Honokiol Protects the Kidney from Renal Ischemia and Reperfusion Injury by Upregulating the Glutathione Biosynthetic Enzymes
Abstract
1. Introduction
2. Experimental Section
2.1. Animals
2.2. Animal Model of Renal IR
2.3. H&E Staining and Immunohistochemistry (IHC)
2.4. Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Assay
2.5. Cell Culture and Treatment
2.6. siRNA-Mediated Transfection
2.7. Cell Viability Assay
2.8. GSH Measurement
2.9. Lipid Peroxidation (Malondialdehyde, MDA) Assay
2.10. Antioxidant Response Element (ARE)-Luciferase Reporter Assay
2.11. Western Blot Analysis
2.12. Reverse Transcription and Quantitative PCR (qPCR) Analysis
2.13. Statistical Analysis
3. Results
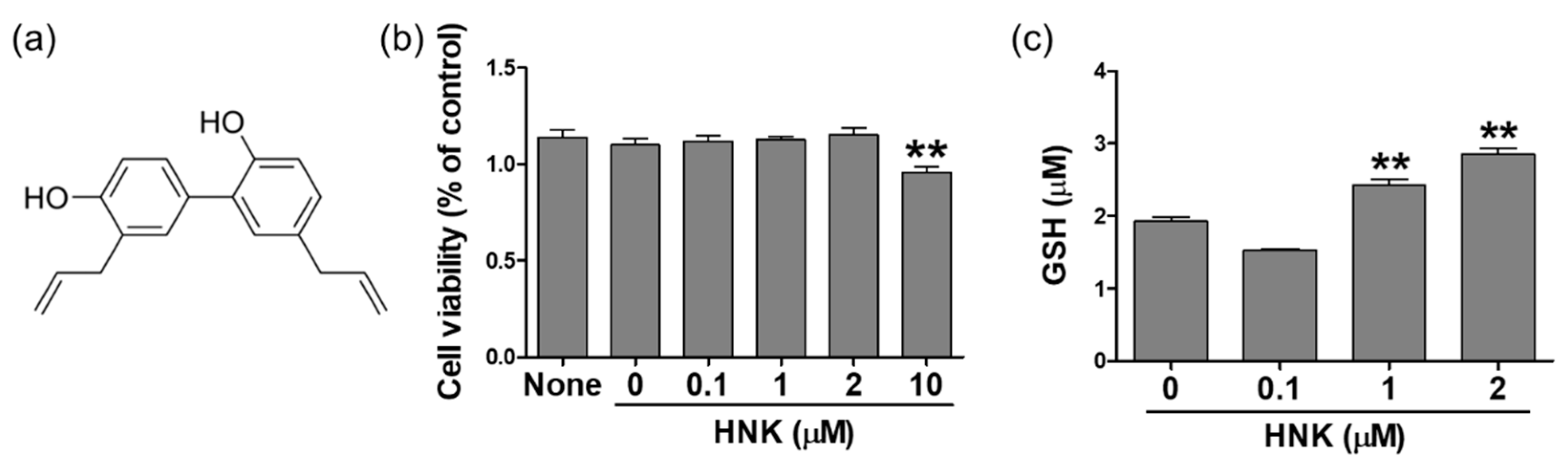
3.1. Honokiol Treatment Increased GSH Levels in HK-2 Cells
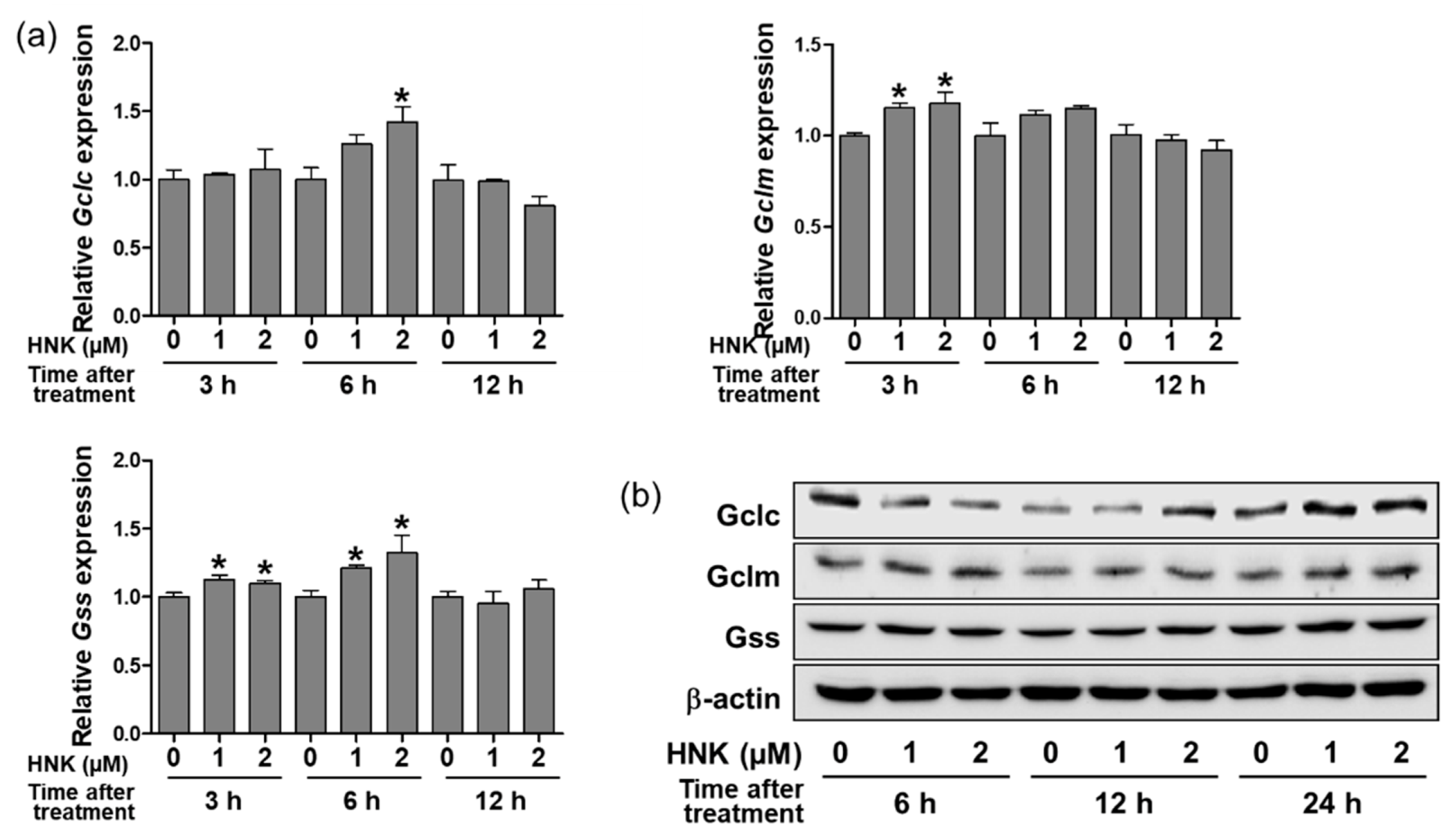
3.2. Honokiol Treatment Upregulated the Expression of GSH Biosynthetic Enzymes in HK-2 Cells
3.3. Honokiol-Induced Upregulation of Gclc Was Regulated by Nuclear Nrf2 Translocation in HK-2 Cells
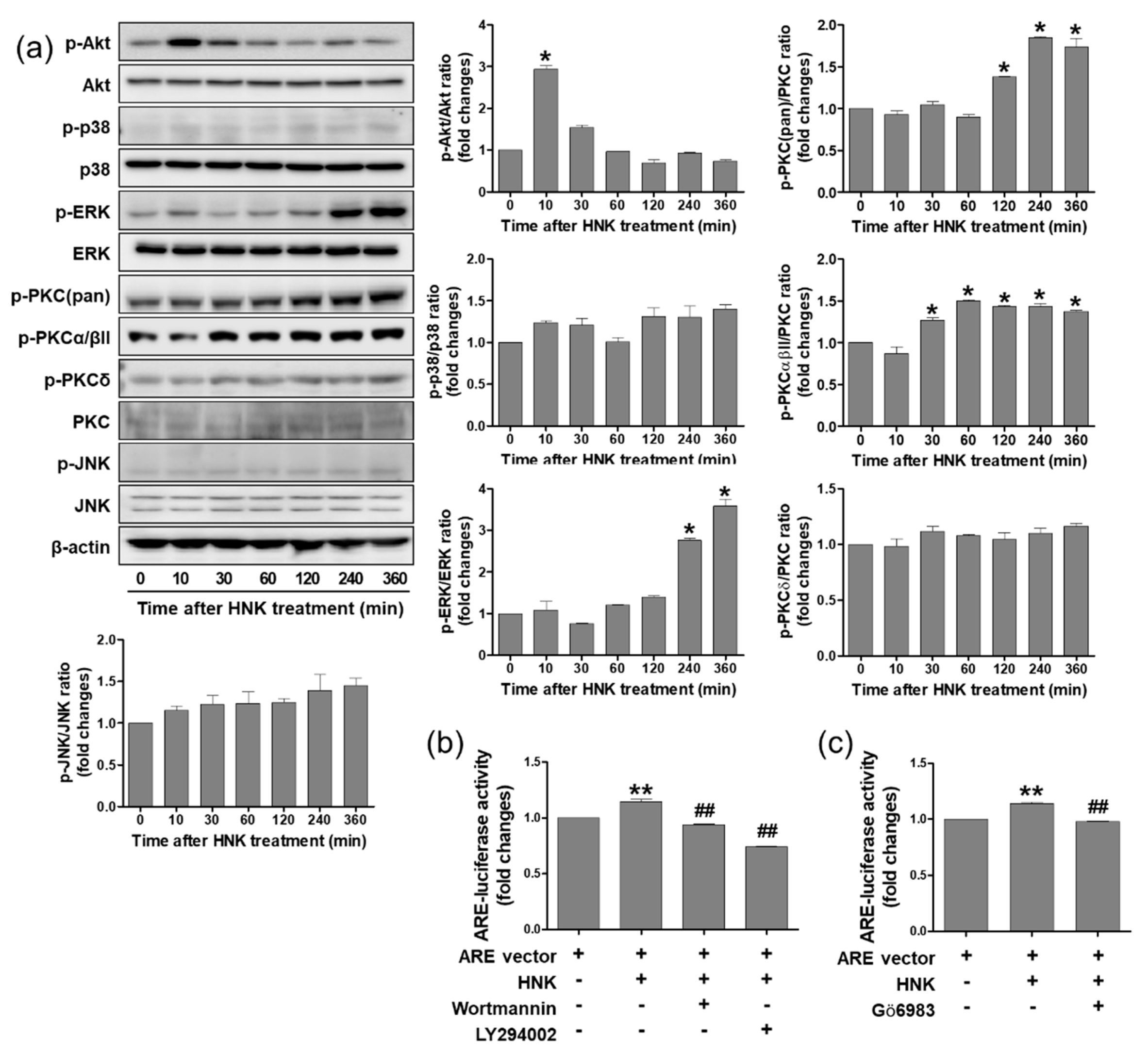
3.4. Honokiol Treatment Increased Nrf2 Transcriptional Activity through PI3K/Akt and PKC Signaling Pathway in HK-2 Cells
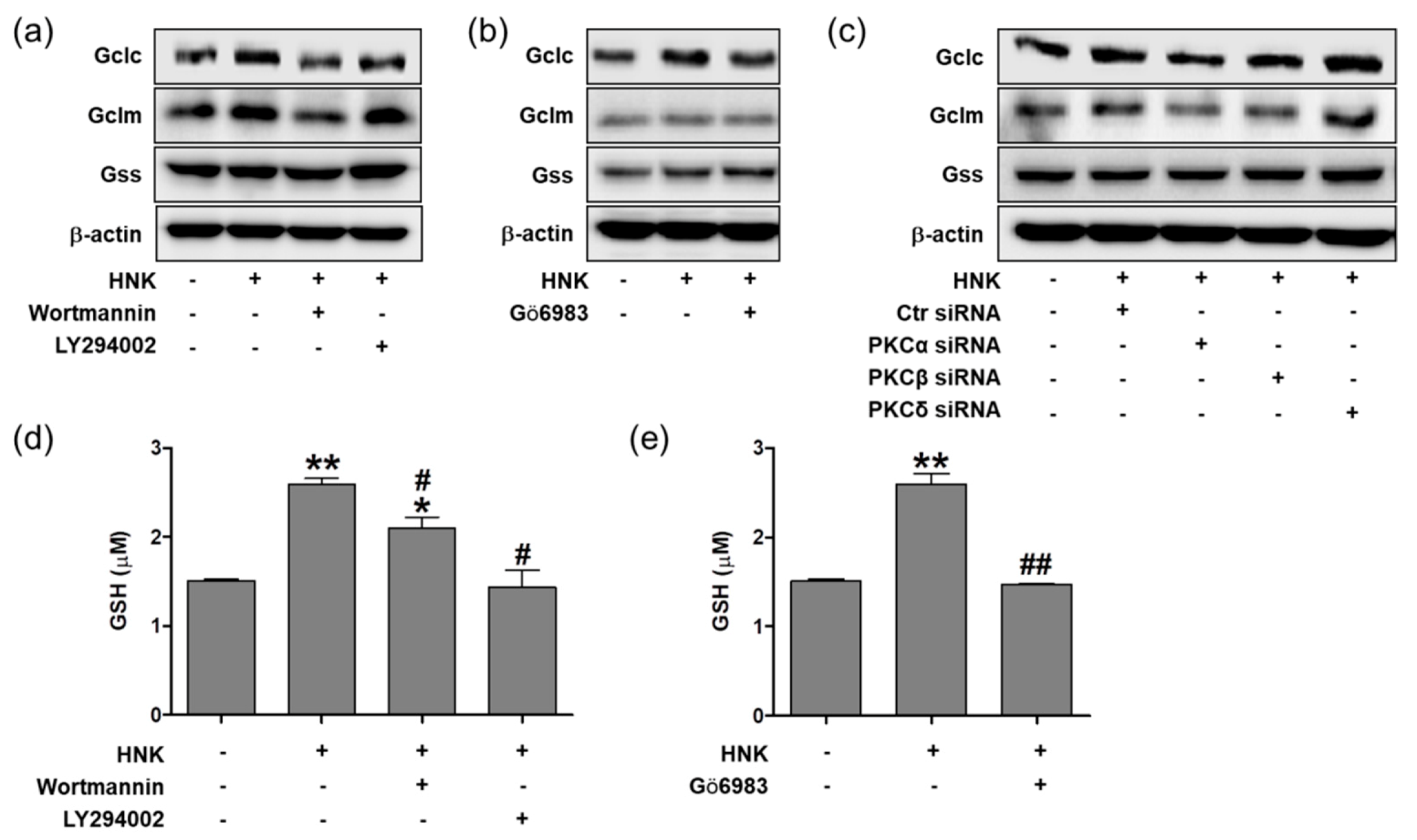
3.5. Honokiol Increases GSH Biosynthesis through PI3K/Akt and PKC Signaling in HK-2 Cells
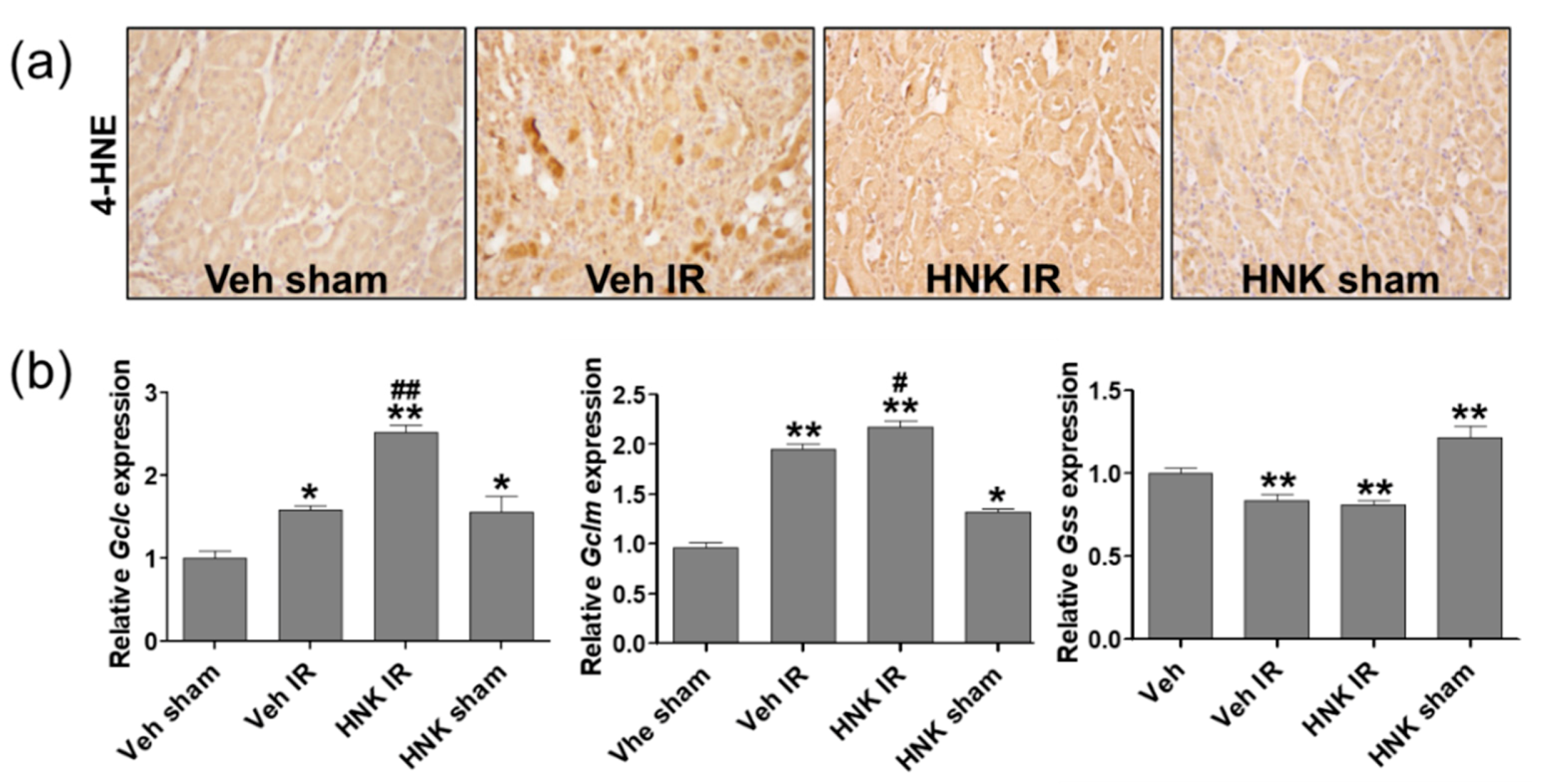
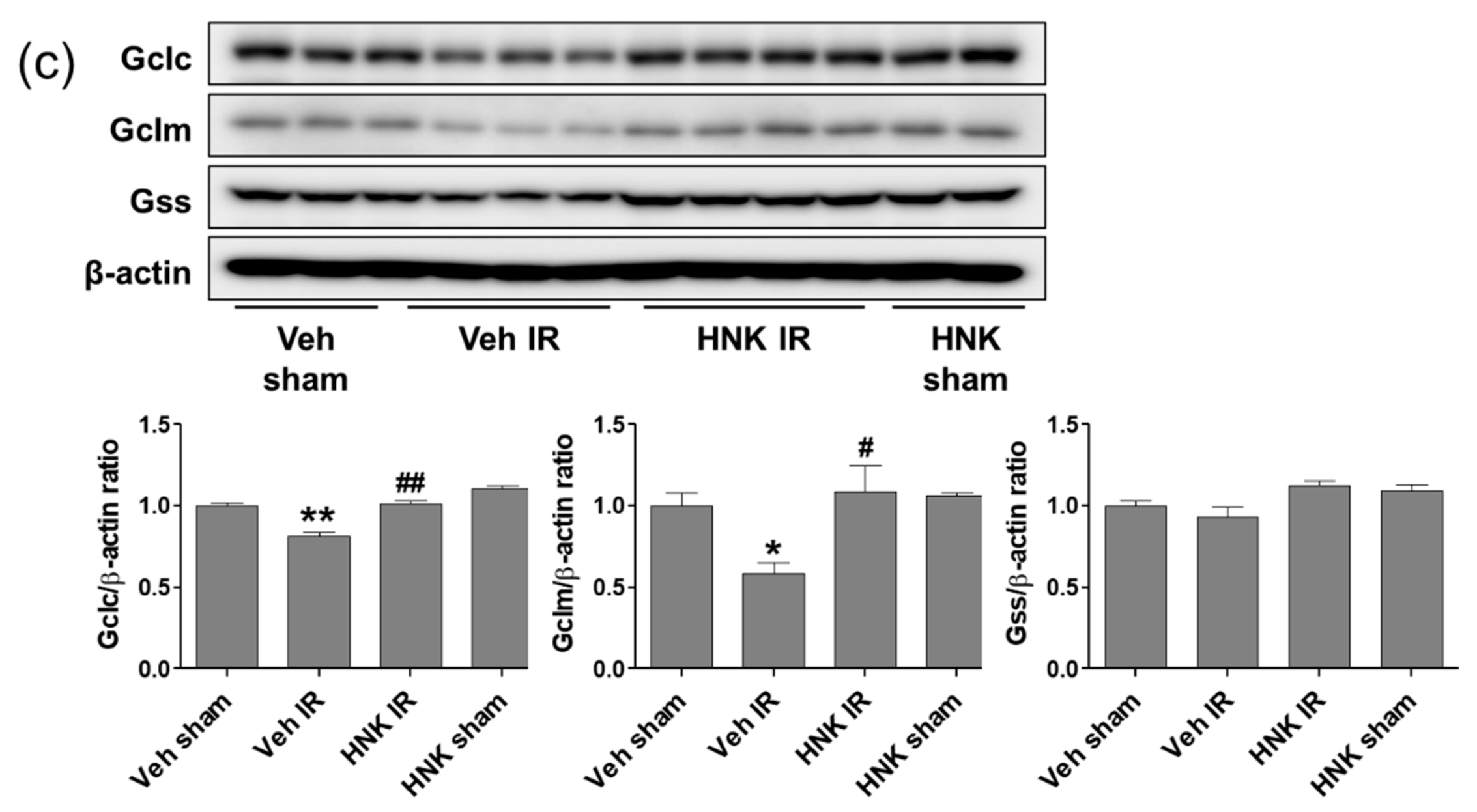
3.6. Honokiol Attenuates Renal Ischemia-Reperfusion Injury by Increasing GSH Biosynthesis
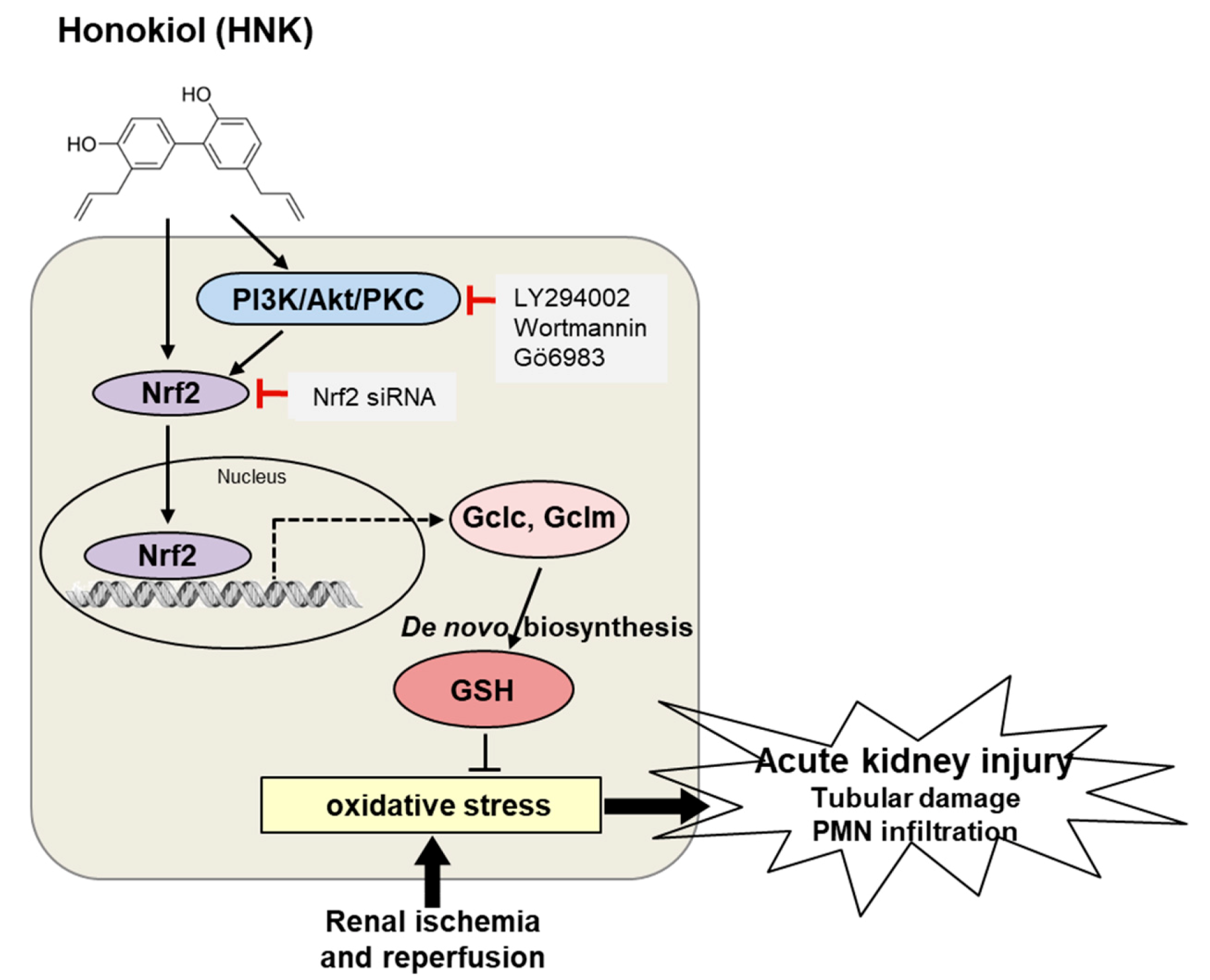
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Granger, D.N.; Kvietys, P.R. Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol. 2015, 6, 524–551. [Google Scholar] [CrossRef] [PubMed]
- Malek, M.; Nematbakhsh, M. Renal ischemia/reperfusion injury; from pathophysiology to treatment. J. Ren. Inj. Prev. 2015, 4, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Yiang, G.T.; Liao, W.T.; Tsai, A.P.; Cheng, Y.L.; Cheng, P.W.; Li, C.Y.; Li, C.J. Current Mechanistic Concepts in Ischemia and Reperfusion Injury. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 46, 1650–1667. [Google Scholar] [CrossRef]
