Airway Bacteria Quantification Using Polymerase Chain Reaction Combined with Neutrophil and Eosinophil Counts Identifies Distinct COPD Endotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Study Design
2.3. Sputum Measurements
2.4. qPCR Detection of Common Respiratory Pathogens
2.5. Statistical Analysis
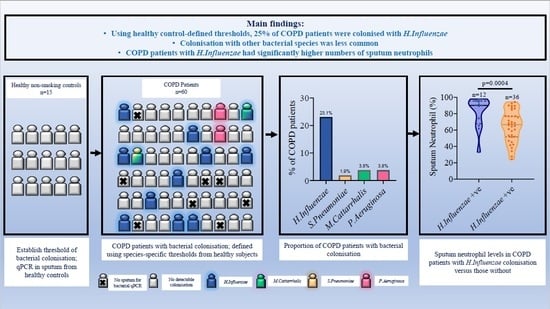
3. Results
3.1. Bacterial Colonisation
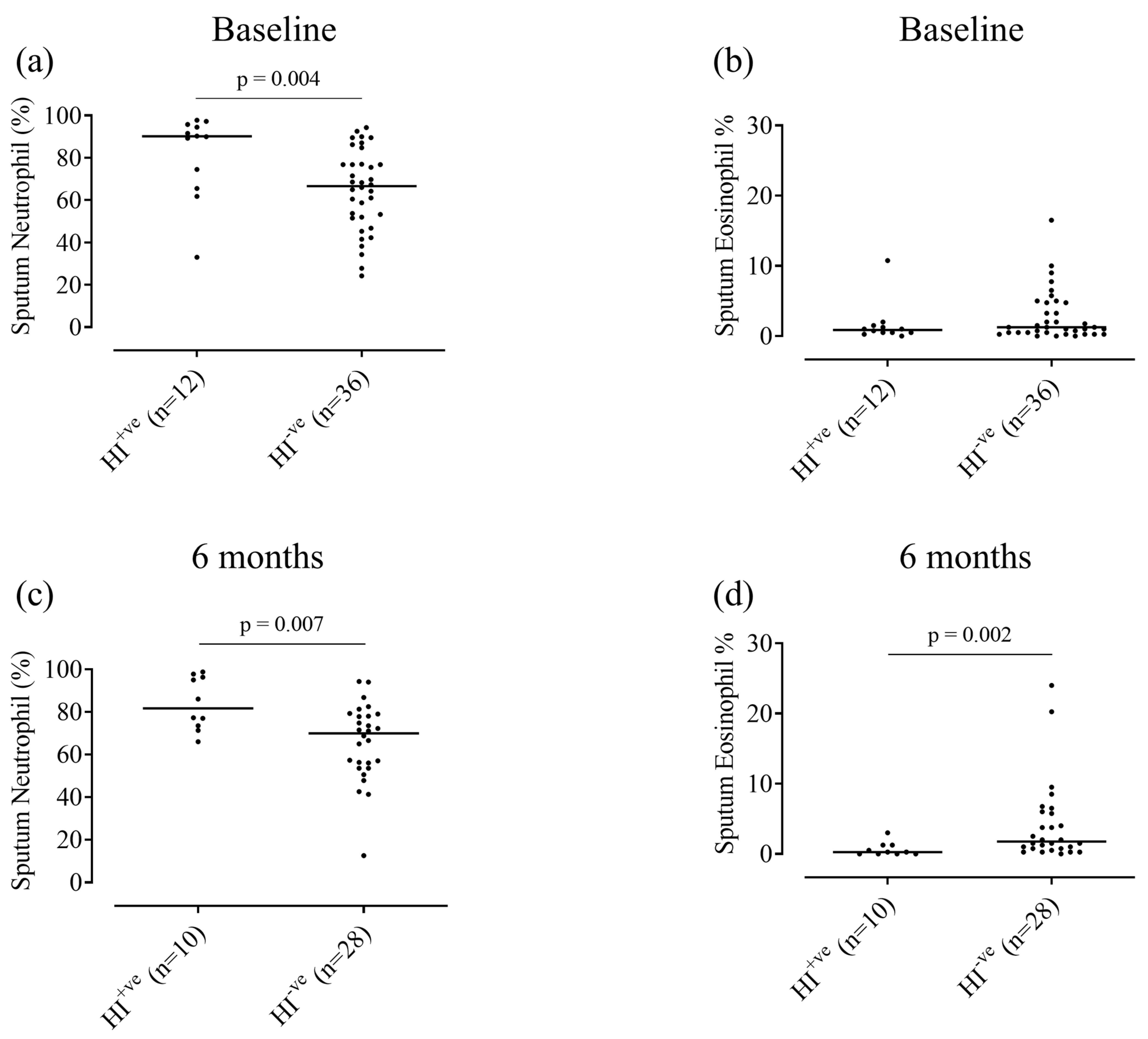
3.2. Relationship between Colonisation and Sputum Cell Counts
4. Discussion
4.1. Prevalence of Bacterial Colonisation
4.2. H. influenzae and Airway Inflammation
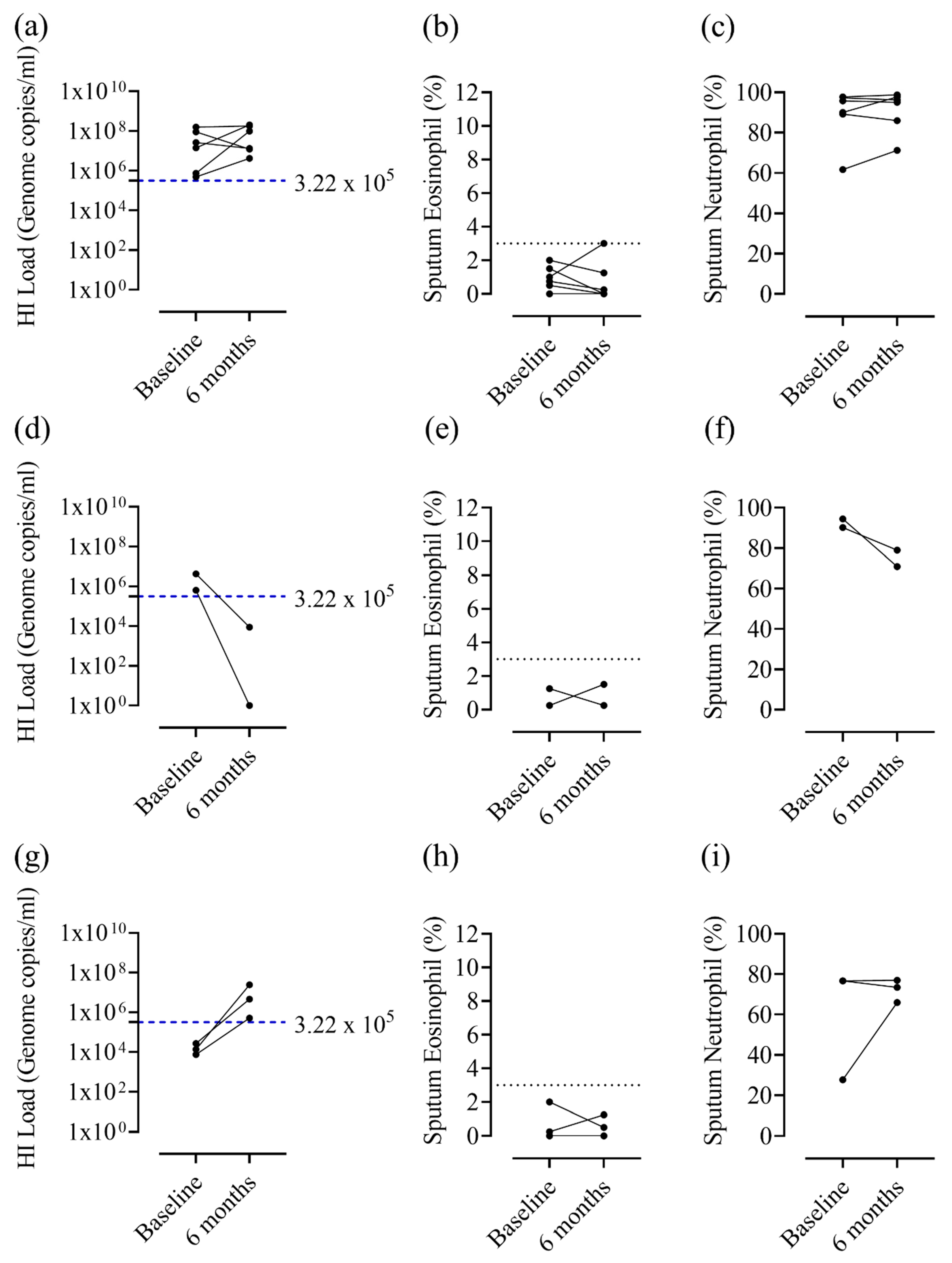
4.3. Eosinophilic Airway Inflammation and Bacterial Colonisation
4.4. Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, D.; Bafadhel, M.; Brightling, C.E.; Sciurba, F.C.; Curtis, J.L.; Martinez, F.J.; Pasquale, C.B.; Merrill, D.D.; Metzdorf, N.; Petruzzelli, S.; et al. Blood Eosinophil Counts in Clinical Trials for Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2020, 202, 660–671. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Locantore, N.; Haldar, K.; Ramsheh, M.Y.; Beech, A.S.; Ma, W.; Brown, J.R.; Tal-Singer, R.; Barer, M.R.; Bafadhel, M.; et al. Inflammatory Endotype-associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 1488–1502. [Google Scholar] [CrossRef] [PubMed]
- Beech, A.S.; Lea, S.; Kolsum, U.; Wang, Z.; Miller, B.E.; Donaldson, G.C.; Wedzicha, J.A.; Brightling, C.E.; Singh, D. Bacteria and sputum inflammatory cell counts; a COPD cohort analysis. Respir. Res. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Edwards, L.; Tal-Singer, R.; Rennard, S. Sputum neutrophils as a biomarker in COPD: Findings from the ECLIPSE study. Respir. Res. 2010, 11, 77. [Google Scholar] [CrossRef]
- Hogg, J.C.; Chu, F.; Utokaparch, S.; Woods, R.; Elliott, W.M.; Buzatu, L.; Cherniack, R.M.; Rogers, R.M.; Sciurba, F.C.; Coxson, H.O.; et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J. Med. 2004, 350, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Higham, A.; Beech, A.; Wolosianka, S.; Jackson, N.; Long, G.; Kolsum, U.; Southworth, T.; Pham, T.H.; Sridhar, S.; McCrae, C.; et al. Type 2 inflammation in eosinophilic chronic obstructive pulmonary disease. Allergy 2021, 76, 1861–1864. [Google Scholar] [CrossRef] [PubMed]
