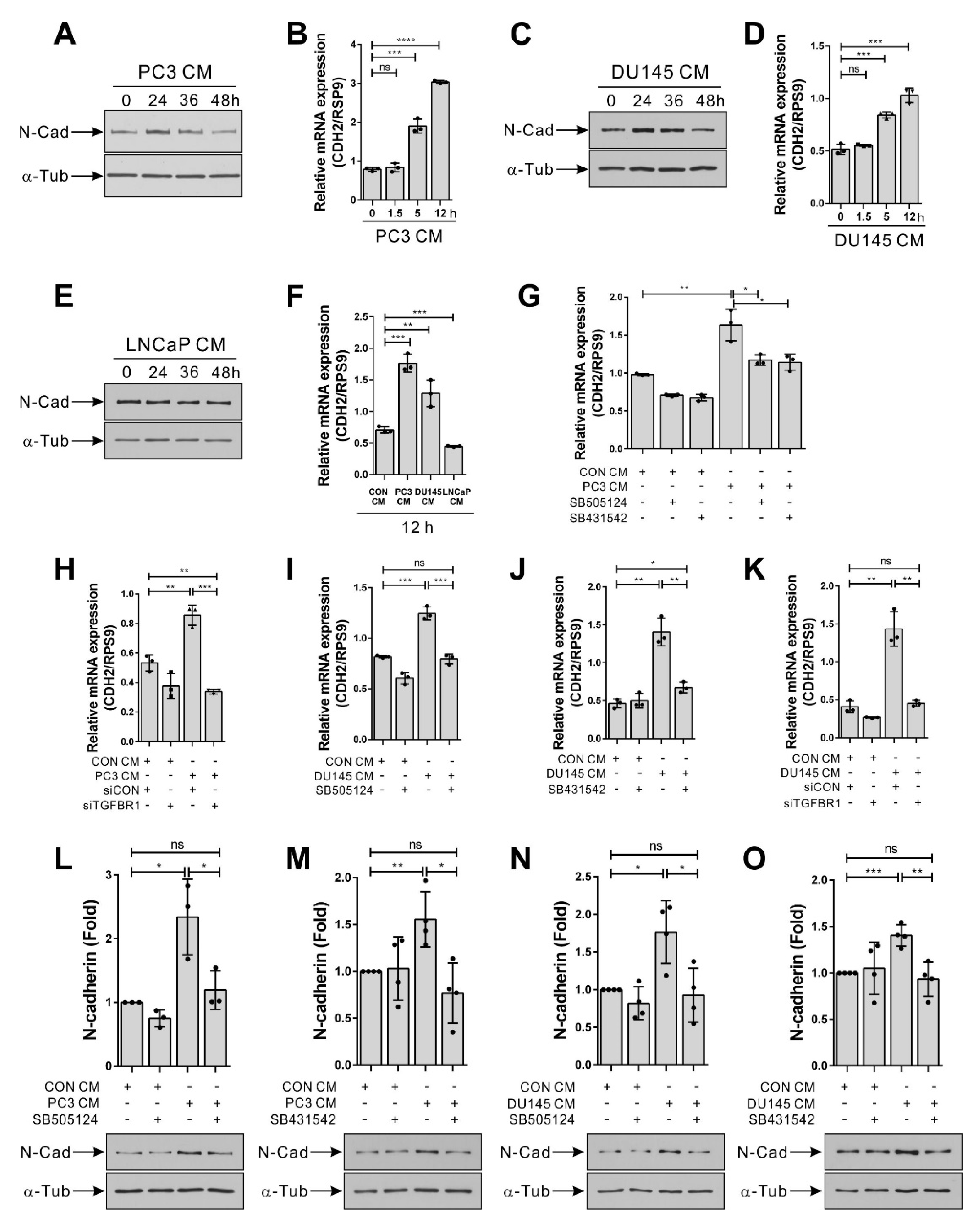
2.5. Western Blot Analysis
2.6. Real-Time Polymerase Chain Reaction (RT-PCR)
2.7. Transfection of Cells with Small Interfering RNAs (siRNAs)
2.8. Immunocytochemistry
2.9. Statistical Analysis
3. Results
3.1. BM-MSCs Migrate in Response to Prostate Tumor Cells
3.2. TGF-β Signal Mediates BM-MSCs’ Migration toward Hormone-Insensitive Prostate Tumor Cells
3.3. Hormone-Insensitive Prostate Tumor Cells Enhance the Expression of N-Cadherin in BM-MSCs via TGF-β Signal
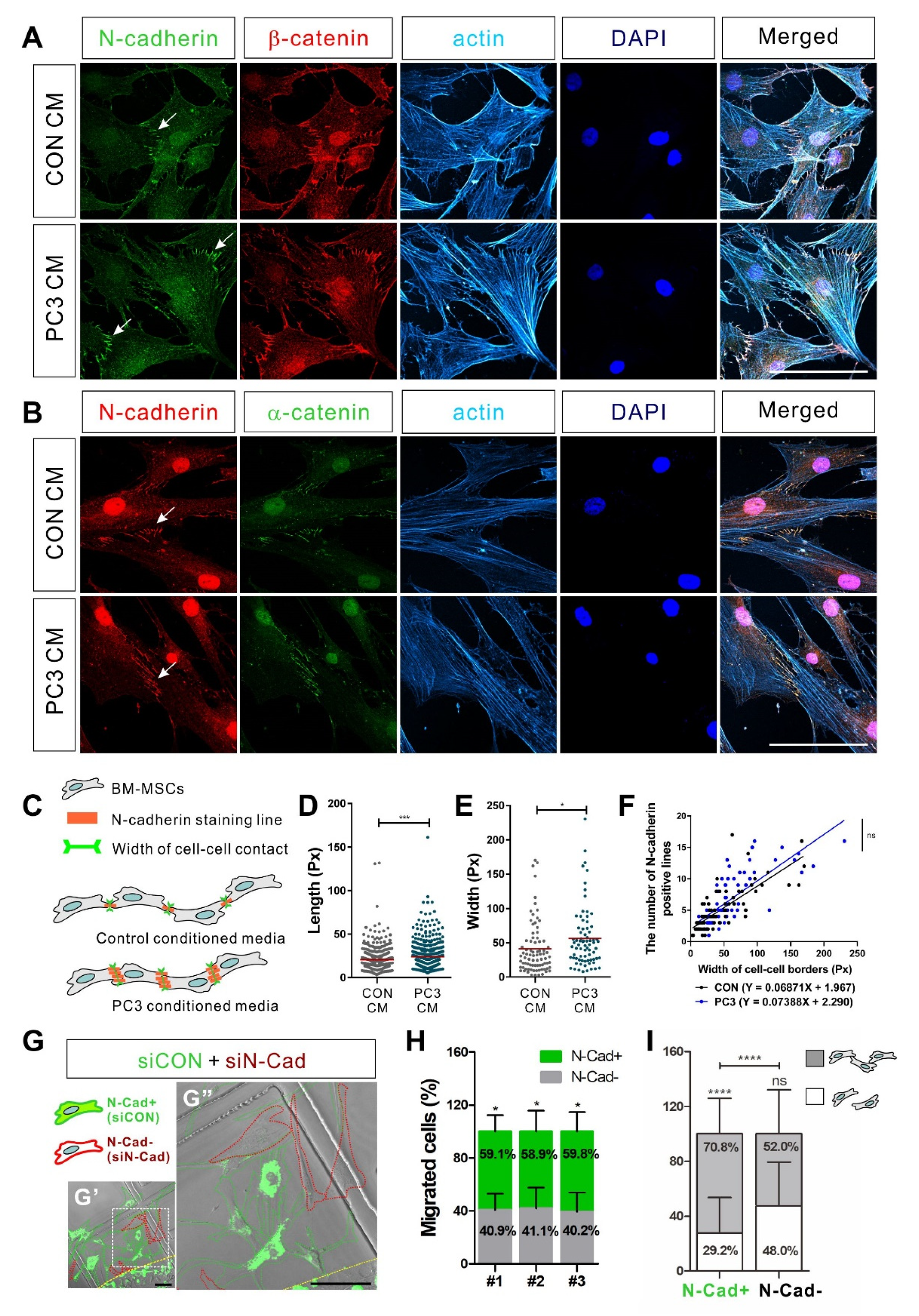
3.4. N-Cadherin Mediates Cell–Cell Adhesion Required for the Collective Migration of BM-MSCs in Response to Hormone-Insensitive Prostate Tumor Cells That Express TGF-β
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bahmad, H.F.; Jalloul, M.; Azar, J.; Moubarak, M.M.; Samad, T.A.; Mukherji, D.; Al-Sayegh, M.; Abou-Kheir, W. Tumor Microenvironment in Prostate Cancer: Toward Identification of Novel Molecular Biomarkers for Diagnosis, Prognosis, and Therapy Development. Front. Genet. 2021, 12, 652747. [Google Scholar] [CrossRef]
- Chandrasekar, T.; Yang, J.C.; Gao, A.C.; Evans, C.P. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl. Androl. Urol. 2015, 4, 365–380. [Google Scholar] [CrossRef]
- Zong, Y.; Goldstein, A.S. Adaptation or selection—Mechanisms of castration-resistant prostate cancer. Nat. Rev. Urol. 2013, 10, 90–98. [Google Scholar] [CrossRef]
- Shiao, S.L.; Chu, G.C.; Chung, L.W. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016, 380, 340–348. [Google Scholar] [CrossRef] [Green Version]
- Placencio, V.R.; Li, X.; Sherrill, T.P.; Fritz, G.; Bhowmick, N.A. Bone marrow derived mesenchymal stem cells incorporate into the prostate during regrowth. PLoS ONE 2010, 5, e12920. [Google Scholar] [CrossRef]
- Yu, Y.; Yang, F.H.; Zhang, W.T.; Guo, Y.D.; Ye, L.; Yao, X.D. Mesenchymal stem cells desensitize castration-resistant prostate cancer to docetaxel chemotherapy via inducing TGF-beta1-mediated cell autophagy. Cell Biosci. 2021, 11, 7. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage potential of adult human mesenchymal stem cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Undale, A.H.; Westendorf, J.J.; Yaszemski, M.J.; Khosla, S. Mesenchymal stem cells for bone repair and metabolic bone diseases. Mayo Clin. Proc. 2009, 84, 893–902. [Google Scholar] [CrossRef] [Green Version]
- Kiel, M.J.; Yilmaz, O.H.; Iwashita, T.; Yilmaz, O.H.; Terhorst, C.; Morrison, S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 2005, 121, 1109–1121. [Google Scholar] [CrossRef] [Green Version]