- Chouchani, E.T.; Pell, V.R.; Gaude, E.; Aksentijevic, D.; Sundier, S.Y.; Robb, E.L.; Logan, A.; Nadtochiy, S.M.; Ord, E.N.J.; Smith, A.C.; et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature 2014, 515, 431–435. [Google Scholar] [CrossRef] [PubMed]
- Paller, M.S.; Hoidal, J.R.; Ferris, T.F. Oxygen free radicals in ischemic acute renal failure in the rat. J. Clin. Investig. 1984, 74, 1156–1164. [Google Scholar] [CrossRef]
- Tsuda, H.; Kawada, N.; Kaimori, J.Y.; Kitamura, H.; Moriyama, T.; Rakugi, H.; Takahara, S.; Isaka, Y. Febuxostat suppressed renal ischemia-reperfusion injury via reduced oxidative stress. Biochem. Biophys. Res. Commun. 2012, 427, 266–272. [Google Scholar] [CrossRef]
- Cho, S.; Yu, S.L.; Kang, J.; Jeong, B.Y.; Lee, H.Y.; Park, C.G.; Yu, Y.B.; Jin, D.C.; Hwang, W.M.; Yun, S.R.; et al. NADPH oxidase 4 mediates TGF-beta1/Smad signaling pathway induced acute kidney injury in hypoxia. PLoS ONE 2019, 14, e0219483. [Google Scholar] [CrossRef]
- Karim, A.S.; Reese, S.R.; Wilson, N.A.; Jacobson, L.M.; Zhong, W.; Djamali, A. Nox2 is a mediator of ischemia reperfusion injury. Am. J. Transplant. Off. J. Am. Soc. Transplant. Am. Soc. Transpl. Surg. 2015, 15, 2888–2899. [Google Scholar] [CrossRef]
- Simone, S.; Rascio, F.; Castellano, G.; Divella, C.; Chieti, A.; Ditonno, P.; Battaglia, M.; Crovace, A.; Staffieri, F.; Oortwijn, B.; et al. Complement-dependent NADPH oxidase enzyme activation in renal ischemia/reperfusion injury. Free Radic. Biol. Med. 2014, 74, 263–273. [Google Scholar] [CrossRef]
- Chen, Q.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Ischemic defects in the electron transport chain increase the production of reactive oxygen species from isolated rat heart mitochondria. American journal of physiology. Cell Physiol. 2008, 294, C460–C466. [Google Scholar] [CrossRef]
- Kezic, A.; Spasojevic, I.; Lezaic, V.; Bajcetic, M. Mitochondria-Targeted Antioxidants: Future Perspectives in Kidney Ischemia Reperfusion Injury. Oxidative Med. Cell. Longev. 2016, 2016, 2950503. [Google Scholar] [CrossRef] [PubMed]