- Kolsum, U.; Damera, G.; Pham, T.H.; Southworth, T.; Mason, S.; Karur, P.; Newbold, P.; Singh, D. Pulmonary inflammation in patients with chronic obstructive pulmonary disease with higher blood eosinophil counts. J. Allergy Clin. Immunol. 2017, 140, 1181–1184.e1187. [Google Scholar] [CrossRef] [PubMed]
- Stockley, R.A.; Halpin, D.M.G.; Celli, B.R.; Singh, D. Chronic Obstructive Pulmonary Disease Biomarkers and Their Interpretation. Am. J. Respir. Crit. Care Med. 2019, 199, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Inflammatory endotypes in COPD. Allergy 2019, 74, 1249–1256. [Google Scholar] [CrossRef] [PubMed]
- Singh, D. Pharmacological treatment of stable chronic obstructive pulmonary disease. Respirology 2021, 26, 643–651. [Google Scholar] [CrossRef]
- Haldar, K.; George, L.; Wang, Z.; Mistry, V.; Ramsheh, M.Y.; Free, R.C.; John, C.; Reeve, N.F.; Miller, B.E.; Tal-Singer, R.; et al. The sputum microbiome is distinct between COPD and health, independent of smoking history. Respir. Res. 2020, 21, 183. [Google Scholar] [CrossRef] [PubMed]
- Ramsheh, M.Y.; Haldar, K.; Esteve-Codina, A.; Purser, L.F.; Richardson, M.; Müller-Quernheim, J.; Greulich, T.; Nowinski, A.; Barta, I.; Stendardo, M.; et al. Lung microbiome composition and bronchial epithelial gene expression in patients with COPD versus healthy individuals: A bacterial 16S rRNA gene sequencing and host transcriptomic analysis. Lancet Microbe 2021, 2, e300–e310. [Google Scholar] [CrossRef]
- Wang, Z.; Maschera, B.; Lea, S.; Kolsum, U.; Michalovich, D.; Van Horn, S.; Traini, C.; Brown, J.R.; Hessel, E.M.; Singh, D. Airway host-microbiome interactions in chronic obstructive pulmonary disease. Respir. Res. 2019, 20, 113. [Google Scholar] [CrossRef] [PubMed]
- Monso, E.; Rosell, A.; Bonet, G.; Manterola, J.; Cardona, P.J.; Ruiz, J.; Morera, J. Risk factors for lower airway bacterial colonization in chronic bronchitis. Eur. Respir. J. 1999, 13, 338–342. [Google Scholar] [CrossRef]
