The Human NUP58 Nucleoporin Can Form Amyloids In Vitro and In Vivo
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis
2.2. Plasmid Construction
2.3. Microbiological Procedures
2.4. Protein Electrophoresis and Hybridization
2.5. C-DAG System
2.6. NUP58 Purification from E. coli and Fibril Preparation
2.7. In Vitro Congo Red Staining
2.8. Transmission Electron Microscopy (TEM)
2.9. Proteinase K Resistance Assay
2.10. Thioflavin T Staining
3. Results and Discussion
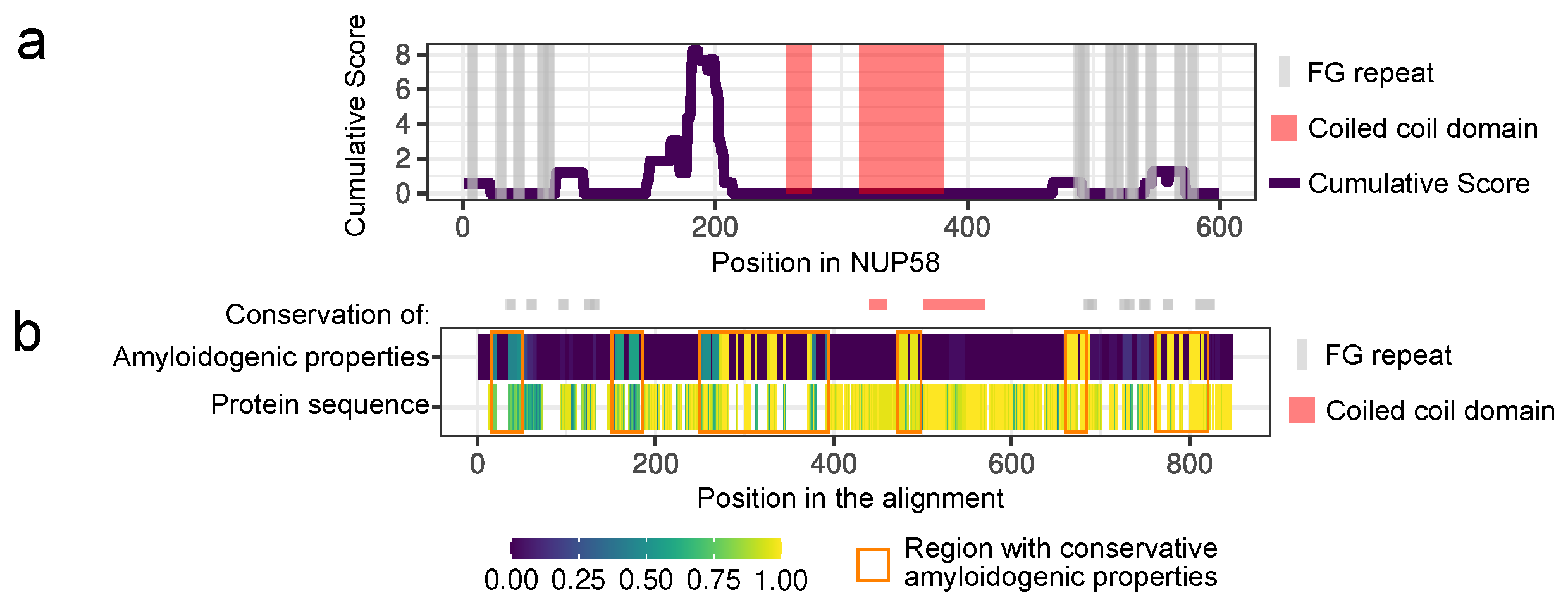
3.1. NUP58 and Its Orthologs Are Potential Amyloids
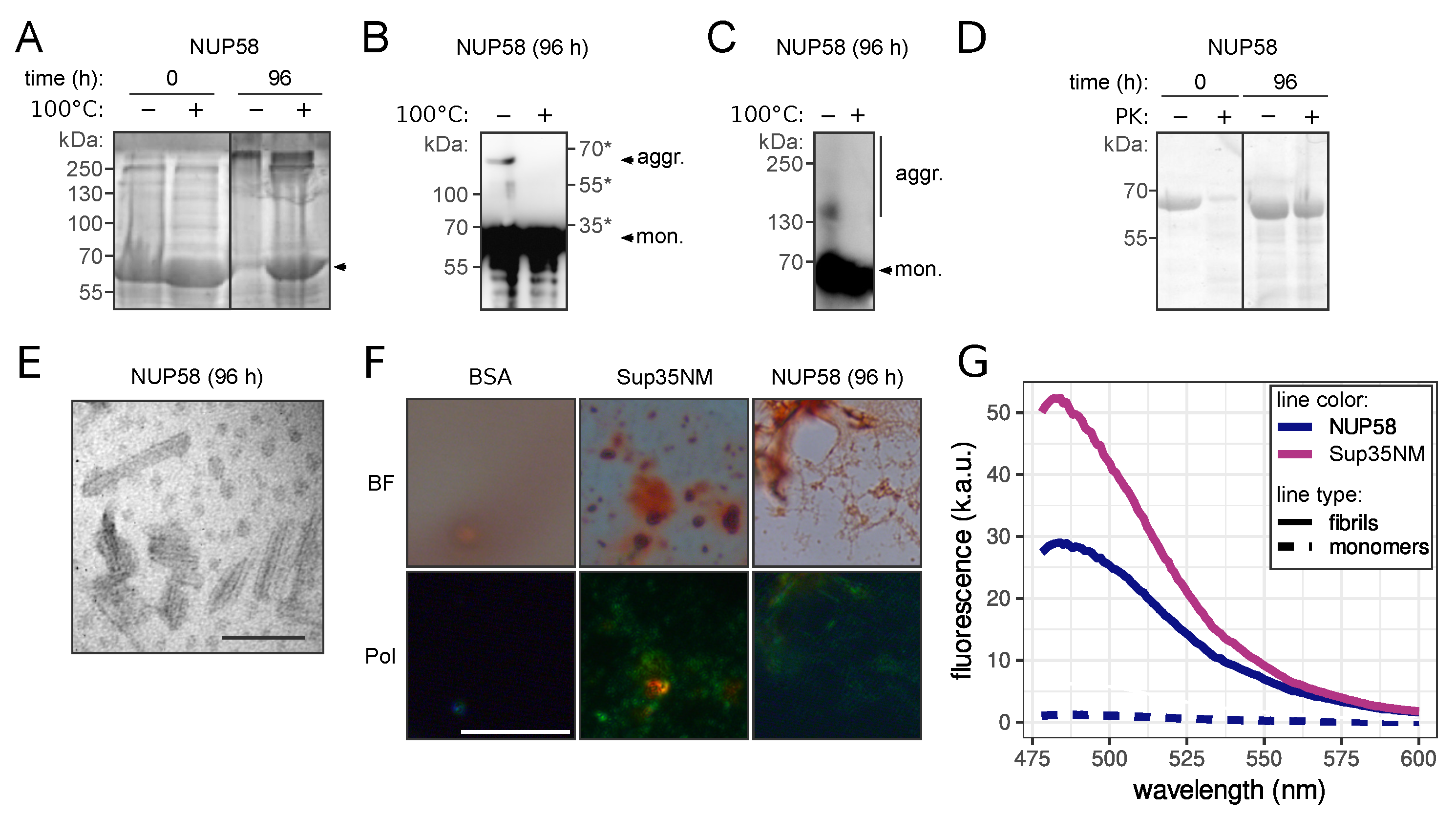
3.2. NUP58 Protein Demonstrates Amyloid Properties In Vitro
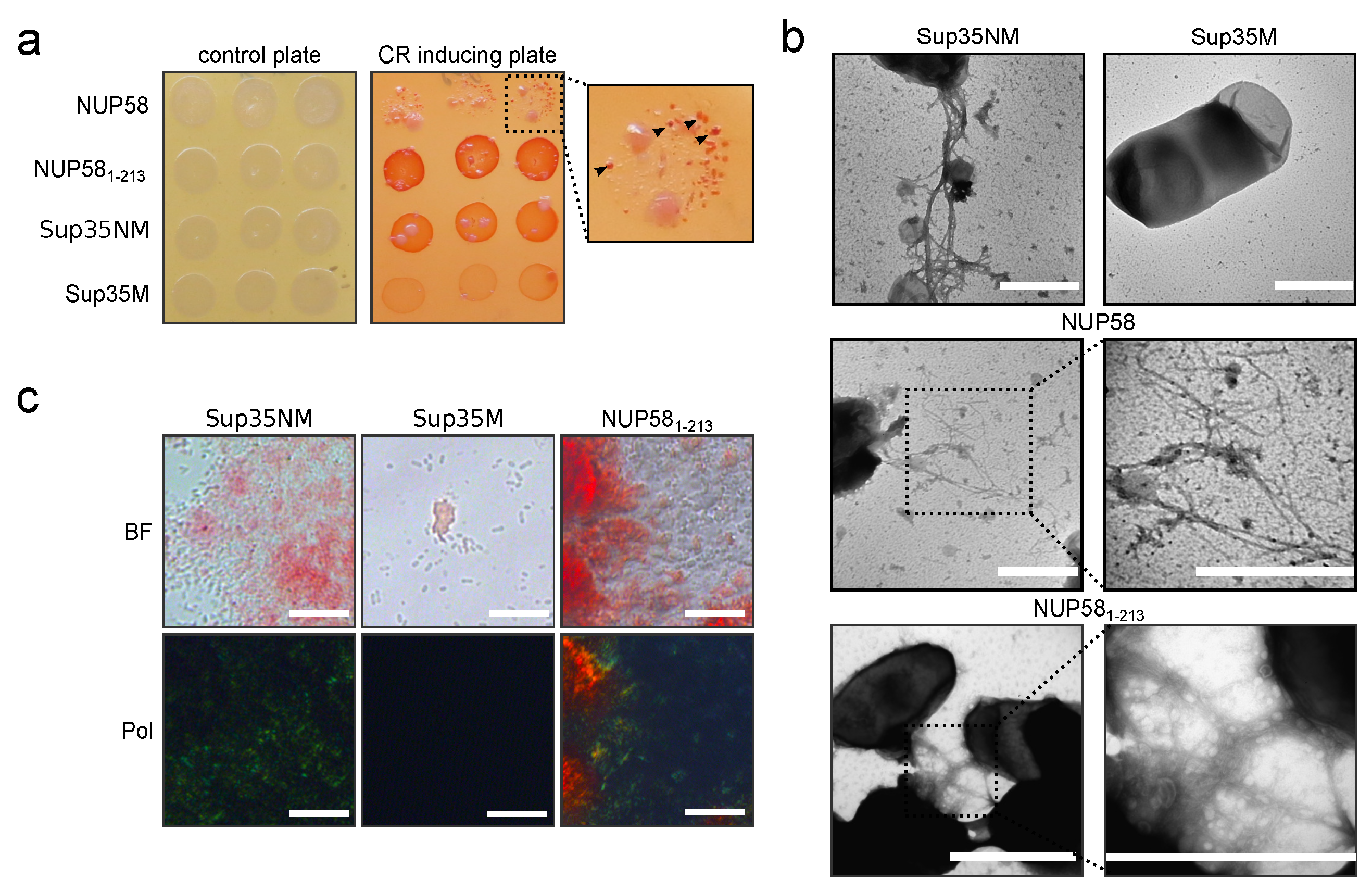
3.3. Human NUP58 Protein Demonstrates Amyloid Properties in C-DAG System
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
Abbreviations
| C-DAG | curli-dependent amyloid generator |
| FG repeat | phenylalanine-glycine repeat |
| NPC | nuclear pore complex |
| Nups | nucleoporins |
| PK | proteinase K |
| BME | β-mercaptoethanol |
| ThT | thioflavin T |
| SDS | sodium dodecyl sulfate |
| TEM | transmission electron microscopy |
Appendix A
Appendix B
| Primers Name | Sequence | Application |
|---|---|---|
| NUPL1-F-1-213 | GGGGACAAGTTTGTACAAAAAAGCAGGCTccatgtccacaggg | Amplification of the NUP58 fragment, corresponding to 1–213 aa, with attB sites. |
| NUPL1-R-1-213 | GGGGACCACTTTGTACAAGAAAGCTGGGTctaattgcctgcagttg | Amplification of the NUP58 fragment, corresponding to 1–213 aa, with attB sites. |
| attB1-SUP35-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTcaacaatgtcggattcaaacc | Amplification of the SUP35 fragment, corresponding to Sup35NM, with attB sites. |
| attB2-SUP35M-stop-R | GGGGACCACTTTGTACAAGAAAGCTGGGTtcaatcgttaacaacttcgtcatcca | Amplification of the SUP35 fragments, corresponding to Sup35NM and Sup35M, with attB sites. |
| attB1-SUP35M-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTcaatgtctttgaacgactttcaaaag | Amplification of the SUP35 fragment, corresponding to Sup35M, with attB sites. |