- Rahal, A.; Kumar, A.; Singh, V.; Yadav, B.; Tiwari, R.; Chakraborty, S.; Dhama, K. Oxidative stress, prooxidants, and antioxidants: The interplay. Biomed. Res. Int. 2014, 2014, 761264. [Google Scholar] [CrossRef] [PubMed]
- Ratliff, B.B.; Abdulmahdi, W.; Pawar, R.; Wolin, M.S. Oxidant Mechanisms in Renal Injury and Disease. Antioxid. Redox Signal 2016, 25, 119–146. [Google Scholar] [CrossRef]
- Hanschmann, E.M.; Godoy, J.R.; Berndt, C.; Hudemann, C.; Lillig, C.H. Thioredoxins, glutaredoxins, and peroxiredoxins--molecular mechanisms and health significance: From cofactors to antioxidants to redox signaling. Antioxid. Redox Signal 2013, 19, 1539–1605. [Google Scholar] [CrossRef]
- Wu, G.; Fang, Y.Z.; Yang, S.; Lupton, J.R.; Turner, N.D. Glutathione metabolism and its implications for health. J. Nutr. 2004, 134, 489–492. [Google Scholar] [CrossRef]
- Dalton, T.P.; Dieter, M.Z.; Yang, Y.; Shertzer, H.G.; Nebert, D.W. Knockout of the mouse glutamate cysteine ligase catalytic subunit (Gclc) gene: Embryonic lethal when homozygous, and proposed model for moderate glutathione deficiency when heterozygous. Biochem. Biophys. Res. Commun. 2000, 279, 324–329. [Google Scholar] [CrossRef]
- Giordano, G.; Afsharinejad, Z.; Guizzetti, M.; Vitalone, A.; Kavanagh, T.J.; Costa, L.G. Organophosphorus insecticides chlorpyrifos and diazinon and oxidative stress in neuronal cells in a genetic model of glutathione deficiency. Toxicol. Appl. Pharmacol. 2007, 219, 181–189. [Google Scholar] [CrossRef]
- McConnachie, L.A.; Mohar, I.; Hudson, F.N.; Ware, C.B.; Ladiges, W.C.; Fernandez, C.; Chatterton-Kirchmeier, S.; White, C.C.; Pierce, R.H.; Kavanagh, T.J. Glutamate cysteine ligase modifier subunit deficiency and gender as determinants of acetaminophen-induced hepatotoxicity in mice. Toxicol. Sci. 2007, 99, 628–636. [Google Scholar] [CrossRef]
- Yang, Y.; Dieter, M.Z.; Chen, Y.; Shertzer, H.G.; Nebert, D.W.; Dalton, T.P. Initial characterization of the glutamate-cysteine ligase modifier subunit Gclm(-/-) knockout mouse. Novel model system for a severely compromised oxidative stress response. J. Biol. Chem. 2002, 277, 49446–49452. [Google Scholar] [CrossRef]
