Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer—A Systematic Review
Abstract
:1. Introduction
1.1. Head and Neck Squamous Cell Carcinoma
1.2. Liquid Biopsy
1.3. Somatic Mutations of Head and Neck Squamous Cell Carcinoma
2. Materials and Methods
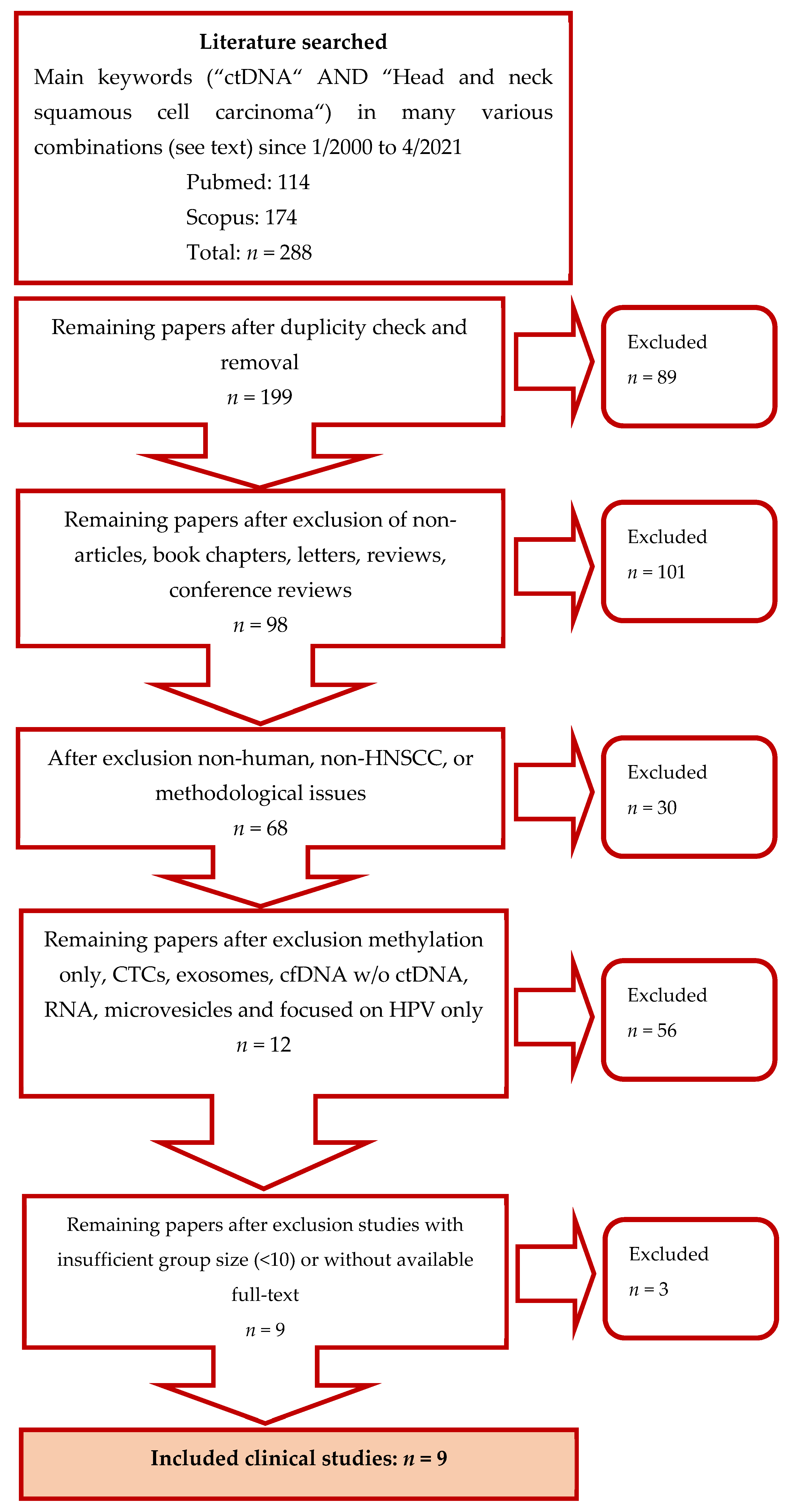
2.1. Search Strategy
2.2. Studies Selection
3. Results
4. Discussion
5. Conclusions and Future Research
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis Primers 2020, 6, 92. [Google Scholar] [CrossRef]
- Marur, S.; Forastiere, A.A. Head and Neck Squamous Cell Carcinoma: Update on Epidemiology, Diagnosis, and Treatment. Mayo Clin. Proc. 2016, 91, 386–396. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, H.; Kulasinghe, A.; Allcock, R.J.N.; Tan, L.Y.; Mokany, E.; Kenny, L.; Punyadeera, C. A Pilot Study to Non-Invasively Track PIK3CA Mutation in Head and Neck Cancer. Diagnostics (Basel) 2018, 8, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajer, C.B.; von Buchwald, C. The role of human papillomavirus in head and neck cancer. APMIS 2010, 118, 510–519. [Google Scholar] [CrossRef]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Chow, L.Q.M. Head and Neck Cancer. N. Engl. J. Med. 2020, 382, 60–72. [Google Scholar] [CrossRef]
- Hwang, I.; Choi, S.H.; Kim, Y.J.; Kim, K.G.; Lee, A.L.; Yun, T.J.; Kim, J.H.; Sohn, C.H. Differentiation of recurrent tumor and posttreatment changes in head and neck squamous cell carcinoma: Application of high b-value diffusion-weighted imaging. AJNR Am. J. Neuroradiol 2013, 34, 2343–2348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Ginkel, J.H.; Huibers, M.M.H.; Noorlag, R.; de Bree, R.; van Es, R.J.J.; Willems, S.M. Liquid Biopsy: A Future Tool for Posttreatment Surveillance in Head and Neck Cancer? Pathobiology 2017, 84, 115–120. [Google Scholar] [CrossRef]
- Schmidt, H.; Kulasinghe, A.; Perry, C.; Nelson, C.; Punyadeera, C. A liquid biopsy for head and neck cancers. Expert Rev. Mol. Diagn. 2016, 16, 165–172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Ribeiro, I.P.; de Melo, J.B.; Carreira, I.M. Head and neck cancer: Searching for genomic and epigenetic biomarkers in body fluids–the state of art. Mol. Cytogenet. 2019, 12, 33. [Google Scholar] [CrossRef]
- Mandel, P. Les acides nucléiques du plasma sanguin chez l’homme. C R Seances Soc. Biol. Fil. 1948, 142, 241–243. [Google Scholar]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar]
- Gormally, E.; Caboux, E.; Vineis, P.; Hainaut, P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. Mutat. Res. 2007, 635, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Lousada-Fernandez, F.; Rapado-Gonzalez, O.; Lopez-Cedrun, J.L.; Lopez-Lopez, R.; Muinelo-Romay, L.; Suarez-Cunqueiro, M.M. Liquid Biopsy in Oral Cancer. Int J. Mol. Sci. 2018, 19, 1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Tolani, B.; Nie, X.; Zhi, X.; Hu, M.; He, B. Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther. Clin. Risk Manag. 2017, 13, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.Y.K.; Zhen, G.; Agrawal, N. The role of tumor DNA as a diagnostic tool for head and neck squamous cell carcinoma. Semin. Cancer Biol. 2019, 55, 1–7. [Google Scholar] [CrossRef]