| Bsp120I-ccdB-F | tataGGGCCCCAcaccatcaccatcaccatagatctg | Amplification of the ccdB cassette. |
| XbaI-ccdB-R | tataTCTAGAgctatctagctagcgatatcaccac | Amplification of the ccdB cassette. |
References
- Benson, M.D.; Buxbaum, J.N.; Eisenberg, D.S.; Merlini, G.; Saraiva, M.J.M.; Sekijima, Y.; Sipe, J.D.; Westermark, P. Amyloid nomenclature 2020: Update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 2020, 27, 217–222. [Google Scholar] [CrossRef]
- Sergeeva, A.V.; Galkin, A.P. Functional amyloids of eukaryotes: Criteria, classification, and biological significance. Curr. Genet. 2020, 66, 849–866. [Google Scholar] [CrossRef]
- Rubel, M.S.; Fedotov, S.A.; Grizel, A.V.; Sopova, J.V.; Malikova, O.A.; Chernoff, Y.O.; Rubel, A.A. Functional mammalian amyloids and amyloid-like proteins. Life 2020, 10, 156. [Google Scholar] [CrossRef]
- Matiiv, A.B.; Trubitsina, N.P.; Matveenko, A.G.; Barbitoff, Y.A.; Zhouravleva, G.A.; Bondarev, S.A. Amyloid and amyloid-like aggregates: Diversity and the term crisis. Biochemistry 2020, 85, 1011–1034. [Google Scholar] [PubMed]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Milles, S.; Huy Bui, K.; Koehler, C.; Eltsov, M.; Beck, M.; Lemke, E.A. Facilitated aggregation of FG nucleoporins under molecular crowding conditions. EMBO Rep. 2013, 14, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Ader, C.; Frey, S.; Maas, W.; Schmidt, H.B.; Görlich, D.; Baldus, M. Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. USA 2010, 107, 6281–6285. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, R.; Wright, J.J.R.; Alberti, S.; Lindquist, S.; Rexach, M. Prion formation by a yeast GLFG nucleoporin. Prion 2012, 6, 391–399. [Google Scholar] [CrossRef]
- Dultz, E.; Ellenberg, J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 2010, 191, 15–22. [Google Scholar] [CrossRef]
- Onischenko, E.; Weis, K. Nuclear pore complex—A coat specifically tailored for the nuclear envelope. Curr. Opin. Cell Biol. 2011, 23, 293–301. [Google Scholar] [CrossRef]