- Lu, S.C. Regulation of glutathione synthesis. Mol. Asp. Med. 2009, 30, 42–59. [Google Scholar] [CrossRef] [PubMed]
- Aquilano, K.; Baldelli, S.; Ciriolo, M.R. Glutathione: New roles in redox signaling for an old antioxidant. Front. Pharmacol. 2014, 5, 196. [Google Scholar] [CrossRef] [PubMed]
- Golab, F.; Kadkhodaee, M.; Zahmatkesh, M.; Hedayati, M.; Arab, H.; Schuster, R.; Zahedi, K.; Lentsch, A.B.; Soleimani, M. Ischemic and non-ischemic acute kidney injury cause hepatic damage. Kidney Int. 2009, 75, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Siow, Y.L.; Isaak, C.K.; Karmin, O. Downregulation of Glutathione Biosynthesis Contributes to Oxidative Stress and Liver Dysfunction in Acute Kidney Injury. Oxidative Med. Cell. Longev. 2016, 2016, 9707292. [Google Scholar] [CrossRef] [PubMed]
- Tonelli, C.; Chio, I.I.C.; Tuveson, D.A. Transcriptional Regulation by Nrf2. Antioxid. Redox Signal 2018, 29, 1727–1745. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Yamamoto, M. Molecular mechanisms activating the Nrf2-Keap1 pathway of antioxidant gene regulation. Antioxid. Redox Signal 2005, 7, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q. Role of nrf2 in oxidative stress and toxicity. Annu. Rev. Pharmacol. Toxicol. 2013, 53, 401–426. [Google Scholar] [CrossRef]
- Kensler, T.W.; Wakabayashi, N.; Biswal, S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007, 47, 89–116. [Google Scholar] [CrossRef]
- Robledinos-Anton, N.; Fernandez-Gines, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxidative Med. Cell Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef]
- Li, N.; Xie, H.; Li, L.; Wang, J.; Fang, M.; Yang, N.; Lin, H. Effects of honokiol on sepsis-induced acute kidney injury in an experimental model of sepsis in rats. Inflammation 2014, 37, 1191–1199. [Google Scholar] [CrossRef]
- Yu, Y.; Li, M.; Su, N.; Zhang, Z.; Zhao, H.; Yu, H.; Xu, Y. Honokiol protects against renal ischemia/reperfusion injury via the suppression of oxidative stress, iNOS, inflammation and STAT3 in rats. Mol. Med. Rep. 2016, 13, 1353–1360. [Google Scholar] [CrossRef]