- Kiel, M.J.; Morrison, S.J. Uncertainty in the niches that maintain haematopoietic stem cells. Nat. Rev. Immunol. 2008, 8, 290–301. [Google Scholar] [CrossRef]
- Rennert, R.C.; Sorkin, M.; Garg, R.K.; Gurtner, G.C. Stem cell recruitment after injury: Lessons for regenerative medicine. Regen. Med. 2012, 7, 833–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, M.; Li, C.; Zhen, G.; Jiao, K.; He, W.; Jia, X.; Wang, W.; Shi, C.; Xing, Q.; Chen, Y.F.; et al. Injury-activated transforming growth factor beta controls mobilization of mesenchymal stem cells for tissue remodeling. Stem Cells 2012, 30, 2498–2511. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Wang, C.; Liu, R.; Wei, C.; Duan, J.; Liu, K.; Li, S.; Zou, H.; Zhao, J.; Wang, L.; et al. Effect of TGF-beta1 on the Migration and Recruitment of Mesenchymal Stem Cells after Vascular Balloon Injury: Involvement of Matrix Metalloproteinase-14. Sci. Rep. 2016, 6, 21176. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Wu, X.; Lei, W.; Pang, L.; Wan, C.; Shi, Z.; Zhao, L.; Nagy, T.R.; Peng, X.; Hu, J.; et al. TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009, 15, 757–765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergfeld, S.A.; DeClerck, Y.A. Bone marrow-derived mesenchymal stem cells and the tumor microenvironment. Cancer Metastasis Rev. 2010, 29, 249–261. [Google Scholar] [CrossRef]
- Mishra, P.J.; Mishra, P.J.; Humeniuk, R.; Medina, D.J.; Alexe, G.; Mesirov, J.P.; Ganesan, S.; Glod, J.W.; Banerjee, D. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008, 68, 4331–4339. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.; Kim, J.K.; Shiozawa, Y.; Wang, J.; Mishra, A.; Joseph, J.; Berry, J.E.; McGee, S.; Lee, E.; Sun, H.; et al. Recruitment of mesenchymal stem cells into prostate tumours promotes metastasis. Nat. Commun. 2013, 4, 1795. [Google Scholar] [CrossRef] [Green Version]
- Barcellos-de-Souza, P.; Comito, G.; Pons-Segura, C.; Taddei, M.L.; Gori, V.; Becherucci, V.; Bambi, F.; Margheri, F.; Laurenzana, A.; Del Rosso, M.; et al. Mesenchymal Stem Cells are Recruited and Activated into Carcinoma-Associated Fibroblasts by Prostate Cancer Microenvironment-Derived TGF-beta1. Stem Cells 2016, 34, 2536–2547. [Google Scholar] [CrossRef] [PubMed]
- Borghese, C.; Cattaruzza, L.; Pivetta, E.; Normanno, N.; De Luca, A.; Mazzucato, M.; Celegato, M.; Colombatti, A.; Aldinucci, D. Gefitinib inhibits the cross-talk between mesenchymal stem cells and prostate cancer cells leading to tumor cell proliferation and inhibition of docetaxel activity. J. Cell. Biochem. 2013, 114, 1135–1144. [Google Scholar] [CrossRef]