- Alix-Panabieres, C.; Pantel, K. Real-time liquid biopsy: Circulating tumor cells versus circulating tumor DNA. Ann. Transl. Med. 2013, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Holdhoff, M.; Schmidt, K.; Donehower, R.; Diaz, L.A., Jr. Analysis of circulating tumor DNA to confirm somatic KRAS mutations. J. Natl. Cancer Inst. 2009, 101, 1284–1285. [Google Scholar] [CrossRef]
- Payne, K.; Spruce, R.; Beggs, A.; Sharma, N.; Kong, A.; Martin, T.; Parmar, S.; Praveen, P.; Nankivell, P.; Mehanna, H. Circulating tumor DNA as a biomarker and liquid biopsy in head and neck squamous cell carcinoma. Head Neck 2018, 40, 1598–1604. [Google Scholar] [CrossRef] [Green Version]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; El Messaoudi, S.; Gahan, P.B.; Anker, P.; Stroun, M. Origins, structures, and functions of circulating DNA in oncology. Cancer Metastasis Rev. 2016, 35, 347–376. [Google Scholar] [CrossRef] [Green Version]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A., Jr.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Ginkel, J.H.; Huibers, M.M.H.; van Es, R.J.J.; de Bree, R.; Willems, S.M. Droplet digital PCR for detection and quantification of circulating tumor DNA in plasma of head and neck cancer patients. BMC Cancer 2017, 17, 428. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.; Kulasinghe, A.; Kenny, L.; Punyadeera, C. The development of a liquid biopsy for head and neck cancers. Oral Oncol. 2016, 61, 8–11. [Google Scholar] [CrossRef]
- Porter, A.; Natsuhara, M.; Daniels, G.A.; Patel, S.P.; Sacco, A.G.; Bykowski, J.; Banks, K.C.; Cohen, E.E.W. Next generation sequencing of cell free circulating tumor DNA in blood samples of recurrent and metastatic head and neck cancer patients. Transl. Cancer Res. 2020, 9, 203–209. [Google Scholar] [CrossRef]
- Chan, K.C.; Yeung, S.W.; Lui, W.B.; Rainer, T.H.; Lo, Y.M. Effects of preanalytical factors on the molecular size of cell-free DNA in blood. Clin. Chem. 2005, 51, 781–784. [Google Scholar] [CrossRef]
- Swinkels, D.W.; Wiegerinck, E.; Steegers, E.A.; de Kok, J.B. Effects of blood-processing protocols on cell-free DNA quantification in plasma. Clin. Chem. 2003, 49, 525–526. [Google Scholar] [CrossRef] [Green Version]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef] [Green Version]
- Perdomo, S.; Avogbe, P.H.; Foll, M.; Abedi-Ardekani, B.; Facciolla, V.L.; Anantharaman, D.; Chopard, P.; Calvez-Kelm, F.L.; Vilensky, M.; Polesel, J.; et al. Circulating tumor DNA detection in head and neck cancer: Evaluation of two different detection approaches. Oncotarget 2017, 8, 72621–72632. [Google Scholar] [CrossRef] [Green Version]