- Seo, M.S.; Kim, H.J.; Kim, H.; Park, S.W. Ethyl Pyruvate Directly Attenuates Active Secretion of HMGB1 in Proximal Tubular Cells via Induction of Heme Oxygenase-1. J. Clin. Med. 2019, 8, 629. [Google Scholar] [CrossRef]
- Birben, E.; Sahiner, U.M.; Sackesen, C.; Erzurum, S.; Kalayci, O. Oxidative stress and antioxidant defense. World Allergy Organ. J. 2012, 5, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Fiser, B.; Jojart, B.; Csizmadia, I.G.; Viskolcz, B. Glutathione--hydroxyl radical interaction: A theoretical study on radical recognition process. PLoS ONE 2013, 8, e73652. [Google Scholar] [CrossRef] [PubMed]
- Masella, R.; Di Benedetto, R.; Vari, R.; Filesi, C.; Giovannini, C. Novel mechanisms of natural antioxidant compounds in biological systems: Involvement of glutathione and glutathione-related enzymes. J. Nutr. Biochem. 2005, 16, 577–586. [Google Scholar] [CrossRef]
- Scharf, G.; Prustomersky, S.; Knasmuller, S.; Schulte-Hermann, R.; Huber, W.W. Enhancement of glutathione and g-glutamylcysteine synthetase, the rate limiting enzyme of glutathione synthesis, by chemoprotective plant-derived food and beverage components in the human hepatoma cell line HepG2. Nutr. Cancer 2003, 45, 74–83. [Google Scholar] [CrossRef]
- Jeon, S.E.; Choi-Kwon, S.; Park, K.A.; Lee, H.J.; Park, M.S.; Lee, J.H.; Kwon, S.B.; Park, K.C. Dietary supplementation of (+)-catechin protects against UVB-induced skin damage by modulating antioxidant enzyme activities. Photodermatol. Photoimmunol. Photomed. 2003, 19, 235–241. [Google Scholar] [CrossRef]
- Ramirez-Mares, M.V.; de Mejia, E.G. Comparative study of the antioxidant effect of ardisin and epigallocatechin gallate in rat hepatocytes exposed to benomyl and 1-nitropyrene. Food Chem. Toxicol. 2003, 41, 1527–1535. [Google Scholar] [CrossRef]
- Tran, P.O.; Parker, S.M.; LeRoy, E.; Franklin, C.C.; Kavanagh, T.J.; Zhang, T.; Zhou, H.; Vliet, P.; Oseid, E.; Harmon, J.S.; et al. Adenoviral overexpression of the glutamylcysteine ligase catalytic subunit protects pancreatic islets against oxidative stress. J. Biol. Chem. 2004, 279, 53988–53993. [Google Scholar] [CrossRef]