- Wilkinson, T.M.A.; Aris, E.; Bourne, S.; Clarke, S.C.; Peeters, M.; Pascal, T.G.; Schoonbroodt, S.; Tuck, A.C.; Kim, V.; Ostridge, K.; et al. A prospective, observational cohort study of the seasonal dynamics of airway pathogens in the aetiology of exacerbations in COPD. Thorax 2017, 72, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Bafadhel, M.; Haldar, K.; Barker, B.; Patel, H.; Mistry, V.; Barer, M.R.; Pavord, I.D.; Brightling, C.E. Airway bacteria measured by quantitative polymerase chain reaction and culture in patients with stable COPD: Relationship with neutrophilic airway inflammation, exacerbation frequency, and lung function. Int. J. Chron Obstruct. Pulmon. Dis. 2015, 10, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Barker, B.L.; Haldar, K.; Patel, H.; Pavord, I.D.; Barer, M.R.; Brightling, C.E.; Bafadhel, M. Association between pathogens detected using quantitative polymerase chain reaction with airway inflammation in COPD at stable state and exacerbations. Chest 2015, 147, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Bafadhel, M.; Haldar, K.; Spivak, A.; Mayhew, D.; Miller, B.E.; Tal-Singer, R.; Johnston, S.L.; Ramsheh, M.Y.; Barer, M.R.; et al. Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 2016, 47, 1082–1092. [Google Scholar] [CrossRef]
- Maddi, S.; Kolsum, U.; Jackson, S.; Barraclough, R.; Maschera, B.; Simpson, K.D.; Pascal, T.G.; Durviaux, S.; Hessel, E.M.; Singh, D. Ampicillin resistance in Haemophilus influenzae from COPD patients in the UK. Int. J. Chron Obstruct. Pulmon. Dis. 2017, 12, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Mackay, A.J.; Patel, A.R.; Garcha, D.S.; Kowlessar, B.S.; Brill, S.E.; Donnelly, L.E.; Barnes, P.J.; Donaldson, G.C.; Wedzicha, J.A. Inflammatory thresholds and the species-specific effects of colonising bacteria in stable chronic obstructive pulmonary disease. Respir. Res. 2014, 15, 114. [Google Scholar] [CrossRef] [PubMed]
- Hajiro, T.; Nishimura, K.; Tsukino, M.; Ikeda, A.; Koyama, H.; Izumi, T. Analysis of clinical methods used to evaluate dyspnea in patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 1998, 158, 1185–1189. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.W.; Harding, G.; Berry, P.; Wiklund, I.; Chen, W.H.; Kline Leidy, N. Development and first validation of the COPD Assessment Test. Eur. Respir. J. 2009, 34, 648–654. [Google Scholar] [CrossRef]
