In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. DNA Isolation from Cell Pellets
2.3. DNA Methylation Arrays in Cell Culture
2.4. Evaluation of the Cell Culture Array Results
2.5. Comparison of In Vitro and In Vivo Array Data
2.6. In Vivo Confirmation in Additional Urine Specimens
2.7. Preparation of DNA from Urine
2.8. Quantitative Mass Spectrometry of DNA Methylation
2.9. Statistics and Modeling
3. Results and Discussion
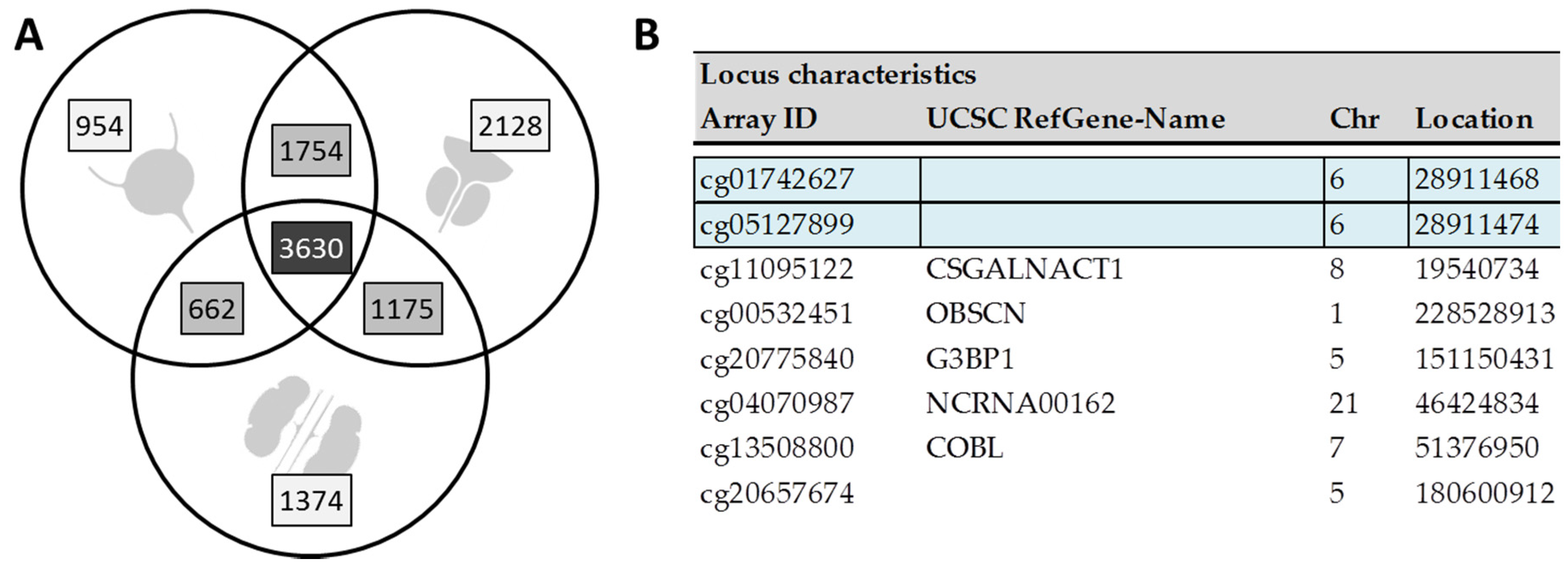
3.1. Cell Line Screening Reveals Large Overlap of Differentially Methylated Sites between Cancer Types
3.2. Few Targets Are Specific for Urothelial Cancer Cell Lines Only
3.3. Comparison with Previous In Vivo Data Identifies Eight Targets of Potential In Vivo Diagnostic Relevance
3.4. DNA Methylation Is Significantly Increased in Urine of UC Patients
3.5. BLSP Methylation Is Largely Independent on Age, Stage and Grade in Healthy Controls and/or UC Patients
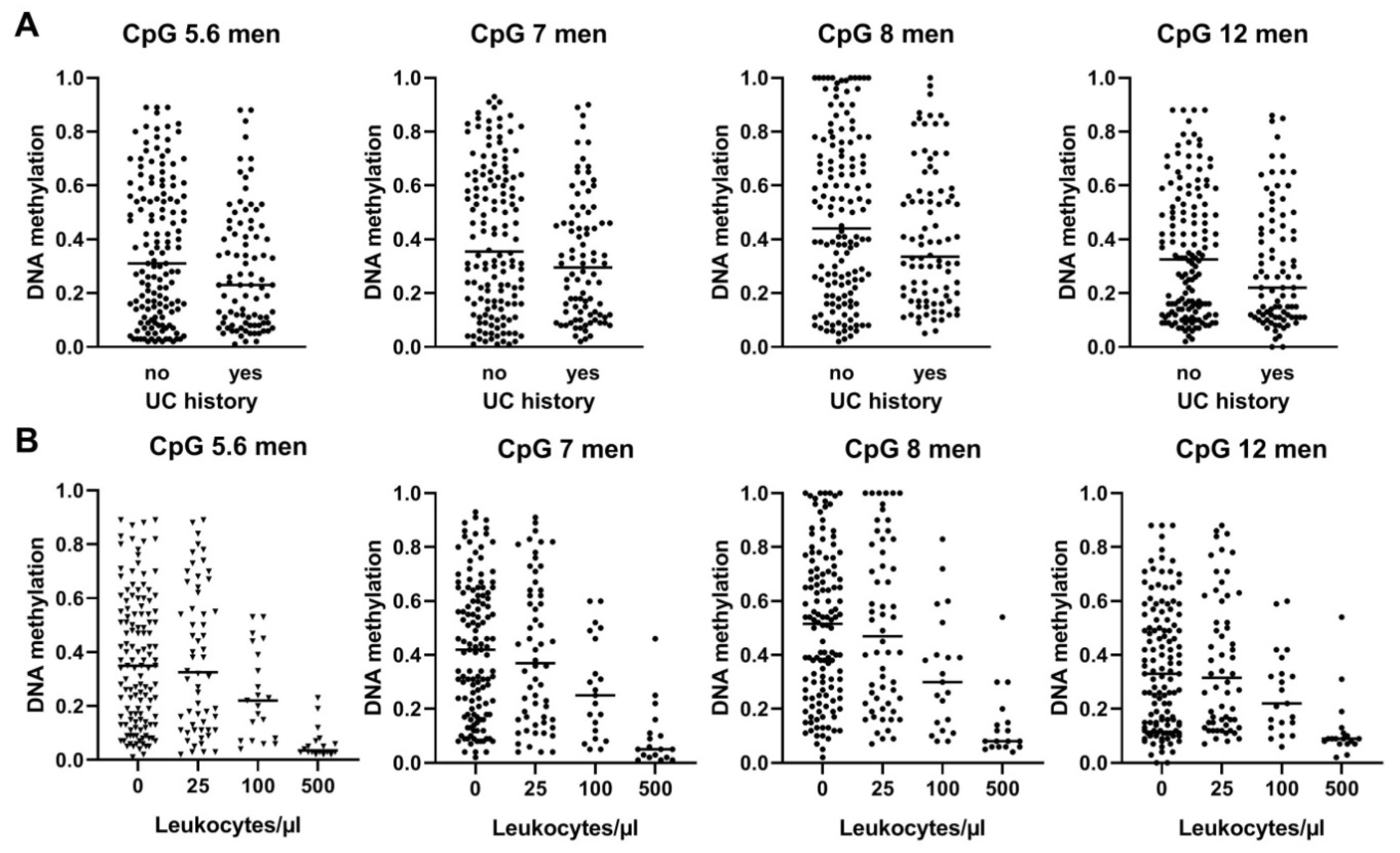
3.6. BLSP Methylation Is also Influenced by UC History and by Leukocyte Counts in Male UC Patients
3.7. The Methylation of the BLSP Amplicon in Urine Is of Diagnostic Relevance in Men
3.8. The Methylation of the BLSP Amplicon in Urine Is of no Diagnostic Use in Women
3.9. Retrospective Analyses of Primary Urothelial Cells Reveal Potential Pre-Malignant and Age-Dependent Methylation Patterns
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Shin, H.R.; Bray, F.; Forman, D.; Mathers, C.; Parkin, D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 2010, 127, 2893–2917. [Google Scholar] [CrossRef] [PubMed]
- Kamat, A.M.; Hahn, N.M.; Efstathiou, J.A.; Lerner, S.P.; Malström, P.-U.; Choi, W.; Guo, C.C.; Lotan, Y.; Kassouf, W. Bladder cancer. Lancet 2016, 388, 2796–2810. [Google Scholar] [CrossRef]
- Burke, D.M.; Shackley, D.C.; O’Reilly, P.H. The community-based morbidity of flexible cystoscopy. BJU Int. 2002, 89, 347–349. [Google Scholar] [CrossRef] [PubMed]
