COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells
Abstract
1. Introduction
2. Experimental Section
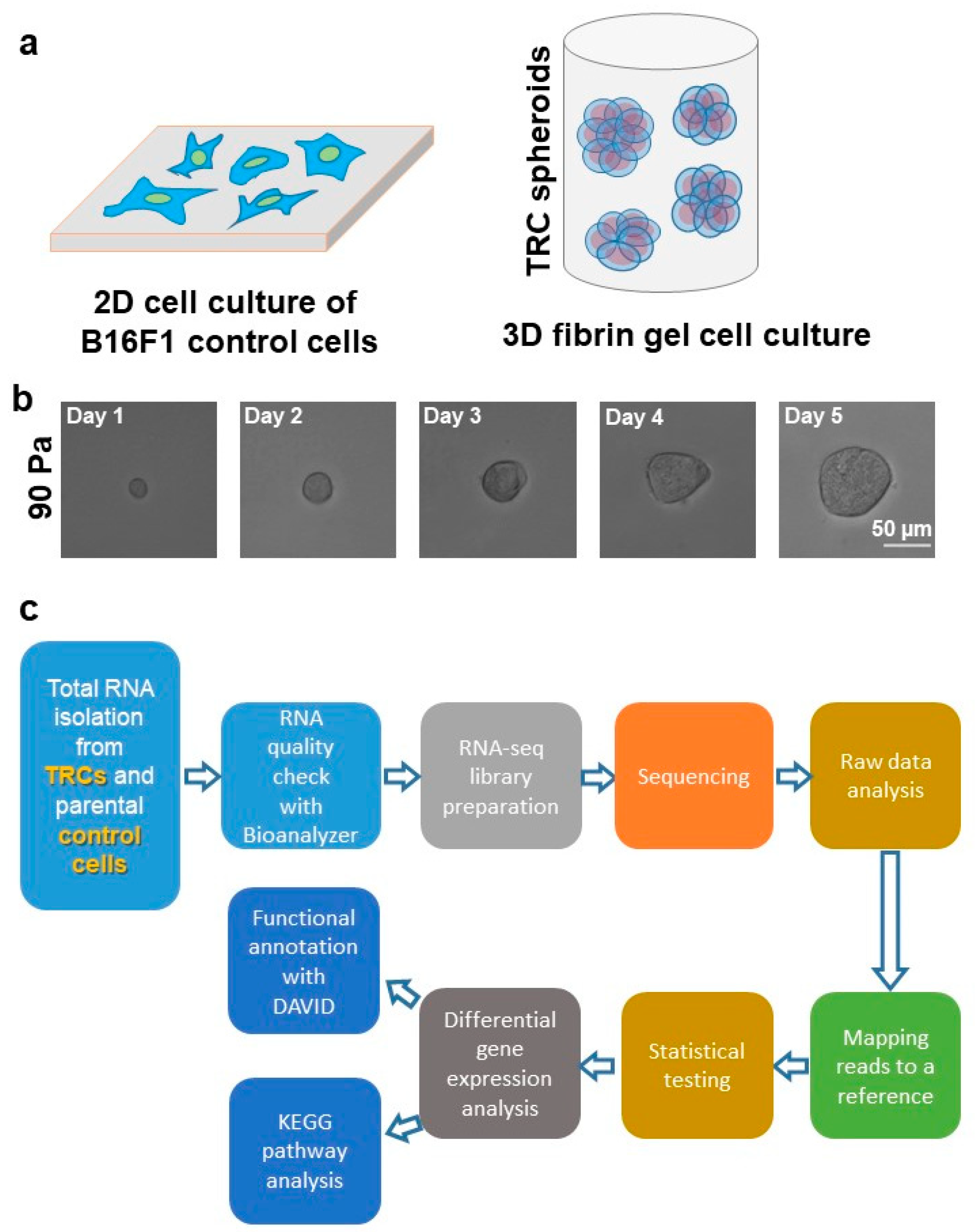
2.1. 3D Fibrin Gel Fabrication
2.2. RNA-seq Data Collection
2.3. RNA-seq Bioinformatics and Differential Expression Analysis
2.4. DAVID, Gene Ontology, and KEGG Pathway Analyses
3. Results
3.1. Growth and Selection of TRC Spheroids by 90 Pa Stiffness 3D Fibrin Gel
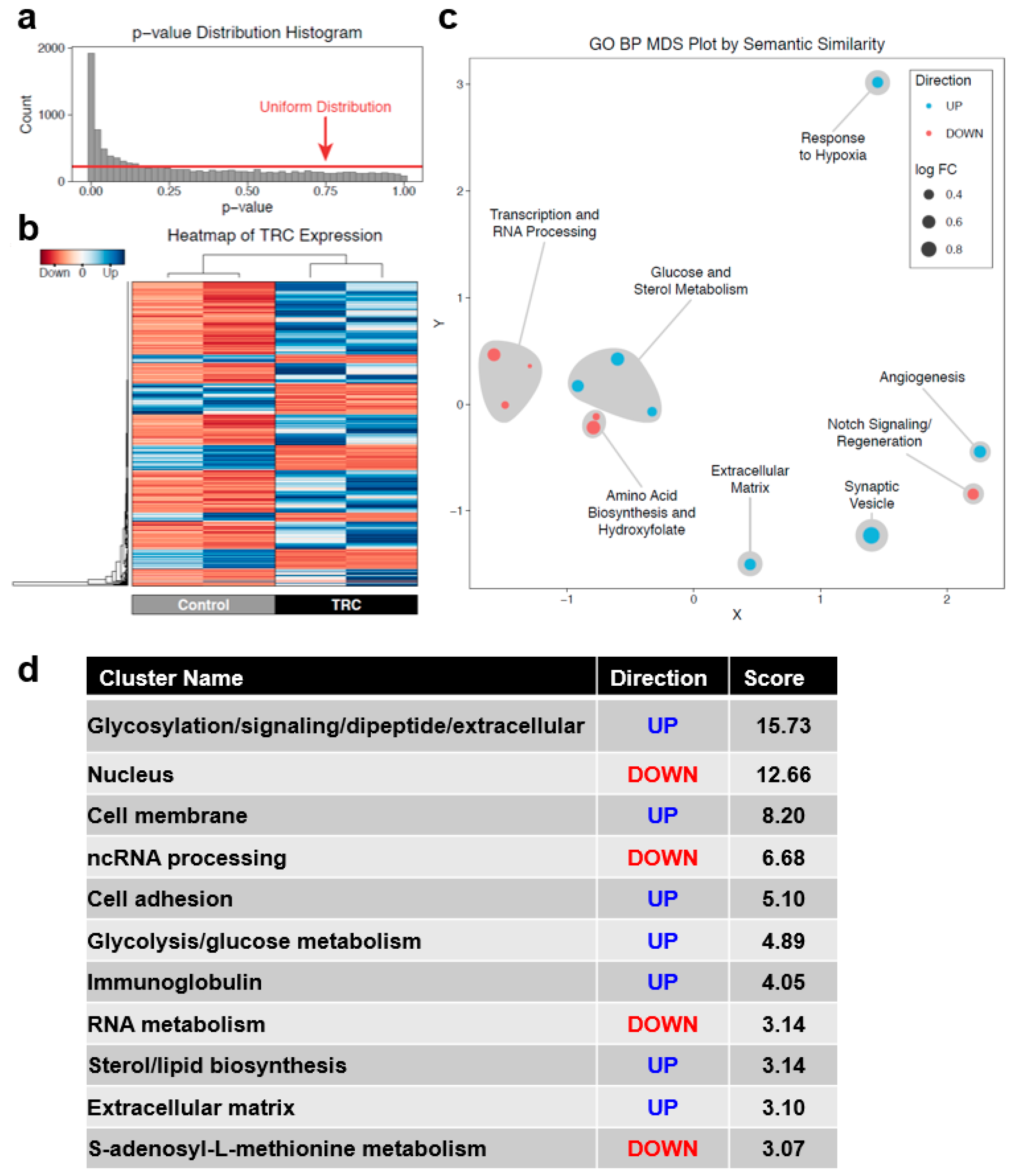
3.2. Identification of DEGs between Parental B16F1 Cells and 3D Fibrin-Derived TRC Spheroids
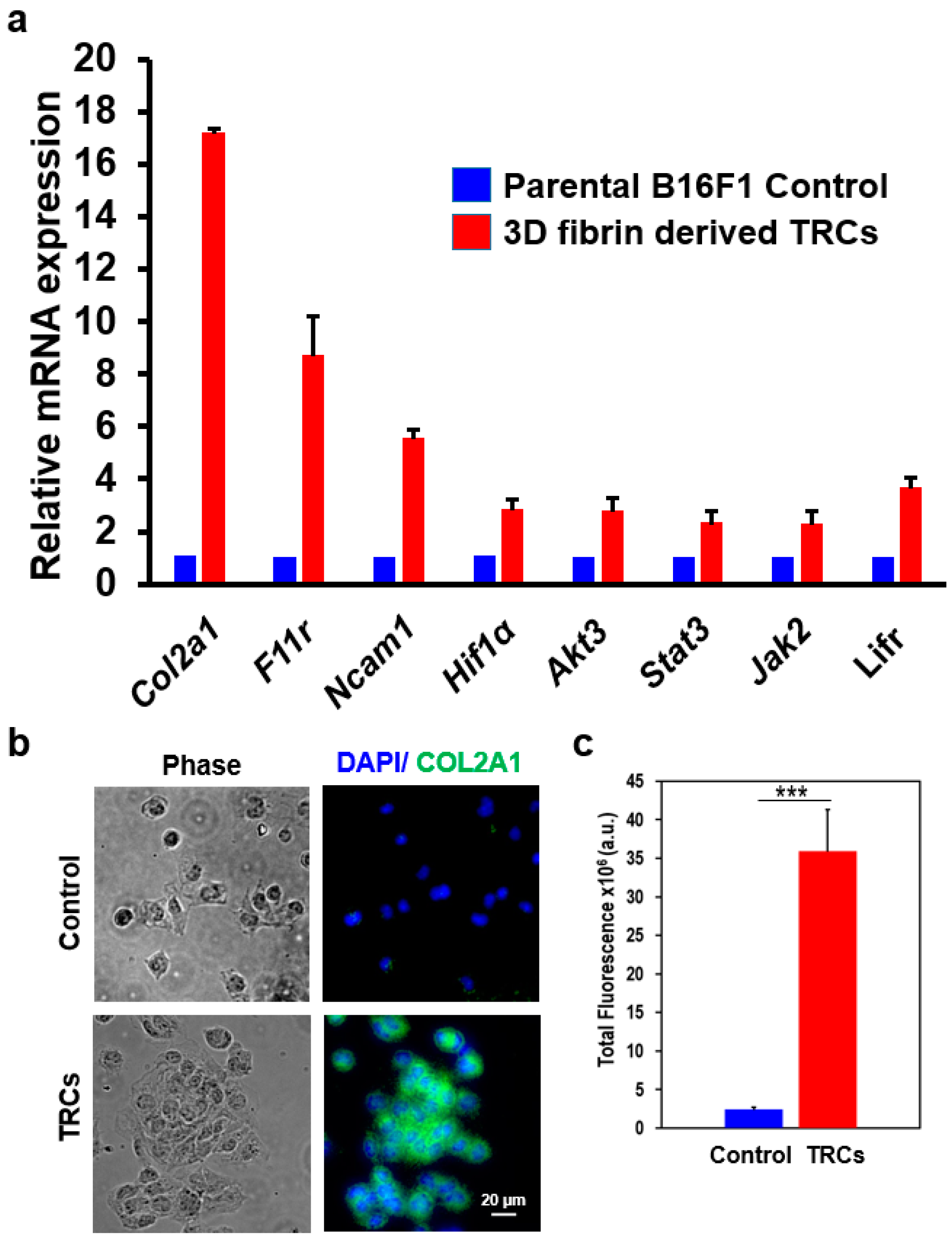
3.3. COL2A1 Is a Highly Specific Biomarker of Melanoma TRCs
3.4. A Unique Set of Gene Regulatory Features Exists in TRCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.C.; Kopetz, S.; Wistuba, I.I.; Zhang, J. Metastasis of cancer: When and how? Ann. Oncol. 2017, 28, 2045–2047. [Google Scholar] [CrossRef] [PubMed]
- Shackleton, M.; Quintana, E.; Fearon, E.R.; Morrison, S.J. Heterogeneity in Cancer: Cancer Stem Cells versus Clonal Evolution. Cell 2009, 138, 822–829. [Google Scholar] [CrossRef]
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta (BBA) Rev. Cancer 2010, 1805, 105–117. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Baccelli, I.; Trumpp, A. The evolving concept of cancer and metastasis stem cells. J. Cell Biol. 2012, 198, 281–293. [Google Scholar] [CrossRef]
- Bao, B.; Ahmad, A.; Azmi, A.S.; Ali, S.; Sarkar, F.H. Overview of Cancer Stem Cells (CSCs) and Mechanisms of Their Regulation: Implications for Cancer Therapy. Curr. Protoc. Pharmacol. 2013, 61, 14–25. [Google Scholar] [CrossRef]