- Jones, P.W.; Quirk, F.H.; Baveystock, C.M.; Littlejohns, P. A self-complete measure of health status for chronic airflow limitation. The St. George’s Respiratory Questionnaire. Am. Rev. Respir. Dis. 1992, 145, 1321–1327. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.R.; Hankinson, J.; Brusasco, V.; Burgos, F.; Casaburi, R.; Coates, A.; Crapo, R.; Enright, P.; van der Grinten, C.P.; Gustafsson, P.; et al. Standardisation of spirometry. Eur. Respir. J. 2005, 26, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Graham, B.L.; Steenbruggen, I.; Miller, M.R.; Barjaktarevic, I.Z.; Cooper, B.G.; Hall, G.L.; Hallstrand, T.S.; Kaminsky, D.A.; McCarthy, K.; McCormack, M.C.; et al. Standardization of Spirometry 2019 Update. An Official American Thoracic Society and European Respiratory Society Technical Statement. Am. J. Respir. Crit. Care Med. 2019, 200, e70–e88. [Google Scholar] [CrossRef]
- Garcha, D.S.; Thurston, S.J.; Patel, A.R.; Mackay, A.J.; Goldring, J.J.; Donaldson, G.C.; McHugh, T.D.; Wedzicha, J.A. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 2012, 67, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Bafadhel, M.; McCormick, M.; Saha, S.; McKenna, S.; Shelley, M.; Hargadon, B.; Mistry, V.; Reid, C.; Parker, D.; Dodson, P.; et al. Profiling of sputum inflammatory mediators in asthma and chronic obstructive pulmonary disease. Respir. Int. Rev. Thorac. Dis. 2012, 83, 36–44. [Google Scholar] [CrossRef]
- McCulloch, E.; Lucas, C.; Ramage, G.; Williams, C. Improved early diagnosis of Pseudomonas aeruginosa by real-time PCR to prevent chronic colonisation in a paediatric cystic fibrosis population. J. Cyst. Fibros. 2011, 10, 21–24. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Morris, A.; Beck, J.M.; Schloss, P.D.; Campbell, T.B.; Crothers, K.; Curtis, J.L.; Flores, S.C.; Fontenot, A.P.; Ghedin, E.; Huang, L.; et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 2013, 187, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Hilty, M.; Burke, C.; Pedro, H.; Cardenas, P.; Bush, A.; Bossley, C.; Davies, J.; Ervine, A.; Poulter, L.; Pachter, L.; et al. Disordered microbial communities in asthmatic airways. PLoS ONE 2010, 5, e8578. [Google Scholar] [CrossRef] [PubMed]