- Koo, K.; Zubkoff, L.; Sirovich, B.E.; Goodney, P.P.; Robertson, D.J.; Seigne, J.D.; Schroeck, F.R. The burden of cystoscopic bladder cancer surveillance: Anxiety, discomfort, and patient preferences for decision making. Urology 2017, 108, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.K.; Lind, G.E.; Guldberg, P.; Dahl, C. DNA-methylation-based detection of urological cancer in urine: Overview of biomarkers and considerations on biomarker design, source of DNA, and detection technologies. Int. J. Mol. Sci. 2019, 20, 2657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandimalla, R.; van Tilborg, A.; Zwarthoff, E.C. DNA methylation-based biomarkers in bladder cancer. Nat. Rev. Urol. 2013, 10, 326–335. [Google Scholar] [CrossRef]
- Schulz, W.A.; Goering, W. DNA methylation in urothelial carcinoma. Epigenomics 2016, 8, 1415–1428. [Google Scholar] [CrossRef]
- Van der Aa, M.N.M.; Steyerberg, E.W.; Bangma, C.; van Rhijn, B.W.G.; Zwarthoff, E.C.; van der Kwast, T.H. Cystoscopy revisited as the gold standard for detecting bladder cancer recurrence: Diagnostic review bias in the randomized, prospective CEFUB trial. J. Urol. 2010, 183, 76–80. [Google Scholar] [CrossRef]
- Stasik, S.; Salomo, K.; Heberling, U.; Froehner, M.; Sommer, U.; Baretton, G.B.; Ehniger, G.; Wirth, M.P.; Thiede, C.; Fuessel, S. Evaluation of TERT promoter mutations in urinary cell-free DNA and sediment DNA for detection of bladder cancer. Clin. Biochem. 2019, 64, 60–63. [Google Scholar] [CrossRef]
- Ward, D.G.; Gordon, N.S.; Boucher, R.H.; Pirrie, S.J.; Baxter, L.; Ott, S.; Silcock, S.; Whalley, C.M.; Stockton, J.D.; Beggs, A.D.; et al. Targeted deep sequencing of urothelial bladder cancers and associated urinary DNA: A 23-gene panel with utility for non-invasive diagnosis and risk stratification. BJU Int. 2019, 124, 532–544. [Google Scholar] [CrossRef] [Green Version]
- Eskra, J.N.; Rabizadeh, D.; Pavlovich, C.P.; Catalona, W.J.; Luo, J. Approaches to urinary detection of prostate cancer. Prostate Cancer Prostatic Dis. 2019, 22, 362–381. [Google Scholar] [CrossRef]
- Heichman, K.A.; Warren, J.D. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin. Chem. Lab. Med. 2012, 50, 1707–1721. [Google Scholar] [CrossRef]