- Galot, R.; van Marcke, C.; Helaers, R.; Mendola, A.; Goebbels, R.M.; Caignet, X.; Ambroise, J.; Wittouck, K.; Vikkula, M.; Limaye, N.; et al. Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol. 2020, 104, 104631. [Google Scholar] [CrossRef] [PubMed]
- Coulet, F.; Blons, H.; Cabelguenne, A.; Lecomte, T.; Lacourreye, O.; Brasnu, D.; Beaune, P.; Zucman, J.; Laurent-Puig, P. Detection of plasma tumor DNA in head and neck squamous cell carcinoma by microsatellite typing and p53 mutation analysis. Cancer Res. 2000, 60, 707–711. [Google Scholar] [PubMed]
- Schwaederle, M.; Chattopadhyay, R.; Kato, S.; Fanta, P.T.; Banks, K.C.; Choi, I.S.; Piccioni, D.E.; Ikeda, S.; Talasaz, A.; Lanman, R.B.; et al. Genomic Alterations in Circulating Tumor DNA from Diverse Cancer Patients Identified by Next-Generation Sequencing. Cancer Res. 2017, 77, 5419–5427. [Google Scholar] [CrossRef] [Green Version]
- Jung, M.; Klotzek, S.; Lewandowski, M.; Fleischhacker, M.; Jung, K. Changes in concentration of DNA in serum and plasma during storage of blood samples. Clin. Chem. 2003, 49, 1028–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez Sayans, M.; Chamorro Petronacci, C.M.; Lorenzo Pouso, A.I.; Padin Iruegas, E.; Blanco Carrion, A.; Suarez Penaranda, J.M.; Garcia Garcia, A. Comprehensive Genomic Review of TCGA Head and Neck Squamous Cell Carcinomas (HNSCC). J. Clin. Med. 2019, 8, 1896. [Google Scholar] [CrossRef] [Green Version]
- Brennan, J.A.; Boyle, J.O.; Koch, W.M.; Goodman, S.N.; Hruban, R.H.; Eby, Y.J.; Couch, M.J.; Forastiere, A.A.; Sidransky, D. Association between cigarette smoking and mutation of the p53 gene in squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 1995, 332, 712–717. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [Green Version]
- Mahoney, P.A.; Weber, U.; Onofrechuk, P.; Biessmann, H.; Bryant, P.J.; Goodman, C.S. The fat tumor suppressor gene in Drosophila encodes a novel member of the cadherin gene superfamily. Cell 1991, 67, 853–868. [Google Scholar] [CrossRef]
- Tanoue, T.; Takeichi, M. New insights into Fat cadherins. J. Cell Sci. 2005, 118, 2347–2353. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science 2011, 333, 1154–1157. [Google Scholar] [CrossRef] [Green Version]
- Miracca, E.C.; Kowalski, L.P.; Nagai, M.A. High prevalence of p16 genetic alterations in head and neck tumours. Br. J. Cancer 1999, 81, 677–683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Mes, S.W.; Brink, A.; Sistermans, E.A.; Straver, R.; Oudejans, C.B.M.; Poell, J.B.; Leemans, C.R.; Brakenhoff, R.H. Comprehensive multiparameter genetic analysis improves circulating tumor DNA detection in head and neck cancer patients. Oral Oncol. 2020, 109, 104852. [Google Scholar] [CrossRef] [PubMed]