- Pragman, A.A.; Knutson, K.A.; Gould, T.J.; Isaacson, R.E.; Reilly, C.S.; Wendt, C.H. Chronic obstructive pulmonary disease upper airway microbiota alpha diversity is associated with exacerbation phenotype: A case-control observational study. Respir. Res. 2019, 20, 114. [Google Scholar] [CrossRef]
- Shimizu, K.; Yoshii, Y.; Morozumi, M.; Chiba, N.; Ubukata, K.; Uruga, H.; Hanada, S.; Saito, N.; Kadota, T.; Ito, S.; et al. Pathogens in COPD exacerbations identified by comprehensive real-time PCR plus older methods. Int. J. Chron Obstruct. Pulmon. Dis. 2015, 10, 2009–2016. [Google Scholar] [CrossRef]
- Winslow, S.; Odqvist, L.; Diver, S.; Riise, R.; Abdillahi, S.; Wingren, C.; Lindmark, H.; Wellner, A.; Lundin, S.; Yrlid, L.; et al. Multi-omics links IL-6 trans-signalling with neutrophil extracellular trap formation and Haemophilus infection in COPD. Eur. Respir. J. 2021. [Google Scholar] [CrossRef]
- George, L.; Taylor, A.R.; Esteve-Codina, A.; Soler Artigas, M.; Thun, G.A.; Bates, S.; Pavlidis, S.; Wagers, S.; Boland, A.; Prasse, A.; et al. Blood eosinophil count and airway epithelial transcriptome relationships in COPD versus asthma. Allergy 2020, 75, 370–380. [Google Scholar] [CrossRef]
- Southworth, T.; Higham, A.; Kolsum, U.; Li, J.; Scott, T.; Dungwa, J.; Sridhar, S.; Pham, T.H.; Newbold, P.; Singh, D. The relationship between airway immunoglobulin activity and eosinophils in COPD. J. Cell Mol. Med. 2021, 25, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Fukuoka, A.; Matsushita, K.; Morikawa, T.; Adachi, T.; Yasuda, K.; Kiyonari, H.; Fujieda, S.; Yoshimoto, T. Human cystatin SN is an endogenous protease inhibitor that prevents allergic rhinitis. J. Allergy Clin. Immunol. 2019, 143, 1153–1162.e1112. [Google Scholar] [CrossRef]
- Gela, A.; Kasetty, G.; Jovic, S.; Ekoff, M.; Nilsson, G.; Morgelin, M.; Kjellstrom, S.; Pease, J.E.; Schmidtchen, A.; Egesten, A. Eotaxin-3 (CCL26) exerts innate host defense activities that are modulated by mast cell proteases. Allergy 2015, 70, 161–170. [Google Scholar] [CrossRef]