- McGuire, J.J.; Frieling, J.S.; Lo, C.H.; Li, T.; Muhammad, A.; Lawrence, H.R.; Lawrence, N.J.; Cook, L.M.; Lynch, C.C. Mesenchymal stem cell-derived interleukin-28 drives the selection of apoptosis resistant bone metastatic prostate cancer. Nat. Commun. 2021, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zhang, Q.; Ma, C.; Yang, X.; Lin, R.; Zhang, H.; Liu, Y.; Han, Z.; Cheng, J. Mesenchymal stem cells recruited by castration-induced inflammation activation accelerate prostate cancer hormone resistance via chemokine ligand 5 secretion. Stem Cell Res. Ther. 2018, 9, 242. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.T.; Pontes-Junior, J.; Antunes, A.A.; Sousa-Canavez, J.M.; Abe, D.K.; Cruz, J.A.; Dall’oglio, M.F.; Crippa, A.; Passerotti, C.C.; Ribeiro-Filho, L.A.; et al. Tgf-beta1 expression as a biomarker of poor prognosis in prostate cancer. Clinics 2011, 66, 1143–1147. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.T.; Chang, Y.H.; Lin, W.Y.; Chen, W.C.; Chen, M.F. TGF Beta1 Expression Correlates with Survival and Tumor Aggressiveness of Prostate Cancer. Ann. Surg. Oncol. 2015, 22 (Suppl. 3), 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Shariat, S.F.; Kattan, M.W.; Traxel, E.; Andrews, B.; Zhu, K.; Wheeler, T.M.; Slawin, K.M. Association of pre- and postoperative plasma levels of transforming growth factor beta(1) and interleukin 6 and its soluble receptor with prostate cancer progression. Clin. Cancer Res. 2004, 10, 1992–1999. [Google Scholar] [CrossRef] [Green Version]
- Shariat, S.F.; Menesses-Diaz, A.; Kim, I.Y.; Muramoto, M.; Wheeler, T.M.; Slawin, K.M. Tissue expression of transforming growth factor-beta1 and its receptors: Correlation with pathologic features and biochemical progression in patients undergoing radical prostatectomy. Urology 2004, 63, 1191–1197. [Google Scholar] [CrossRef]
- Jones, E.; Pu, H.; Kyprianou, N. Targeting TGF-beta in prostate cancer: Therapeutic possibilities during tumor progression. Expert Opin. Ther. Targets 2009, 13, 227–234. [Google Scholar] [CrossRef]
- Barrett, C.S.; Millena, A.C.; Khan, S.A. TGF-beta Effects on Prostate Cancer Cell Migration and Invasion Require FosB. Prostate 2017, 77, 72–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danielpour, D. Functions and regulation of transforming growth factor-beta (TGF-beta) in the prostate. Eur. J. Cancer 2005, 41, 846–857. [Google Scholar] [CrossRef]