- Braig, F.; Voigtlaender, M.; Schieferdecker, A.; Busch, C.J.; Laban, S.; Grob, T.; Kriegs, M.; Knecht, R.; Bokemeyer, C.; Binder, M. Liquid biopsy monitoring uncovers acquired RAS-mediated resistance to cetuximab in a substantial proportion of patients with head and neck squamous cell carcinoma. Oncotarget 2016, 7, 42988–42995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hieronymus, H.; Murali, R.; Tin, A.; Yadav, K.; Abida, W.; Moller, H.; Berney, D.; Scher, H.; Carver, B.; Scardino, P.; et al. Tumor copy number alteration burden is a pan-cancer prognostic factor associated with recurrence and death. Elife 2018, 7, e37294. [Google Scholar] [CrossRef]
- Fiala, C.; Kulasingam, V.; Diamandis, E.P. Circulating Tumor DNA for Early Cancer Detection. J. Appl. Lab. Med. 2018, 3, 300–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, H.L.; D’Agostino, R.B., Jr.; Meegalla, N.; Petro, R.; Commander, S.; Topaloglu, U.; Zhang, W.; Porosnicu, M. The Prognostic and Therapeutic Value of the Mutational Profile of Blood and Tumor Tissue in Head and Neck Squamous Cell Carcinoma. Oncologist 2021, 26, e279–e289. [Google Scholar] [CrossRef]
- Tamkovich, S.N.; Cherepanova, A.V.; Kolesnikova, E.V.; Rykova, E.Y.; Pyshnyi, D.V.; Vlassov, V.V.; Laktionov, P.P. Circulating DNA and DNase activity in human blood. Ann. N. Y. Acad. Sci. 2006, 1075, 191–196. [Google Scholar] [CrossRef]
- Cherepanova, A.V.; Tamkovich, S.N.; Bryzgunova, O.E.; Vlassov, V.V.; Laktionov, P.P. Deoxyribonuclease activity and circulating DNA concentration in blood plasma of patients with prostate tumors. Ann. N. Y. Acad. Sci. 2008, 1137, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Kim, Y.H.; Song, Y.; Kim, H.S.; Sim, H.W.; Poojan, S.; Eom, B.W.; Kook, M.C.; Joo, J.; Hong, K.M. Monitoring circulating tumor DNA by analyzing personalized cancer-specific rearrangements to detect recurrence in gastric cancer. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Sarkozy, C.; Huet, S.; Carlton, V.E.; Fabiani, B.; Delmer, A.; Jardin, F.; Delfau-Larue, M.H.; Hacini, M.; Ribrag, V.; Guidez, S.; et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget 2017, 8, 8765–8774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsao, S.C.; Weiss, J.; Hudson, C.; Christophi, C.; Cebon, J.; Behren, A.; Dobrovic, A. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Sci. Rep. 2015, 5, 11198. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [Green Version]
- Dawson, S.J.; Rosenfeld, N.; Caldas, C. Circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 369, 93–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Study | Ref. | Groups | Tumor | HPV + Patients | DNA Source | Genes | Assay Type | Focus of the Study | Results | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Controls | Site | STAGE | TP53 | PIK3CA | NOTCH1 | CDKN2A | CASP8 | Other | ||||||||