- Botta, D.; Franklin, C.C.; White, C.C.; Krejsa, C.M.; Dabrowski, M.J.; Pierce, R.H.; Fausto, N.; Kavanagh, T.J. Glutamate-cysteine ligase attenuates TNF-induced mitochondrial injury and apoptosis. Free Radic. Biol. Med. 2004, 37, 632–642. [Google Scholar] [CrossRef]
- Matzinger, M.; Fischhuber, K.; Heiss, E.H. Activation of Nrf2 signaling by natural products-can it alleviate diabetes? Biotechnol. Adv. 2018, 36, 1738–1767. [Google Scholar] [CrossRef]
- Nezu, M.; Suzuki, N. Roles of Nrf2 in Protecting the Kidney from Oxidative Damage. Int. J. Mol. Sci. 2020, 21, 2951. [Google Scholar] [CrossRef] [PubMed]
- Surh, Y.J.; Kundu, J.K.; Na, H.K.; Lee, J.S. Redox-sensitive transcription factors as prime targets for chemoprevention with anti-inflammatory and antioxidative phytochemicals. J. Nutr. 2005, 135, 2993S–3001S. [Google Scholar] [CrossRef]
- Moskaug, J.O.; Carlsen, H.; Myhrstad, M.C.; Blomhoff, R. Polyphenols and glutathione synthesis regulation. Am. J. Clin. Nutr. 2005, 81, 277S–283S. [Google Scholar] [CrossRef] [PubMed]
- Myhrstad, M.C.; Carlsen, H.; Nordstrom, O.; Blomhoff, R.; Moskaug, J.O. Flavonoids increase the intracellular glutathione level by transactivation of the gamma-glutamylcysteine synthetase catalytical subunit promoter. Free Radic. Biol. Med. 2002, 32, 386–393. [Google Scholar] [CrossRef]
- Surh, Y.J.; Kundu, J.K.; Na, H.K. Nrf2 as a master redox switch in turning on the cellular signaling involved in the induction of cytoprotective genes by some chemopreventive phytochemicals. Planta Med. 2008, 74, 1526–1539. [Google Scholar] [CrossRef]
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Gokila Vani, M.; Kumar, K.J.; Liao, J.W.; Chien, S.C.; Mau, J.L.; Chiang, S.S.; Lin, C.C.; Kuo, Y.H.; Wang, S.Y. Antcin C from Antrodia cinnamomea Protects Liver Cells Against Free Radical-Induced Oxidative Stress and Apoptosis In Vitro and In Vivo through Nrf2-Dependent Mechanism. Evid. Based Complement. Altern. Med. 2013, 2013, 296082. [Google Scholar] [CrossRef]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Regulation of the antioxidant response element by protein kinase C-mediated phosphorylation of NF-E2-related factor 2. Proc. Natl. Acad. Sci. USA 2000, 97, 12475–12480. [Google Scholar] [CrossRef]