- Croker, A.K.; Allan, A.L. Cancer stem cells: Implications for the progression and treatment of metastatic disease. J. Cell. Mol. Med. 2008, 12, 374–390. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Ailles, L.E.; Weissman, I.L. Cancer stem cells in solid tumors. Curr. Opin. Biotechnol. 2007, 18, 460–466. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Tan, Y.; Zhang, H.; Zhang, Y.; Xu, P.; Chen, J.; Poh, Y.-C.; Tang, K.; Wang, N.; Huang, B. Soft fibrin gels promote selection and growth of tumorigenic cells. Nat. Mater. 2012, 11, 734–741. [Google Scholar] [CrossRef]
- Butcher, D.T.; Alliston, T.; Weaver, V.M. A tense situation: Forcing tumour progression. Nat. Rev. Cancer 2009, 9, 108–122. [Google Scholar] [CrossRef]
- Tan, Y.; Tajik, A.; Chen, J.; Jia, Q.; Chowdhury, F.; Wang, L.; Chen, J.; Zhang, S.; Hong, Y.; Yi, H.; et al. Matrix softness regulates plasticity of tumour-repopulating cells via H3K9 demethylation and Sox2 expression. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Chowdhury, F.; Doğanay, S.; Leslie, B.J.; Singh, R.; Amar, K.; Talluri, B.; Park, S.; Wang, N.; Ha, T. Cdc42-dependent modulation of rigidity sensing and cell spreading in tumor repopulating cells. Biochem. Biophys. Res. Commun. 2018, 500, 557–563. [Google Scholar] [CrossRef]
- Chen, J.; Zhou, W.; Jia, Q.; Chen, J.; Zhang, S.; Yao, W.; Wei, F.; Zhang, Y.; Yang, F.; Huang, W.; et al. Efficient extravasation of tumor-repopulating cells depends on cell deformability. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate—A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.D.; Yamada, K.M. Mechanosensing via cell-matrix adhesions in 3D microenvironments. Exp. Cell Res. 2016, 343, 60–66. [Google Scholar] [CrossRef]