- Benaim, E.A.; Pace, C.M.; Roehrborn, C.G. Gleason score predicts androgen independent progression after androgen deprivation therapy. Eur. Urol. 2002, 42, 12–17. [Google Scholar] [CrossRef]
- Moses, H.L.; Roberts, A.B.; Derynck, R. The Discovery and Early Days of TGF-beta: A Historical Perspective. Cold Spring Harb. Perspect. Biol. 2016, 8, a021865. [Google Scholar] [CrossRef] [Green Version]
- Massague, J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Yu, J.; Kim, W.; Park, K.S. N-cadherin mediates the migration of bone marrow-derived mesenchymal stem cells toward breast tumor cells. Theranostics 2021, 11, 6786–6799. [Google Scholar] [CrossRef]
- Dubon, M.J.; Yu, J.; Choi, S.; Park, K.S. Transforming growth factor beta induces bone marrow mesenchymal stem cell migration via noncanonical signals and N-cadherin. J. Cell Physiol. 2018, 233, 201–213. [Google Scholar] [CrossRef]
- Gumbiner, B.M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Alimperti, S.; Andreadis, S.T. CDH2 and CDH11 act as regulators of stem cell fate decisions. Stem Cell Res. 2015, 14, 270–282. [Google Scholar] [CrossRef] [Green Version]
- Tilley, W.D.; Wilson, C.M.; Marcelli, M.; McPhaul, M.J. Androgen receptor gene expression in human prostate carcinoma cell lines. Cancer Res. 1990, 50, 5382–5386. [Google Scholar]
- Lim, D.J.; Liu, X.L.; Sutkowski, D.M.; Braun, E.J.; Lee, C.; Kozlowski, J.M. Growth of an androgen-sensitive human prostate cancer cell line, LNCaP, in nude mice. Prostate 1993, 22, 109–118. [Google Scholar] [CrossRef]
- Cao, J.; Chiarelli, C.; Richman, O.; Zarrabi, K.; Kozarekar, P.; Zucker, S. Membrane type 1 matrix metalloproteinase induces epithelial-to-mesenchymal transition in prostate cancer. J. Biol. Chem. 2008, 283, 6232–6240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chipuk, J.E.; Cornelius, S.C.; Pultz, N.J.; Jorgensen, J.S.; Bonham, M.J.; Kim, S.J.; Danielpour, D. The androgen receptor represses transforming growth factor-beta signaling through interaction with Smad3. J. Biol. Chem. 2002, 277, 1240–1248. [Google Scholar] [CrossRef] [Green Version]
- Whiteside, T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [Green Version]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [Green Version]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef]
- Quail, D.F.; Joyce, J.A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 2013, 19, 1423–1437. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Raz, Y.; Cohen, N.; Shani, O.; Bell, R.E.; Novitskiy, S.V.; Abramovitz, L.; Levy, C.; Milyavsky, M.; Leider-Trejo, L.; Moses, H.L.; et al. Bone marrow-derived fibroblasts are a functionally distinct stromal cell population in breast cancer. J. Exp. Med. 2018, 215, 3075–3093. [Google Scholar] [CrossRef] [Green Version]