| Schmidt et al., 2018 | [3] | 29 | 10 | OPSCC, OSCC, HPSCC, LSCC | III-IV | 14 | plasma | x | no | Plex-PCRTM | Usability of Plex-PCRTM technology PIK3CA mutation detection p.E545K in HNSCC samples | The results of the pilot study support the applicability of using allele-specific technologies for ctDNA testing in HNSCC. | |||||
| Porter et al., 2019 | [27] | 60 (48 HNSCC) | x | OPSCC, SG, TG, other HNC | III, IV | 15 of OPSCC | blood samples | x | x | x | ARID1A, panel Guardant360 (73-gene ctDNA NGS platform) | NGS | Characterizing a ctDNA blood sample in advanced patients with HNSCC to identify a useful mutation and elucidate a potential role in treatment. | Of the patients with usable mutations, 13% (n = 8) received appropriate targeted therapy (MTT). Result: 3 stable diseases (37.5%), 3 progressive diseases (37.5%) and 2 (25%) were not evaluated at follow-up | |||
| Wang et al., 2015 | [31] | 47 | x | OSCC, OPSCC, HPSCC, LSCC | I-II: 20, III-IV: 3 | 30 | plasma, oral rinses | x | x | x | FBXW7, NRAS, HRAS | PCR | Detect genetically modified DNA in the saliva or plasma of HNSCC patients with tumours of various stages and anatomical sites. | Tumour DNA was found in 76% of saliva (93 pts). Tumour DNA was found in 87% of plasma (47 pts.). | |||
| Perdomo et al., 2017 | [32] | ARCAGE | 36 | x | OSCC, OPSCC, LSCC | I: 6, II: 8, III: 8, IV: 14 | 0 | plasma | x | x | x | x | PTEN | sequencing | To provide a comprehensive evaluation of the presence of ctDNA in plasma and oral washes in HNSCC patients in early and late stages. Two strategies for detecting ctDNA mutations. | ARCAGE: Tumour-specific mutations in 5 genes in plasma (42% of cases), most of them (67%) were early-stage cases | |
| LA | 37 | 49 | OSCC, OPSCC, LSCC, over. | III-IV | 1 | plasma, oral rinses | x | no | LA: mutation concordance in tumour tissue, plasma and mouthwash only in 1 sample; in 4 cases, the conformity of the mutation in the oral lavage and tumour sample | ||||||||
| Galot et al., 2020 | [33] | 39 | x | OSCC, OPSCC, HPSCC, LSCC | x | 5 of OPSCC | plasma | x | x | x | x | EGFR, ERBB3, AXL, CSF1R, RET, PIK3R1, AKT1, MAPK1, NF1, STK11, AURKA, MYC, NFE2L2, KMT2C, KMT2B, CREBBP, NSD1, SMARCA4, SMARCA2, ARID1A, BRCA1, MLH1, MSH2, CTNNB1, FAT1, FAT4, NOTCH2, PTCH1 | NGS | Applicability of liquid biopsy for characterization of the mutational environment of recurrent/metastatic HNSCC | Significantly higher probability of ctDNA detection in patients with metastatic disease than patients with locoregional recurrence alone (70% vs 30%). | ||
| Coulet et al., 2000 | [34] | 11 | x | x | x | x | plasma | x | no | PCR | Quantification cfDNA, evaluation ctDNA and analysis clinical significance. | Plasma DNA concentration was measurable at 35%. Tumour DNA detected in plasma in 18% (ctDNA) | |||||