- Emon, B.; Bauer, J.; Jain, Y.; Jung, B.; Saif, T. Biophysics of Tumor Microenvironment and Cancer Metastasis—A Mini Review. Comput. Struct. Biotechnol. J. 2018, 16, 279–287. [Google Scholar] [CrossRef]
- Plodinec, M.; Loparic, M.; Monnier, C.A.; Obermann, E.C.; Zanetti-Dallenbach, R.; Oertle, P.; Hyotyla, J.T.; Aebi, U.; Bentires-Alj, M.; Lim, R.Y.H.; et al. The nanomechanical signature of breast cancer. Nat. Nanotechnol. 2012, 7, 757–765. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef]
- Wang, J.; Sullenger, B.A.; Rich, J.N. Notch Signaling in Cancer Stem Cells. In Notch Signaling in Embryology and Cancer; Springer: New York, NY, USA, 2012; pp. 174–185. [Google Scholar] [CrossRef]
- Guo, G.; Marrero, L.; Rodriguez, P.; Del Valle, L.; Ochoa, A.; Cui, Y. Trp53 Inactivation in the Tumor Microenvironment Promotes Tumor Progression by Expanding the Immunosuppressive Lymphoid-like Stromal Network. Cancer Res. 2013, 73, 1668–1675. [Google Scholar] [CrossRef]
- Melnikova, V.O.; Bolshakov, S.V.; Walker, C.; Ananthaswamy, H.N. Genomic alterations in spontaneous and carcinogen-induced murine melanoma cell lines. Oncogene 2004, 23, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Arbouzova, N.I. JAK/STAT signalling in Drosophila: Insights into conserved regulatory and cellular functions. Development 2006, 133, 2605–2616. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, F.; Na, S.; Li, D.; Poh, Y.-C.; Tanaka, T.S.; Wang, F.; Wang, N. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat. Mater. 2009, 9, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, F.; Li, Y.; Poh, Y.-C.; Yokohama-Tamaki, T.; Wang, N.; Tanaka, T.S. Soft Substrates Promote Homogeneous Self-Renewal of Embryonic Stem Cells via Downregulating Cell-Matrix Tractions. PLoS ONE 2010, 5, e15655. [Google Scholar] [CrossRef]