- Costa, V.L.; Henrique, R.; Danielsen, S.A.; Eknaes, M.; Patrício, P.; Morais, A.; Oliveira, J.; Lothe, R.A.; Teixeira, M.R.; Lind, G.E. TCF21 and PCDH17 methylation: An innovative panel of biomarkers for a simultaneous detection of urological cancers. Epigenetics 2011, 6, 1120–1130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, A.M.; Cairns, P. Epigenetics of kidney cancer and bladder cancer. Epigenomics 2011, 3, 19–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köhler, C.U.; Bonberg, N.; Ahrens, M.; Behrens, T.; Hovanec, J.; Eisenacher, M.; Noldus, J.; Deix, T.; Braun, K.; Gohlke, H.; et al. Noninvasive diagnosis of urothelial cancer in urine using DNA hypermethylation signatures—Gender matters. Int. J. Cancer 2019, 145, 2861–2872. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Kibbe, W.A.; Lin, S.M. lumi: A pipeline for processing Illumina microarray. Bioinformatics 2008, 24, 1547–1548. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Zhang, X.; Huang, C.C.; Jafari, N.; Kibbe, W.A.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [Green Version]
- Gentleman, R.; Carey, V.J.; Dudoit, S.; Huber, W.; Irizarry, R.A. Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: New York, NY, USA, 2005. [Google Scholar]
- Smyth, G.K.; Michaud, J.; Scott, H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 2005, 21, 2067–2075. [Google Scholar] [CrossRef]
- Lang, K.; Kahveci, S.; Bonberg, N.; Wichert, K.; Behrens, T.; Hovanec, J.; Roghmann, F.; Noldus, J.; Tam, Y.C.; Tannapfel, A.; et al. TGFBI protein is increased in the urine of patients with high-grade urothelial carcinomas, and promotes cell proliferation and migration. Int. J. Mol. Sci. 2019, 20, 4483. [Google Scholar] [CrossRef] [Green Version]
- Ehrich, M.; Nelson, M.R.; Stanssens, P.; Zabeau, M.; Liloglou, T.; Xinarianos, G.; Cantor, C.R.; Field, J.K.; van den Boom, D. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl. Acad. Sci. USA 2005, 102, 15785–15790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coolen, M.W.; Statham, A.L.; Gardiner-Garden, M.; Clark, S.J. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry. Critical evaluation and improvements. Nucleic Acids Res. 2007, 35, e119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Karnovsky, A.; Mahavisno, V.; Weymouth, T.; Pande, M.; Dolinoy, D.C.; Rozek, L.S.; Sartor, M.A. LRpath analysis reveals common pathways dysregulated via DNA methylation across cancer types. BMC Genom. 2012, 13, 526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, R.; Gore, J.L.; Buckley, D.; Fu, R.; Gustafson, K.; Griffin, J.C.; Grusing, S.; Selph, S. Urinary biomarkers for diagnosis of bladder cancer: A systematic review and meta-analysis. Ann. Intern. Med. 2015, 163, 922–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvatore, S.; Salvatore, S.; Cattoni, E.; Siesto, G.; Serati, M.; Sorice, P.; Torella, M. Urinary tract infections in women. Eur. J. Obs. Gynecol. Reprod. Biol. 2011, 156, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Pietrusinski, M.; Kepczynski, L.; Jedrzejczyk, A.; Borkowska, E.; Traczyk-Borszynska, M.; Constantinou, M.; Kaluzewski, B.; Borowiec, M. Detection of bladder cancer in urine sediments by a hypermethylation panel of selected tumor suppressor genes. Cancer Biomark. 2017, 18, 47–59. [Google Scholar] [CrossRef]
- Besaratinia, A.; Cockburn, M.; Tommasi, S. Alterations of DNA methylome in human bladder cancer. Epigenetics 2013, 8, 1013–1022. [Google Scholar] [CrossRef]




| Cells | Supplier | Age 1 | Origin and Characteristics [Medium, Additives and Kits] |
|---|---|---|---|
| RT4 | CLS 2 | 63 | Bladder transitional cell papilloma, G1-2, T1 [McCoy’s 5A, Pan-Biotec P04-06500, 10% FCS] |
| 5637 | DSMZ 3 | 68 | Bladder transitional cell papilloma, G2 [RPMI 1640, Pan-Biotec P04-18500, 10% FCS] |
| J82 | ATCC 4 | 58 | Bladder transitional cell carcinoma, G3 T3 [MEM Eagle, Pan Biotech P04-09500, 10% FCS] |
| LNCAP | DSMZ 3 | 50 | Prostate adenocarcinoma, lymph node metastasis, low metastatic potential [RPMI 1640, Pan Biotech P04-18500, 20% FCS] |
| DU-145 | DSMZ 3 | 69 | Prostate adenocarcinoma, brain metastasis, moderate metastatic potential [RPMI 1640, Pan Biotech P04-18500, 10% FCS] |
| PC-3 | DSMZ 3 | 62 | Prostate adenocarcinoma, bone metastasis, G4, high metastatic potential [Ham’s F12 Pan Biotech P04-15500/RPMI 1640, Pan Biotech P04-18500 1:1, 10% FCS] |