| Schwaederle et al., 2017 | [35] | 25 | x | HNC | x | x | plasma | x | x | EGFR, BRECA2, APC, MET, BRAF, ERBB2, MYC, NF1, ARID1A, SMAD4, BRCA2, FGFR2, BRCA1, PDGFRA, ALK, AR, FGFR1 | NGS | The study specifically examines mutations in the ctDNA of various cancers. Included 25 patients with HNSCC | 88% had a mutation. 40% have 1–3 mutations, 48% have > 3 mutations | ||||
| Mes et al., 2020 | [45] | 40 | 20 | OSCC, OPSCC, HSCC, LSCC, other | I = 2, II = 4, III = 6, IV = 28 | 10 of OPSCC or unknown primary locality | plasma | x | x | x | x | x | AJUBA, FAT1, FBXW7, HRAS, KMT2D, NSD1, PTEN | deep sequencing | Combined detection of somatic mutations, HPV-DNA and CNA using the same sequencing library and recovery assessment in pre-treatment HNSCC patient plasma samples. | The combination of analysis of CNA (copy number aberrations), HPV and somatic mutations in plasma contributes to higher sensitivity than individual modalities. | |
| Braig et al., 2016 | [46] | 20 | x | HNSCC | II: 2, III: 3, IV: 15 | 2 | serum | RAS | NGS | Responses and acquired mutations in the RAS ctDNA gene of HNSCC patients treated with cetuximab plus chemotherapy | Patients with the progressive disease showed a RAS mutation. Patients without progression did not show any additional RAS mutation | ||||||
| Study | Ref. | No of Patients | Overall Percentage Independent on HPV Typing | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ctDNA | Investigated Gene Mutations | TP53 | PIK3CA | NOTCH1 | CDKN2A | CASP8 | ||||
| Schmidt et al., 2018 | [3] | 29 | 31% | PIK3CA | 31% | |||||
| Wang et al., 2015 | [31] | 47 | 87% | HPV16/18, FBXW7, NRAS, HRAS, PIK3CA, TP53, CDKN2A | no data available | |||||
| Perdomo et al., 2017 | ARCAGE | [32] | 36 | 42% | TP53, NOTCH1, CDKN2A, CASP8, PTEN | 31% | 3% | 6% | 3% | |
| LA | 37 | 8% | TP53 | 8% | ||||||
| Coulet et al., 2000 | [34] | 11 | 18% | TP53 | 18% | |||||
| Mes et al., 2020 | [45] | 40 | 67% | AJUBA, CASP8, CDKN2A, FAT1, FBXW7, HRAS, KMT2D, NOTCH1, NSD1, PIK3CA, PTEN, TP53 | no data available | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hudečková, M.; Koucký, V.; Rottenberg, J.; Gál, B. Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer—A Systematic Review. Biomedicines 2021, 9, 1548. https://doi.org/10.3390/biomedicines9111548
Hudečková M, Koucký V, Rottenberg J, Gál B. Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer—A Systematic Review. Biomedicines. 2021; 9(11):1548. https://doi.org/10.3390/biomedicines9111548
Chicago/Turabian StyleHudečková, Markéta, Vladimír Koucký, Jan Rottenberg, and Břetislav Gál. 2021. "Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer—A Systematic Review" Biomedicines 9, no. 11: 1548. https://doi.org/10.3390/biomedicines9111548
APA StyleHudečková, M., Koucký, V., Rottenberg, J., & Gál, B. (2021). Gene Mutations in Circulating Tumour DNA as a Diagnostic and Prognostic Marker in Head and Neck Cancer—A Systematic Review. Biomedicines, 9(11), 1548. https://doi.org/10.3390/biomedicines9111548






