A Novel Smad7 Genetic Variant Mapping on the Genomic Region Targeted by Mongersen Is Associated with Crohn’s Disease
Abstract
1. Introduction
2. Experimental Section
2.1. Study Population
2.2. DNA Extraction and Genotyping
2.3. PBMCs Isolation and Culture
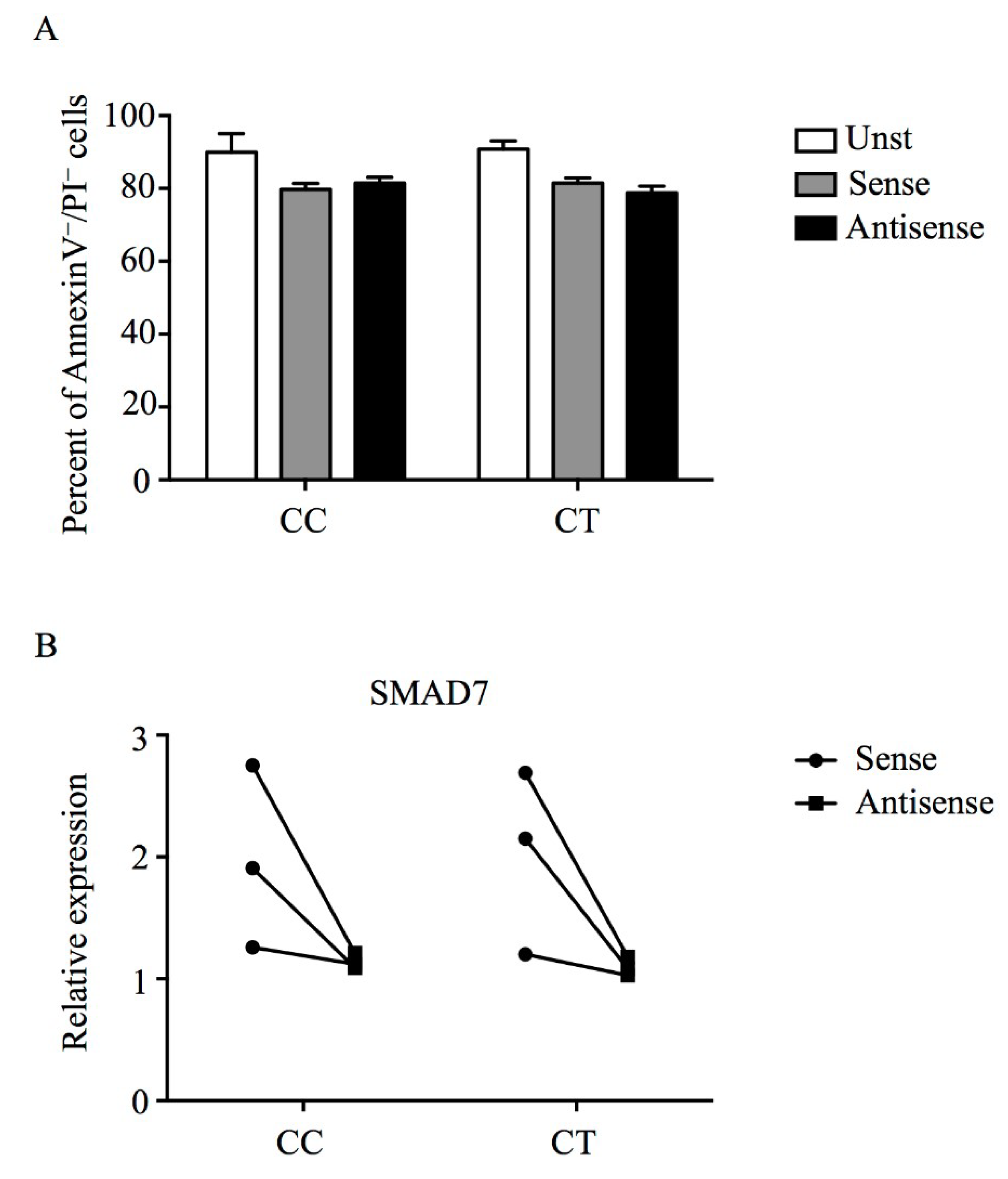
2.4. Assessment of Cell Death
2.5. RNA Extraction and Real-Time PCR
2.6. Statistical Analysis
3. Results
3.1. Frequency of Variant Allele of rs144204026 in CD
3.2. Relationship between Frequency of the T Variant Allele and the Demographic/Clinical Characteristic of CD Patients
3.3. Mongersen Down-Regulates Smad7 in PBMCs of Individuals Carrying T Variant Allele
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cosnes, J.; Gower-Rousseau, C.; Seksik, P.; Cortot, A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011, 140, 1785–1794. [Google Scholar] [CrossRef] [PubMed]
- Digby-Bell, J.L.; Atreya, R.; Monteleone, G.; Powell, N. Interrogating host immunity to predict treatment response in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, G.; Del Vecchio Blanco, G.; Monteleone, I.; Fina, D.; Caruso, R.; Gioia, V.; Ballerini, S.; Federici, G.; Bernardini, S.; Pallone, F.; et al. Post-transcriptional regulation of Smad7 in the gut of patients with inflammatory bowel disease. Gastroenterology 2005, 129, 1420–1429. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, G.; Kumberova, A.; Croft, N.M.; McKenzie, C.; Steer, H.W.; MacDonald, T.T. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J. Clin. Investig. 2001, 108, 601–609. [Google Scholar] [CrossRef]
- Monteleone, G.; Mann, J.; Monteleone, I.; Vavassori, P.; Bremner, R.; Fantini, M.; Del Vecchio Blanco, G.; Tersigni, R.; Alessandroni, L.; Mann, D.; et al. A failure of transforming growth factor-beta1 negative regulation maintains sustained NF-kappaB activation in gut inflammation. J. Biol. Chem. 2004, 279, 3925–3932. [Google Scholar] [CrossRef]
- Marafini, I.; Troncone, E.; Salvatori, S.; Monteleone, G. TGF-beta activity restoration and phosphodiesterase 4 inhibition as therapeutic options for inflammatory bowel diseases. Pharmacol. Res. 2020, 155, 104757. [Google Scholar] [CrossRef]
- Sedda, S.; Marafini, I.; Dinallo, V.; Di Fusco, D.; Monteleone, G. The TGF-beta/Smad System in IBD Pathogenesis. Inflamm. Bowel. Dis. 2015, 21, 2921–2925. [Google Scholar] [CrossRef] [PubMed]
- Boirivant, M.; Pallone, F.; Di Giacinto, C.; Fina, D.; Monteleone, I.; Marinaro, M.; Caruso, R.; Colantoni, A.; Palmieri, G.; Sanchez, M.; et al. Inhibition of Smad7 with a specific antisense oligonucleotide facilitates TGF-beta1-mediated suppression of colitis. Gastroenterology 2006, 131, 1786–1798. [Google Scholar] [CrossRef] [PubMed]