- Ying, Q.-L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef]
- Tuncer, E.; Calçada, R.R.; Zingg, D.; Varum, S.; Cheng, P.; Freiberger, S.N.; Deng, C.-X.; Kleiter, I.; Levesque, M.P.; Dummer, R.; et al. SMAD signaling promotes melanoma metastasis independently of phenotype switching. J. Clin. Investig. 2019, 129, 2702–2716. [Google Scholar] [CrossRef]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef]
- Laklai, H.; Miroshnikova, Y.A.; Pickup, M.W.; Collisson, E.A.; Kim, G.E.; Barrett, A.S.; Hill, R.C.; Lakins, J.N.; Schlaepfer, D.D.; Mouw, J.K.; et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016, 22, 497–505. [Google Scholar] [CrossRef]
- Hanker, A.B.; Estrada, M.V.; Bianchini, G.; Moore, P.D.; Zhao, J.; Cheng, F.; Koch, J.P.; Gianni, L.; Tyson, D.R.; Sánchez, V.; et al. Extracellular Matrix/Integrin Signaling Promotes Resistance to Combined Inhibition of HER2 and PI3K in HER2+ Breast Cancer. Cancer Res. 2017, 77, 3280–3292. [Google Scholar] [CrossRef]
- Jia, Q.; Zhou, W.; Yao, W.; Yang, F.; Zhang, S.; Singh, R.; Chen, J.; Chen, J.J.; Zhang, Y.; Wei, F.; et al. Downregulation of YAP-dependent Nupr1 promotes tumor-repopulating cell growth in soft matrices. Oncogenesis 2016, 5, e220. [Google Scholar] [CrossRef]
- Altomare, D.A.; Testa, J.R. Perturbations of the AKT signaling pathway in human cancer. Oncogene 2005, 24, 7455–7464. [Google Scholar] [CrossRef] [PubMed]