- Terry, L.J.; Wente, S.R. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 2009, 8, 1814–1827. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Ahmed, A.B.; Znassi, N.; Château, M.T.; Kajava, A.V. A structure-based approach to predict predisposition to amyloidosis. Alzheimers Dement. 2015, 11, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Dosztányi, Z.; Csizmók, V.; Tompa, P.; Simon, I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J. Mol. Biol. 2005, 347, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Dosztányi, Z.; Csizmok, V.; Tompa, P.; Simon, I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 2005, 21, 3433–3434. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; The UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: Efficient Manipulation of Biological Strings; R Package Version 2.56.0; 2020. Available online: https://bioconductor.org/packages/release/bioc/html/Biostrings.html (accessed on 15 September 2020).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Sivanathan, V.; Hochschild, A. A bacterial export system for generating extracellular amyloid aggregates. Nat. Protoc. 2013, 8, 1381–1390. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbour, NY, USA, 1989. [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Alexandrov, I.M.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003, 278, 49636–49643. [Google Scholar] [CrossRef] [PubMed]
- Drozdova, P.B.; Barbitoff, Y.A.; Belousov, M.V.; Skitchenko, R.K.; Rogoza, T.M.; Leclercq, J.Y.; Kajava, A.V.; Matveenko, A.G.; Zhouravleva, G.A.; Bondarev, S.A. Estimation of amyloid aggregate sizes with semi-denaturing detergent agarose gel electrophoresis and its limitations. Prion 2020, 14, 118–128. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Alexandrov, I.M.; Mitkevich, O.V.; Shkundina, I.S.; Ter-Avanesyan, M.D. Purification and analysis of prion and amyloid aggregates. Methods 2006, 39, 50–55. [Google Scholar] [CrossRef] [PubMed]
- Studier, F.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Biol. Chem. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Serio, T.R.; Cashikar, A.G.; Moslehi, J.J.; Kowal, A.S.; Lindquist, S.L. Yeast prion [PSI+] and its determinant, Sup35p. Methods Enzymol. 1999, 309, 649–673. [Google Scholar] [PubMed]