- Feagan, B.G.; Sands, B.E.; Rossiter, G.; Li, X.; Usiskin, K.; Zhan, X.; Colombel, J.F. Effects of Mongersen (GED-0301) on Endoscopic and Clinical Outcomes in Patients With Active Crohn’s Disease. Gastroenterology 2018, 154, 61.e66–64.e66. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, G.; Fantini, M.C.; Onali, S.; Zorzi, F.; Sancesario, G.; Bernardini, S.; Calabrese, E.; Viti, F.; Monteleone, I.; Biancone, L.; et al. Phase I clinical trial of Smad7 knockdown using antisense oligonucleotide in patients with active Crohn’s disease. Mol. Ther. 2012, 20, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Monteleone, G.; Neurath, M.F.; Ardizzone, S.; Di Sabatino, A.; Fantini, M.C.; Castiglione, F.; Scribano, M.L.; Armuzzi, A.; Caprioli, F.; Sturniolo, G.C.; et al. Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn’s disease. N. Engl. J. Med. 2015, 372, 1104–1113. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E.; Feagan, B.G.; Sandborn, W.J.; Schreiber, S.; Peyrin-Biroulet, L.; Frederic Colombel, J.; Rossiter, G.; Usiskin, K.; Ather, S.; Zhan, X.; et al. Mongersen (GED-0301) for Active Crohn’s Disease: Results of a Phase 3 Study. Am. J. Gastroenterol. 2020, 115, 738–745. [Google Scholar] [CrossRef] [PubMed]
- Di Fusco, D.; Dinallo, V.; Marafini, I.; Figliuzzi, M.M.; Romano, B.; Monteleone, G. Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease. Front. Pharmacol. 2019, 10, 305. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.W.; Matteucci, M.D.; Lewis, J.G.; Gutierrez, A.J.; Moulds, C.; Froehler, B.C. Antisense gene inhibition by oligonucleotides containing C-5 propyne pyrimidines. Science 1993, 260, 1510–1513. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.K.; Maxwell, J.E.; Qian, Q.; Bellizzi, A.M.; Braun, T.A.; Iannettoni, M.D.; Darbro, B.W.; Howe, J.R. Esophageal cancer in a family with hamartomatous tumors and germline PTEN frameshift and SMAD7 missense mutations. Cancer Genet. 2015, 208, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.A.; Boucher, G.; Lees, C.W.; Franke, A.; D’Amato, M.; Taylor, K.D.; Lee, J.C.; Goyette, P.; Imielinski, M.; Latiano, A.; et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011, 43, 246–252. [Google Scholar] [CrossRef]
- Franke, A.; McGovern, D.P.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; Roberts, R.; et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 2010, 42, 1118–1125. [Google Scholar] [CrossRef]