- Boyton, R.J.; Reynolds, C.J.; Quigley, K.J.; Altmann, D.M. Immune mechanisms and the impact of the disrupted lung microbiome in chronic bacterial lung infection and bronchiectasis. Clin. Exp. Immunol. 2013, 171, 117–123. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Huffnagle, G.B. The role of the bacterial microbiome in lung disease. Expert Rev. Respir. Med. 2013, 7, 245–257. [Google Scholar] [CrossRef]
- Thomas, R.A.; Green, R.H.; Brightling, C.E.; Birring, S.S.; Parker, D.; Wardlaw, A.J.; Pavord, I.D. The influence of age on induced sputum differential cell counts in normal subjects. Chest 2004, 126, 1811–1814. [Google Scholar] [CrossRef]
- Lim, M.Y.; Yoon, H.S.; Rho, M.; Sung, J.; Song, Y.M.; Lee, K.; Ko, G. Analysis of the association between host genetics, smoking, and sputum microbiota in healthy humans. Sci. Rep. 2016, 6, 23745. [Google Scholar] [CrossRef] [PubMed]
- Erb-Downward, J.R.; Thompson, D.L.; Han, M.K.; Freeman, C.M.; McCloskey, L.; Schmidt, L.A.; Young, V.B.; Toews, G.B.; Curtis, J.L.; Sundaram, B.; et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS ONE 2011, 6, e16384. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.A.; Faner, R.; Oscullo, G.; de la Rosa, D.; Soler-Cataluna, J.J.; Ballester, M.; Agusti, A. Inhaled Steroids, Circulating Eosinophils, Chronic Airway Infection, and Pneumonia Risk in Chronic Obstructive Pulmonary Disease. A Network Analysis. Am. J. Respir. Crit. Care Med. 2020, 201, 1078–1085. [Google Scholar] [CrossRef] [PubMed]
- Contoli, M.; Pauletti, A.; Rossi, M.R.; Spanevello, A.; Casolari, P.; Marcellini, A.; Forini, G.; Gnesini, G.; Marku, B.; Barnes, N.; et al. Long-term effects of inhaled corticosteroids on sputum bacterial and viral loads in COPD. Eur. Respir. J. 2017, 50, 1700451. [Google Scholar] [CrossRef]

| Characteristic | COPD n = 60 | Healthy Non-Smokers n = 15 | p-Value |
|---|---|---|---|
| Gender (% Male) | 58.3 | 60.0 | 0.91 |
| Age | 64.9 (7.3) | 59.0 (10.4) | 0.02 |
| Smoking status (Current %) | 43.3 | 0.0 | n/a |
| Pack years | 43.9 (18.9) | n/a | n/a |