- Kajava, A.V.; Baxa, U.; Steven, A.C. β Arcades: Recurring Motifs in Naturally Occurring and Disease-Related Amyloid Fibrils. FASEB J. 2010, 24, 1311–1319. [Google Scholar] [CrossRef]
- Bondarev, S.A.; Zhouravleva, G.A.; Belousov, M.V.; Kajava, A.V. Structure-based view on [PSI+] prion properties. Prion 2015, 9, 190–199. [Google Scholar] [CrossRef][Green Version]
- Roche, D.B.; Villain, E.; Kajava, A.V. Usage of a dataset of NMR resolved protein structures to test aggregation vs. solubility prediction algorithms. Protein Sci. 2017, 26, 1864–1869. [Google Scholar] [CrossRef]
- Dewangan, P.S.; Sonawane, P.J.; Chouksey, A.R.; Chauhan, R. The Nup62 coiled-coil motif provides plasticity for triple-helix bundle formation. Biochemistry 2017, 56, 2803–2811. [Google Scholar] [CrossRef]
- King, C.Y.; Tittmann, P.; Gross, H.; Gebert, R.; Aebi, M.; Wüthrich, K. Prion-inducing domain 2–114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 1997, 94, 6618–6622. [Google Scholar] [CrossRef]
- Eichner, T.; Radford, S.E. Understanding the complex mechanisms of β2-microglobulin amyloid assembly. FEBS J. 2011, 278, 3868–3883. [Google Scholar] [CrossRef]
- Chiti, F.; Dobson, C.M. Amyloid formation by globular proteins under native conditions. Nat. Chem. Biol. 2009, 5, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Milles, S.; Mercadante, D.; Aramburu, I.V.; Jensen, M.R.; Banterle, N.; Koehler, C.; Tyagi, S.; Clarke, J.; Shammas, S.L.; Blackledge, M.; et al. Plasticity of an Ultrafast Interaction between Nucleoporins and Nuclear Transport Receptors. Cell 2015, 163, 734–745. [Google Scholar] [CrossRef]
- Labokha, A.A.; Gradmann, S.; Frey, S.; Hülsmann, B.B.; Urlaub, H.; Baldus, M.; Görlich, D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013, 32, 204–218. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, H.B.; Görlich, D. Transport selectivity of nuclear pores, phase separation, and membraneless organelles. Trends Biochem. Sci. 2016, 41, 46–61. [Google Scholar] [CrossRef] [PubMed]