- Bedogni, B.; Powell, M.B. Hypoxia, melanocytes and melanoma—Survival and tumor development in the permissive microenvironment of the skin. Pigment. Cell Melanoma Res. 2009, 22, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Sharma, A.K.; Madhunapantula, S.V.; Desai, D.; Huh, S.J.; Mosca, P.; Amin, S.; Robertson, G.P. Targeting Akt3 Signaling in Malignant Melanoma Using Isoselenocyanates. Clin. Cancer Res. 2009, 15, 1674–1685. [Google Scholar] [CrossRef] [PubMed]
- Stahl, J.M.; Sharma, A.; Cheung, M.; Zimmerman, M.; Cheng, J.Q.; Bosenberg, M.W.; Kester, M.; Sandirasegarane, L.; Robertson, G.P. Deregulated Akt3 Activity Promotes Development of Malignant Melanoma. Cancer Res. 2004, 64, 7002–7010. [Google Scholar] [CrossRef] [PubMed]
- Jin, W. Role of JAK/STAT3 Signaling in the Regulation of Metastasis, the Transition of Cancer Stem Cells, and Chemoresistance of Cancer by Epithelial–Mesenchymal Transition. Cells 2020, 9, 217. [Google Scholar] [CrossRef]
- Lesinski, G.B. The potential for targeting the STAT3 pathway as a novel therapy for melanoma. Future Oncol. 2013, 9, 925–927. [Google Scholar] [CrossRef]
- Guo, H.; Cheng, Y.; Martinka, M.; McElwee, K. High LIFr expression stimulates melanoma cell migration and is associated with unfavorable prognosis in melanoma. Oncotarget 2015, 6, 25484–25498. [Google Scholar] [CrossRef]
- Ohanna, M.; Cheli, Y.; Bonet, C.; Bonazzi, V.F.; Allegra, M.; Giuliano, S.; Bille, K.; Bahadoran, P.; Giacchero, D.; Lacour, J.P.; et al. Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget 2013, 4, 2212–2224. [Google Scholar] [CrossRef]
- Nicholas, C.; Lesinski, G.B. The Jak-STAT Signal Transduction Pathway in Melanoma. In Breakthroughs in Melanoma Research; IntechOpen: London, UK, 2011. [Google Scholar] [CrossRef]