- Craft, N.; Shostak, Y.; Carey, M.; Sawyers, C.L. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nat. Med. 1999, 5, 280–285. [Google Scholar] [CrossRef]
- Den, R.B.; George, D.; Pieczonka, C.; McNamara, M. Ra-223 Treatment for Bone Metastases in Castrate-Resistant Prostate Cancer: Practical Management Issues for Patient Selection. Am. J. Clin. Oncol. 2019, 42, 399–406. [Google Scholar] [CrossRef]
- Logothetis, C.J.; Gallick, G.E.; Maity, S.N.; Kim, J.; Aparicio, A.; Efstathiou, E.; Lin, S.H. Molecular classification of prostate cancer progression: Foundation for marker-driven treatment of prostate cancer. Cancer Discov. 2013, 3, 849–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.M.; Sixt, M. Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol. 2019, 20, 738–752. [Google Scholar] [CrossRef] [PubMed]
- Scarpa, E.; Mayor, R. Collective cell migration in development. J. Cell Biol. 2016, 212, 143–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theveneau, E.; Mayor, R. Cadherins in collective cell migration of mesenchymal cells. Curr. Opin. Cell Biol. 2012, 24, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theveneau, E.; Marchant, L.; Kuriyama, S.; Gull, M.; Moepps, B.; Parsons, M.; Mayor, R. Collective chemotaxis requires contact-dependent cell polarity. Dev. Cell 2010, 19, 39–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noh, J.; Yu, J.; Kim, W.; Park, A.; Park, K.-S. Bone Marrow-Derived Mesenchymal Stem Cells Migrate toward Hormone-Insensitive Prostate Tumor Cells Expressing TGF-β via N-Cadherin. Biomedicines 2021, 9, 1572. https://doi.org/10.3390/biomedicines9111572
Noh J, Yu J, Kim W, Park A, Park K-S. Bone Marrow-Derived Mesenchymal Stem Cells Migrate toward Hormone-Insensitive Prostate Tumor Cells Expressing TGF-β via N-Cadherin. Biomedicines. 2021; 9(11):1572. https://doi.org/10.3390/biomedicines9111572
Chicago/Turabian StyleNoh, Jinok, Jinyeong Yu, Wootak Kim, Aran Park, and Ki-Sook Park. 2021. "Bone Marrow-Derived Mesenchymal Stem Cells Migrate toward Hormone-Insensitive Prostate Tumor Cells Expressing TGF-β via N-Cadherin" Biomedicines 9, no. 11: 1572. https://doi.org/10.3390/biomedicines9111572
APA StyleNoh, J., Yu, J., Kim, W., Park, A., & Park, K.-S. (2021). Bone Marrow-Derived Mesenchymal Stem Cells Migrate toward Hormone-Insensitive Prostate Tumor Cells Expressing TGF-β via N-Cadherin. Biomedicines, 9(11), 1572. https://doi.org/10.3390/biomedicines9111572