- Celetti, G.; Paci, G.; Caria, J.; VanDelinder, V.; Bachand, G.; Lemke, E.A. The liquid state of FG-nucleoporins mimics permeability barrier properties of nuclear pore complexes. J. Cell Biol. 2020, 219, e201907157. [Google Scholar] [CrossRef]
- Li, C.; Goryaynov, A.; Yang, W. The selective permeability barrier in the nuclear pore complex. Nucleus 2016, 7, 430–446. [Google Scholar] [CrossRef]
- Fragasso, A.; de Vries, H.W.; Andersson, J.; van der Sluis, E.O.; van der Giessen, E.; Dahlin, A.; Onck, P.R.; Dekker, C. A designer FG-Nup that reconstitutes the selective transport barrier of the nuclear pore complex. Nat. Commun. 2021, 12, 2010. [Google Scholar] [CrossRef]
- Onischenko, E.; Tang, J.H.; Andersen, K.R.; Knockenhauer, K.E.; Vallotton, P.; Derrer, C.P.; Kralt, A.; Mugler, C.F.; Chan, L.Y.; Schwartz, T.U.; et al. Natively unfolded FG repeats stabilize the structure of the nuclear pore complex. Cell 2017, 171, 904–917. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Danilov, L.G.; Moskalenko, S.E.; Matveenko, A.G.; Sukhanova, X.V.; Belousov, M.V.; Zhouravleva, G.A.; Bondarev, S.A. The Human NUP58 Nucleoporin Can Form Amyloids In Vitro and In Vivo. Biomedicines 2021, 9, 1451. https://doi.org/10.3390/biomedicines9101451
Danilov LG, Moskalenko SE, Matveenko AG, Sukhanova XV, Belousov MV, Zhouravleva GA, Bondarev SA. The Human NUP58 Nucleoporin Can Form Amyloids In Vitro and In Vivo. Biomedicines. 2021; 9(10):1451. https://doi.org/10.3390/biomedicines9101451
Chicago/Turabian StyleDanilov, Lavrentii G., Svetlana E. Moskalenko, Andrew G. Matveenko, Xenia V. Sukhanova, Mikhail V. Belousov, Galina A. Zhouravleva, and Stanislav A. Bondarev. 2021. "The Human NUP58 Nucleoporin Can Form Amyloids In Vitro and In Vivo" Biomedicines 9, no. 10: 1451. https://doi.org/10.3390/biomedicines9101451
APA StyleDanilov, L. G., Moskalenko, S. E., Matveenko, A. G., Sukhanova, X. V., Belousov, M. V., Zhouravleva, G. A., & Bondarev, S. A. (2021). The Human NUP58 Nucleoporin Can Form Amyloids In Vitro and In Vivo. Biomedicines, 9(10), 1451. https://doi.org/10.3390/biomedicines9101451