- Rufini, S.; Ciccacci, C.; Di Fusco, D.; Ruffa, A.; Pallone, F.; Novelli, G.; Biancone, L.; Borgiani, P. Autophagy and inflammatory bowel disease: Association between variants of the autophagy-related IRGM gene and susceptibility to Crohn’s disease. Dig. Liver. Dis. 2015, 47, 744–750. [Google Scholar] [CrossRef]
- Tam, V.; Patel, N.; Turcotte, M.; Bosse, Y.; Pare, G.; Meyre, D. Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 2019, 20, 467–484. [Google Scholar] [CrossRef]
- Cirulli, E.T.; Goldstein, D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 2010, 11, 415–425. [Google Scholar] [CrossRef]
- Bewtra, M.; Lichtenstein, G.R. Mongersen and SMAD-7 Inhibition, Not a Lucky 7 for Patients With IBD: When Trial Design Is as Important as Disease Therapy. Am. J. Gastroenterol. 2020, 115, 687–688. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.Z.; van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.C.; Jostins, L.; Shah, T.; et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef] [PubMed]
| Diagnosis | CD_1 N (%) | UC_1 N (%) | CD_2 N (%) | UC_2 N (%) | CD_Tot N (%) | UC_Tot N (%) |
|---|---|---|---|---|---|---|
| N | 235 | 198 | 122 | 142 | 357 | 340 |
| Gender Female | 123 (52.3) | 86 (43.4) | 49 (40.2) | 49 (34.5) | 172 (48.2) | 135 (39.7) |
| Mean Age | 48 | 55 | 51 | 51 | 50 | 52 |
| Smoking habit | ||||||
| Current | 64 (27.2) | 24 (12.1) | 18 (14.8) | 2 (1.4) | 82 (23) | 26 (7.6) |
| Former | 47 (20) | 44 (22.2) | 13 (10.7) | 15 (10.6) | 60 (16.8) | 59 (17.4) |
| Family history of IBD | 48 (20.4) | 27 (13.6) | 7 (5.7) | 3 (2.1) | 55 (15.4) | 30 (8.8) |
| CD location * | ||||||
| Ileum | 136 (57.9) | - | 39 (32) | - | 175 (49) | - |
| Colon | 15 (6.4) | - | 19 (15.6) | - | 34 (9.5) | - |
| Ileo-colon | 64 (27.2) | - | 64 (52.5) | - | 128 (35.9) | - |
| Isolated upper disease | 5 (2.1) | - | 5 (1.4) | - | ||
| CD behaviour ** | ||||||
| Non-structuring/ Non-penetrating | 63 (26.8) | - | 60 (49.2) | - | 123 (34.5) | - |
| Stricturing | 100 (42.6) | - | 26 (21.3) | - | 126 (35.3) | - |
| Penetrating | 57 (24.3) | - | 23 (18.9) | - | 80 (22.4) | - |
| Perianal disease | 73 (31.1) | - | 22 (18.0) | - | 95 (26.6) | - |
| Colitis extent *** | ||||||
| Distal | - | 95 (47.9) | - | 49 (34.5) | - | 144 (42.3) |
| Left sided | - | 28 (14.1) | - | 38 (26.7) | - | 66 (19.4) |
| Extensive | - | 60 (30.3) | - | 55 (38.7) | - | 115 (33.8) |
| Controls N = 363 (%) | CD_1 N = 235 (%) | UC_1 N = 198 (%) | CD_2 N = 122 (%) | UC_2 N = 142 (%) | CD_Tot N = 357 (%) | UC_Tot N = 340 (%) | |
|---|---|---|---|---|---|---|---|
| CC | 357 (98.3) | 224 (95.3) | 191 (96.5) | 114 (93.4) | 137 (96.5) | 338 (94.7) | 328 (96.5) |
| CT | 6 (1.7) | 11 (4.7) | 7 (3.5) | 8 (6.6) | 5 (3.5) | 19 (5.3) | 12 (3.5) |