| BMI (kg/m2) | 28.4 (5.7) | 26.2 (3.2) | 0.12 |
| Retrospective Exacerbation rate (1-year period) | 1.1 (1.3) | n/a | n/a |
| 0 (%) | 41.6 | n/a | n/a |
| 1 (%) | 31.7 | n/a | n/a |
| ≥2 (%) | 26.7 | n/a | n/a |
| Prospective annualised exacerbation rate (annualised to a 1-year period) | 1.2 (1.8) | n/a | n/a |
| a FEV1 (L) | 1.8 (0.6) | 3.1 (0.8) | <0.01 |
| a FEV1 (%) | 66.7 (16.6) | 105.5 (11.9) | <0.01 |
| a FEV1/FVC Ratio (%) | 54.0 (11.3) | 75.9 (4.7) | <0.01 |
| GOLD Category (%) | |||
| 1 | 26.7 | n/a | n/a |
| 2 | 55.0 | n/a | n/a |
| 3 | 18.3 | n/a | n/a |
| 4 | 0 | n/a | n/a |
| CAT | 22.3 (5.6) | n/a | n/a |
| mMRC | 4.0 [2.0–4.0] | n/a | n/a |
| SGRQ-C (Total) | 54.2 (16.1) | n/a | n/a |
| Atopy (%) | 12.1 | 20.0 | 0.42 |
| Chronic bronchitis (%) | 83.3 | n/a | n/a |
| ICS Use (%) | 71.7 | n/a | n/a |
| LABA + LAMA + ICS (%) | 58.3 | n/a | n/a |
| LABA + LAMA (%) | 0.0 | n/a | n/a |
| ICS only (%) | 1.7 | n/a | n/a |
| LABA only (%) | 0.0 | n/a | n/a |
| LAMA only (%) | 15.0 | n/a | n/a |
| No inhaled medication (%) | 5.0 | n/a | n/a |
| Sputum characteristics | |||
| Sputum total cell count × 106/g | 8.25 [0.62–100.9] | 7.60 [2.81–20.48] | 0.39 |
| Sputum Neutrophil (%) | 69.13 [24.25–97.75] | 70.50 [37.50–88.50] | 0.32 |
| Sputum Eosinophil (%) | 1.00 [0.00–16.50] | 0.00 [0.00–4.25] | <0.01 |
| Sputum Lymphocyte (%) | 0.50 [0.00–4.75] | 0.50 [0.00–3.00] | 0.49 |
| Sputum Macrophage (%) | 21.00 [1.00–68.00] | 27.00 [6.25–58.50] | 0.17 |
| Sputum Epithelial Cells (%) | 1.63 [0.00–16.50] | 2.75 [0.00–14.25] | 0.35 |
| Sputum Neutrophil cell count × 106/g | 5.22 [0.32–98.08] | 5.04 [1.24–14.74] | 0.33 |
| Sputum Eosinophil cell count × 106/g | 0.08 [0.00–2.45] | 0.00 [0.00–0.79] | <0.01 |
| Sputum Lymphocyte cell count × 106/g | 0.03 [0.00–0.64] | 0.04 [0.00–0.33] | 0.92 |
| Sputum Macrophage cell count × 106/g | 1.28 [0.20–7.57] | 2.06 [0.38–5.53] | 0.30 |
| Sputum Epithelial cell count × 106/g | 0.16 [0.00–1.59] | 0.17 [0.00–1.42] | 0.78 |
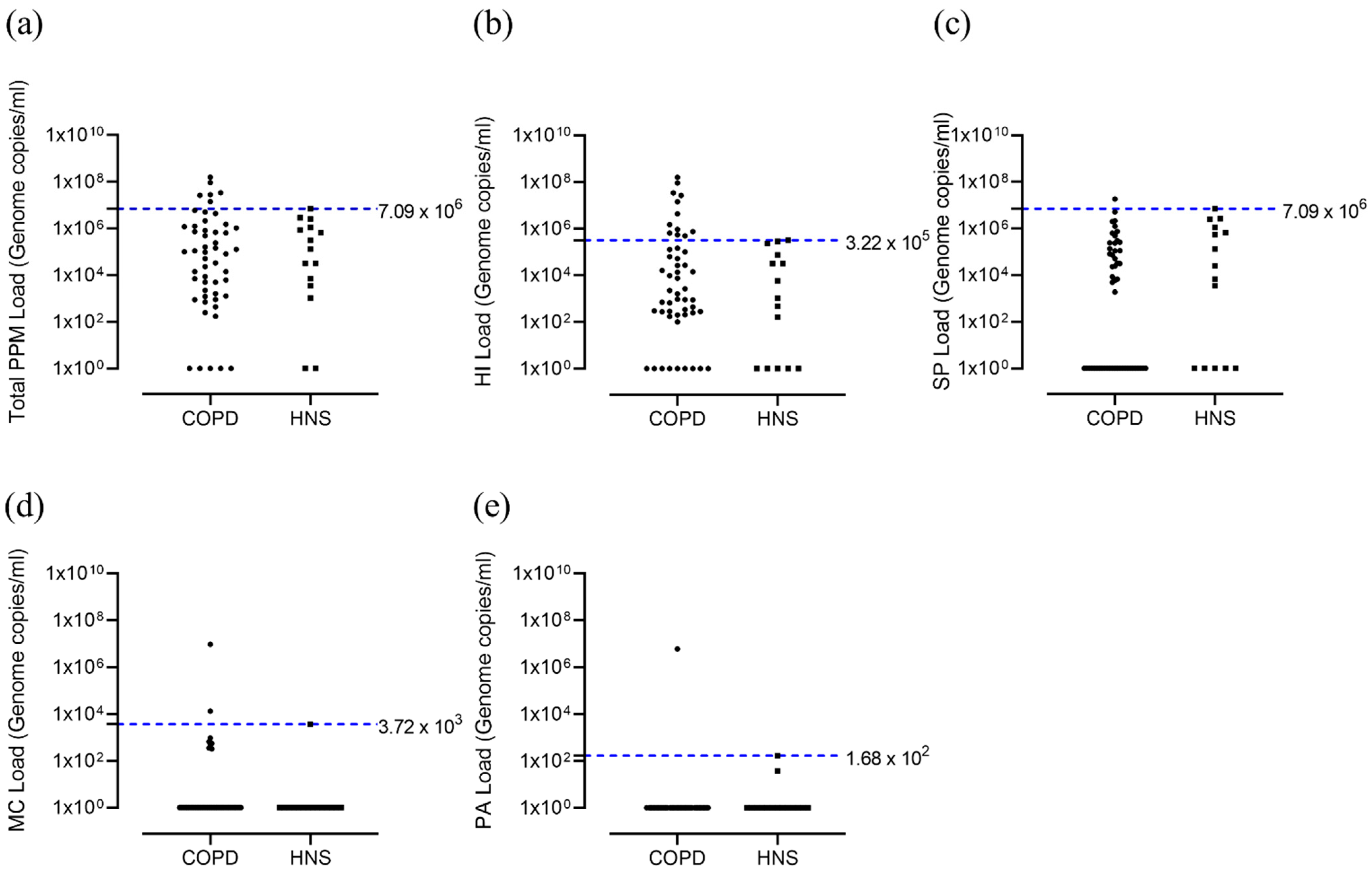
| Total PPM Load (genome copies/mL) | 9.01 × 104 [0.00–1.58 × 108] | 1.31 × 105 [0.00–7.09 × 106] | 0.86 |