- Liu, Y.; Lv, J.; Liu, J.; Liang, X.; Jin, X.; Xie, J.; Zhang, L.; Chen, D.; Fiskesund, R.; Tang, K.; et al. STAT3/p53 pathway activation disrupts IFN-β–induced dormancy in tumor-repopulating cells. J. Clin. Investig. 2018, 128, 1057–1073. [Google Scholar] [CrossRef]
| Gene Name | Fold Change | |
|---|---|---|
| Cell surface biomarkers | Col2a1 | 20.82 |
| Ncam1 | 7.7 | |
| F11r | 4.41 | |
| Col11a2 | 3.45 | |
| Negr1 | 2.64 | |
| Cd47 | 2.31 | |
| p53 pathway | Trp53 | −2.07 |
| JAK/ STAT pathway | Lifr | 4.47 |
| Jak2 | 1.51 | |
| Stat3 | 1.97 | |
| Hypoxia related genes | Hif1a | 1.96 |
| Angiogenesis | Vegfa | 4.36 |
| PI3K/ Akt pathway | Akt1 | 1.04 |
| Akt2 | 1.23 | |
| Akt3 | 2.25 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Talluri, B.; Amar, K.; Saul, M.; Shireen, T.; Konjufca, V.; Ma, J.; Ha, T.; Chowdhury, F. COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells. Biomedicines 2020, 8, 360. https://doi.org/10.3390/biomedicines8090360
Talluri B, Amar K, Saul M, Shireen T, Konjufca V, Ma J, Ha T, Chowdhury F. COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells. Biomedicines. 2020; 8(9):360. https://doi.org/10.3390/biomedicines8090360
Chicago/Turabian StyleTalluri, Bhavana, Kshitij Amar, Michael Saul, Tasnim Shireen, Vjollca Konjufca, Jian Ma, Taekjip Ha, and Farhan Chowdhury. 2020. "COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells" Biomedicines 8, no. 9: 360. https://doi.org/10.3390/biomedicines8090360
APA StyleTalluri, B., Amar, K., Saul, M., Shireen, T., Konjufca, V., Ma, J., Ha, T., & Chowdhury, F. (2020). COL2A1 Is a Novel Biomarker of Melanoma Tumor Repopulating Cells. Biomedicines, 8(9), 360. https://doi.org/10.3390/biomedicines8090360