| TT | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| p = 0.029 | p = 0.15 | p = 0.01 | p = 0.19 | p = 0.007 | p = 0.11 |
| Controls N = 726 (%) | CD_Tot N = 714 (%) | UC_Tot N = 680 (%) | |
|---|---|---|---|
| C | 720 (99.2) | 695 (97.3) | 668 (98.2) |
| T | 6 (0.8) | 19 (2.7) | 12 (1.7) |
| p = 0.008 OR = 3.28 (1.3–8.3) | p = 0.12 OR = 2.16 (0.8–5.8) |
| Diagnosis | CC N = 338 (%) | CT N = 19 (%) | p Value |
|---|---|---|---|
| Gender Female | 163 (48.2) | 10 (55.5) | p = 0.54 |
| Mean Age | 50 | 50 | |
| Smoking habit | |||
| Current | 78 (23.1) | 4 (22.2) | p = 0.93 |
| Former | 57 (16.9) | 3 (16.7) | p = 0.98 |
| Family history of IBD | 52 (15.4) | 3 (16.7) | |
| CD location * | |||
| Ileum | 166 (49) | 9 (50) | p = 0.94 |
| Colon | 33 (9.5) | 1 (5.6) | p = 0.55 |
| Ileo-colon | 120 (35.9) | 8 (44.4) | p = 0.44 |
| Isolated upper disease | 5 (1.4) | - | |
| CD behaviour ** | |||
| Non structuring/ Non penetrating | 116 (34.5) | 7 (38.9) | p = 0.69 |
| Stricturing | 121 (35.3) | 5 (27.8) | p = 0.48 |
| Penetrating | 74 (22.4) | 6 (33.3) | p = 0.25 |
| Perianal disease | 92 (27.2) | 3 (16.7) | p = 0.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Fusco, D.; Marafini, I.; Stolfi, C.; Troncone, E.; Onali, S.; Lolli, E.; Caprioli, F.; Mazza, S.; Raffaella, C.; Manzo, L.; et al. A Novel Smad7 Genetic Variant Mapping on the Genomic Region Targeted by Mongersen Is Associated with Crohn’s Disease. Biomedicines 2020, 8, 234. https://doi.org/10.3390/biomedicines8080234
Di Fusco D, Marafini I, Stolfi C, Troncone E, Onali S, Lolli E, Caprioli F, Mazza S, Raffaella C, Manzo L, et al. A Novel Smad7 Genetic Variant Mapping on the Genomic Region Targeted by Mongersen Is Associated with Crohn’s Disease. Biomedicines. 2020; 8(8):234. https://doi.org/10.3390/biomedicines8080234
Chicago/Turabian StyleDi Fusco, Davide, Irene Marafini, Carmine Stolfi, Edoardo Troncone, Sara Onali, Elisabetta Lolli, Flavio Caprioli, Stefano Mazza, Cascella Raffaella, Laura Manzo, and et al. 2020. "A Novel Smad7 Genetic Variant Mapping on the Genomic Region Targeted by Mongersen Is Associated with Crohn’s Disease" Biomedicines 8, no. 8: 234. https://doi.org/10.3390/biomedicines8080234
APA StyleDi Fusco, D., Marafini, I., Stolfi, C., Troncone, E., Onali, S., Lolli, E., Caprioli, F., Mazza, S., Raffaella, C., Manzo, L., Borgiani, P., Giuffrida, P., Di Sabatino, A., Monteleone, I., & Monteleone, G. (2020). A Novel Smad7 Genetic Variant Mapping on the Genomic Region Targeted by Mongersen Is Associated with Crohn’s Disease. Biomedicines, 8(8), 234. https://doi.org/10.3390/biomedicines8080234