- Huang, H.C.; Nguyen, T.; Pickett, C.B. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002, 277, 42769–42774. [Google Scholar] [CrossRef]
- Xu, C.; Yuan, X.; Pan, Z.; Shen, G.; Kim, J.H.; Yu, S.; Khor, T.O.; Li, W.; Ma, J.; Kong, A.N. Mechanism of action of isothiocyanates: The induction of ARE-regulated genes is associated with activation of ERK and JNK and the phosphorylation and nuclear translocation of Nrf2. Mol. Cancer Ther. 2006, 5, 1918–1926. [Google Scholar] [CrossRef]
- Cho, J.H.; Jeon, Y.J.; Park, S.M.; Shin, J.C.; Lee, T.H.; Jung, S.; Park, H.; Ryu, J.; Chen, H.; Dong, Z.; et al. Multifunctional effects of honokiol as an anti-inflammatory and anti-cancer drug in human oral squamous cancer cells and xenograft. Biomaterials 2015, 53, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Peng, S.; Li, X.; Yao, J.; Xu, J.; Fang, J. Honokiol Alleviates Oxidative Stress-Induced Neurotoxicity via Activation of Nrf2. ACS Chem. Neurosci. 2018, 9, 3108–3116. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.R.; Park, K.K.; Chun, K.S.; Chung, W.Y. Honokiol inhibits the progression of collagen-induced arthritis by reducing levels of pro-inflammatory cytokines and matrix metalloproteinases and blocking oxidative tissue damage. J. Pharmacol. Sci. 2010, 114, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, J.; Jung, E.; Park, Y.; Kim, K.; Park, B.; Jung, K.; Park, E.; Kim, J.; Park, D. In vitro antibacterial and anti-inflammatory effects of honokiol and magnolol against Propionibacterium sp. Eur. J. Pharmacol. 2004, 496, 189–195. [Google Scholar] [CrossRef]
- Sulakhiya, K.; Kumar, P.; Gurjar, S.S.; Barua, C.C.; Hazarika, N.K. Beneficial effect of honokiol on lipopolysaccharide induced anxiety-like behavior and liver damage in mice. Pharm. Biochem. Behav. 2015, 132, 79–87. [Google Scholar] [CrossRef]