| HI Load (genome copies/mL) | 1.94 × 103 [0.00–1.58 × 108] | 1.05 × 103 [0.00–3.22 × 105] | 0.17 |
| SP Load (genome copies/mL) | 3.41 × 103 [0.00–1.82 × 107] | 2.52 × 104 [0.00–7.09 × 106] | 0.23 |
| MC Load (genome copies/mL) | 0.00 [0.00–9.22 × 106] | 0.00 [0.00–3.72 × 103] | 0.39 |
| PA Load (genome copies/mL) | 0.00 [0.00–5.88 × 106] | 0.00 [0.00–1.68 × 102] | 0.12 |
| Sputum Eos Persistently ≥3% (n = 7) | Sputum Eos ≥3% at One Visit Only (n = 8) | Sputum Eos <3% at Both Visits (n = 33) | ||||
|---|---|---|---|---|---|---|
| Baseline | 6 months | Baseline | 6 months | Baseline | 6 months | |
| No PPM | 6 | 6 | 6 | 6 | 12 | 11 |
| HI | 0 | 0 | 1 | 1 | 7 | 7 |
| PA | 0 | 0 | 0 | 0 | 1 | 0 |
| SP | 0 | 0 | 0 | 0 | 0 | 1 |
| MC | 0 | 1 | 0 | 1 | 0 | 1 |
| a >1 PPM | 0 | 0 | 0 | 0 | 1 | 2 |
| Any bacterial colonisation | 0/6 (0.0%) | 1/7 (14.3%) | 1/7 (14.3%) | 2/8 (25.0%) | 9/21 (42.9%) | 11/22 (50.0%) |
| * No data | 1 | 0 | 1 | 0 | 12 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beech, A.; Lea, S.; Li, J.; Jackson, N.; Mulvanny, A.; Singh, D. Airway Bacteria Quantification Using Polymerase Chain Reaction Combined with Neutrophil and Eosinophil Counts Identifies Distinct COPD Endotypes. Biomedicines 2021, 9, 1337. https://doi.org/10.3390/biomedicines9101337
Beech A, Lea S, Li J, Jackson N, Mulvanny A, Singh D. Airway Bacteria Quantification Using Polymerase Chain Reaction Combined with Neutrophil and Eosinophil Counts Identifies Distinct COPD Endotypes. Biomedicines. 2021; 9(10):1337. https://doi.org/10.3390/biomedicines9101337
Chicago/Turabian StyleBeech, Augusta, Simon Lea, Jian Li, Natalie Jackson, Alex Mulvanny, and Dave Singh. 2021. "Airway Bacteria Quantification Using Polymerase Chain Reaction Combined with Neutrophil and Eosinophil Counts Identifies Distinct COPD Endotypes" Biomedicines 9, no. 10: 1337. https://doi.org/10.3390/biomedicines9101337
APA StyleBeech, A., Lea, S., Li, J., Jackson, N., Mulvanny, A., & Singh, D. (2021). Airway Bacteria Quantification Using Polymerase Chain Reaction Combined with Neutrophil and Eosinophil Counts Identifies Distinct COPD Endotypes. Biomedicines, 9(10), 1337. https://doi.org/10.3390/biomedicines9101337