| RCC-KP | CLS 2 | 59 | Renal clear-cell carcinoma, G3, pT3b, M1 [RPMI 1640 Pan Biotech P04-18500, 10% FCS] |
| CAKI-2 | DSMZ 3 | 69 | Renal papillary carcinoma [McCoy’s 5A, Pan Biotec P04-06500, 10% FCS] |
| CAL-54 | DSMZ 3 | 75 | Renal clear-cell carcinoma, metastatic pleura effusion [DMEM. Pan Biotech P04-01550, 20% FCS+, 0.04 µg/mL hydrocortisone + 10 ng/mL EGF] |
| HBIEpC | PeloBiotech 5 | 69 | Bladder, primary epithelial cells [Epi growth medium, PeloBiotech PB 215-500, subculture Reagent Kit, PeloBiotech PB-090K] |
| HprEpC | PeloBiotech 5 | 69 | Prostate, primary epithelial cells [Epi growth medium, PeloBiotech PB 215-500, subculture Reagent Kit, PeloBiotech PB-090K] |
| HREpC | PromoCell 6 | 77 | Renal, primary epithelial cells [Renal epithelial cell growth medium 2, PromoCell C-26030, DetachKit C-41210] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Köhler, C.U.; Walter, M.; Lang, K.; Plöttner, S.; Roghmann, F.; Noldus, J.; Tannapfel, A.; Tam, Y.C.; Käfferlein, H.U.; Brüning, T. In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer. Biomedicines 2020, 8, 233. https://doi.org/10.3390/biomedicines8080233
Köhler CU, Walter M, Lang K, Plöttner S, Roghmann F, Noldus J, Tannapfel A, Tam YC, Käfferlein HU, Brüning T. In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer. Biomedicines. 2020; 8(8):233. https://doi.org/10.3390/biomedicines8080233
Chicago/Turabian StyleKöhler, Christina U., Michael Walter, Kerstin Lang, Sabine Plöttner, Florian Roghmann, Joachim Noldus, Andrea Tannapfel, Yu Chun Tam, Heiko U. Käfferlein, and Thomas Brüning. 2020. "In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer" Biomedicines 8, no. 8: 233. https://doi.org/10.3390/biomedicines8080233
APA StyleKöhler, C. U., Walter, M., Lang, K., Plöttner, S., Roghmann, F., Noldus, J., Tannapfel, A., Tam, Y. C., Käfferlein, H. U., & Brüning, T. (2020). In-Vitro Identification and In-Vivo Confirmation of DNA Methylation Biomarkers for Urothelial Cancer. Biomedicines, 8(8), 233. https://doi.org/10.3390/biomedicines8080233