- Wu, F.; Yao, H.; Zheng, F.; Tang, S.; Lin, X.; Li, L.; Zhou, J.; Li, H. Protective effects of honokiol against oxidative stress-induced apoptotic signaling in mouse podocytes treated with H2O2. Exp. Ther. Med. 2018, 16, 1278–1284. [Google Scholar] [CrossRef]


| Genes | Primers (5′-3′) |
|---|---|
| Human Gclc | Forward: CCCAAACCATCCTACCCTTT |
| Reverse: CATGTTGGCCTCAACTGTATTG | |
| Human Gclm | Forward: CCTGTTCAGTCCTTGGAGTTG |
| Reverse: CCTCCCAGTAAGGCTGTAAATG | |
| Human Gss | Forward: CCTGTTCAGTCCTTGGAGTTG |
| Reverse: CCTCCCAGTAAGGCTGTAAATG | |
| Human GAPDH | Forward: GGTGTGAACCATGAGAAGTATGA |
| Reverse: GAGTCCTTCCACGATACCAAAG | |
| Mouse Gclc | Forward: CATCGACCTGACCATCGATAAG |
| Reverse: AGGGTGAGTGGGTCTCTAATAA | |
| Mouse Gclm | Forward: GTATCAGTGGGCACAGGTAAA |
| Reverse: CGGGTCATTGTGAGTCAGTAG | |
| Mouse Gss | Forward: ACCTTTGCTGGCCTCTATTC |
| Reverse: TTGTTACCTCCACCCTCTCT | |
| Mouse GAPDH | Forward: GTGGCAAAGTGGAGATTGTTG |
| Reverse: TTGACTGTGCCGTTGAATTTG |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, E.J.; Dusabimana, T.; Je, J.; Jeong, K.; Yun, S.P.; Kim, H.J.; Kim, H.; Park, S.W. Honokiol Protects the Kidney from Renal Ischemia and Reperfusion Injury by Upregulating the Glutathione Biosynthetic Enzymes. Biomedicines 2020, 8, 352. https://doi.org/10.3390/biomedicines8090352
Park EJ, Dusabimana T, Je J, Jeong K, Yun SP, Kim HJ, Kim H, Park SW. Honokiol Protects the Kidney from Renal Ischemia and Reperfusion Injury by Upregulating the Glutathione Biosynthetic Enzymes. Biomedicines. 2020; 8(9):352. https://doi.org/10.3390/biomedicines8090352
Chicago/Turabian StylePark, Eun Jung, Theodomir Dusabimana, Jihyun Je, Kyuho Jeong, Seung Pil Yun, Hye Jung Kim, Hwajin Kim, and Sang Won Park. 2020. "Honokiol Protects the Kidney from Renal Ischemia and Reperfusion Injury by Upregulating the Glutathione Biosynthetic Enzymes" Biomedicines 8, no. 9: 352. https://doi.org/10.3390/biomedicines8090352
APA StylePark, E. J., Dusabimana, T., Je, J., Jeong, K., Yun, S. P., Kim, H. J., Kim, H., & Park, S. W. (2020). Honokiol Protects the Kidney from Renal Ischemia and Reperfusion Injury by Upregulating the Glutathione Biosynthetic Enzymes. Biomedicines, 8(9), 352. https://doi.org/10.3390/biomedicines8090352







