Hyaluronan Metabolism is Associated with DNA Repair Genes in Breast and Colorectal Cancer. Screening of Potential Progression Markers Using qPCR
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Sample Processing
2.2.1. Tumor and Non-Tumor Tissue
2.2.2. Blood Samples
2.3. Quantification of RNA by Real Time PCR
2.4. Immunohistochemical Analysis
2.4.1. Hematoxylin and Eosin Staining
2.4.2. HA Staining
2.4.3. ER, PR, HER2 and KI67 Staining
2.5. Plasma HA Analysis
2.6. Statistical Analysis
3. Results
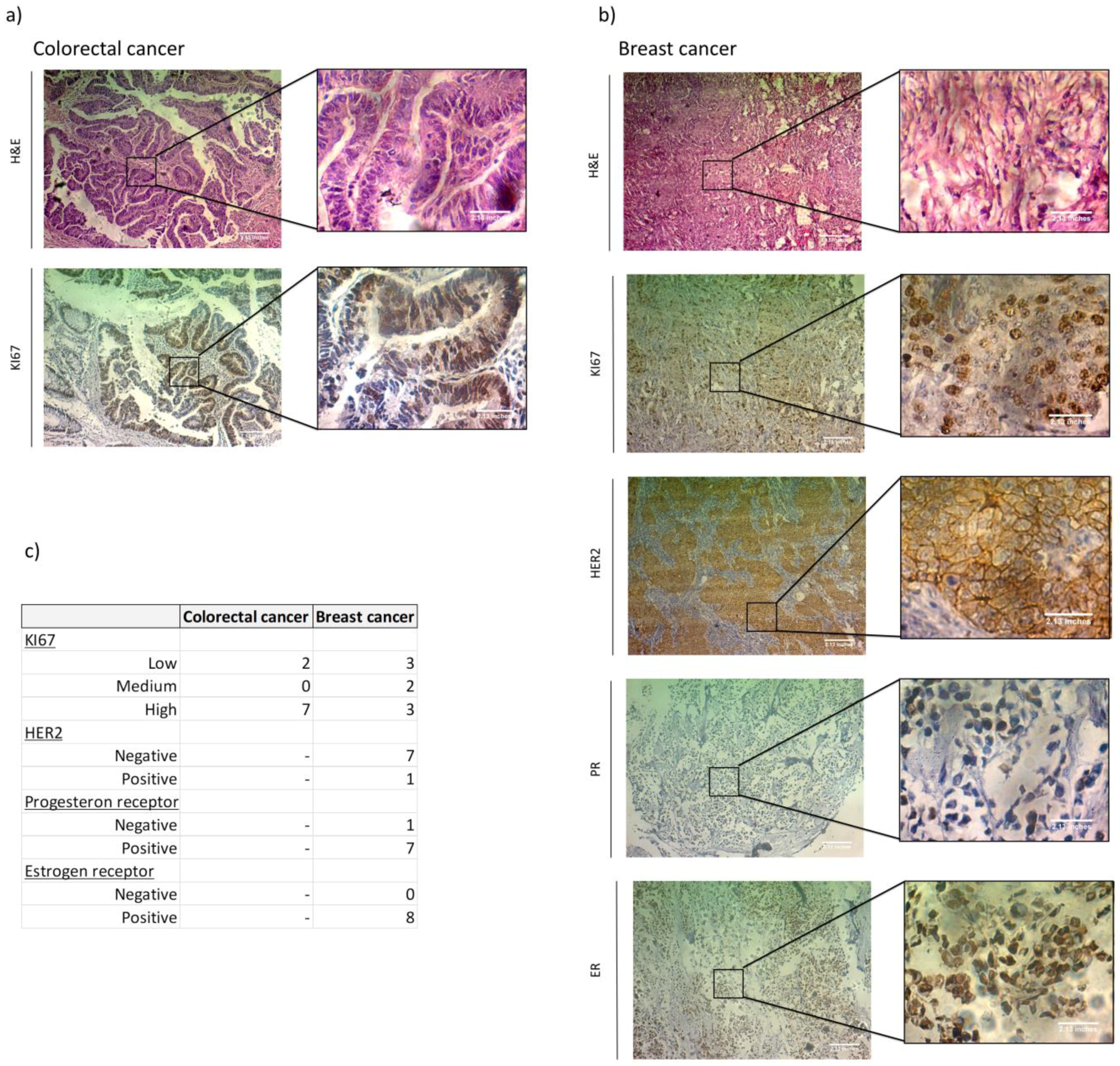
3.1. ER, PR, HER2 and KI67 Analysis
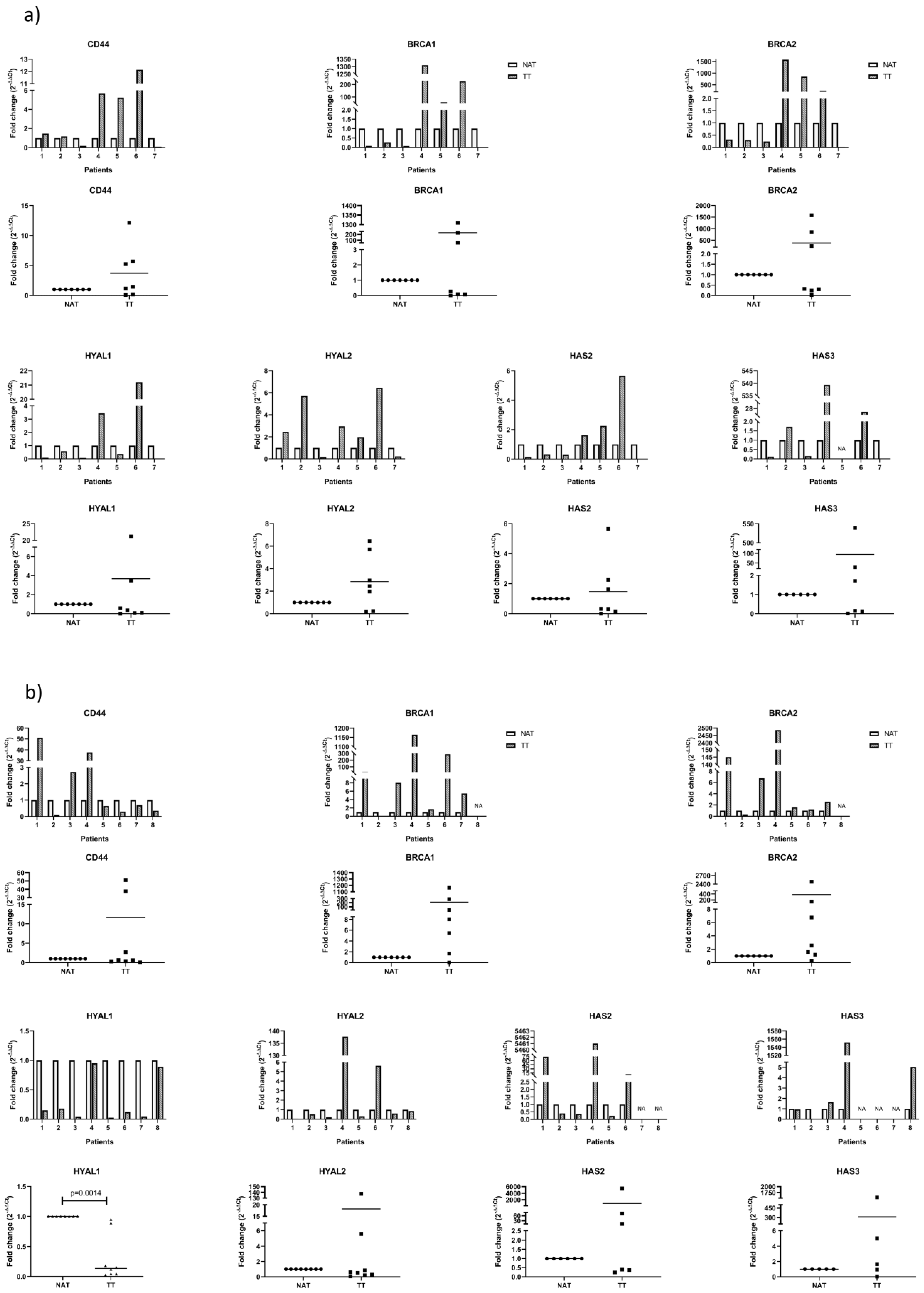
3.2. mRNA Analysis of Proposed Genes by qPCR
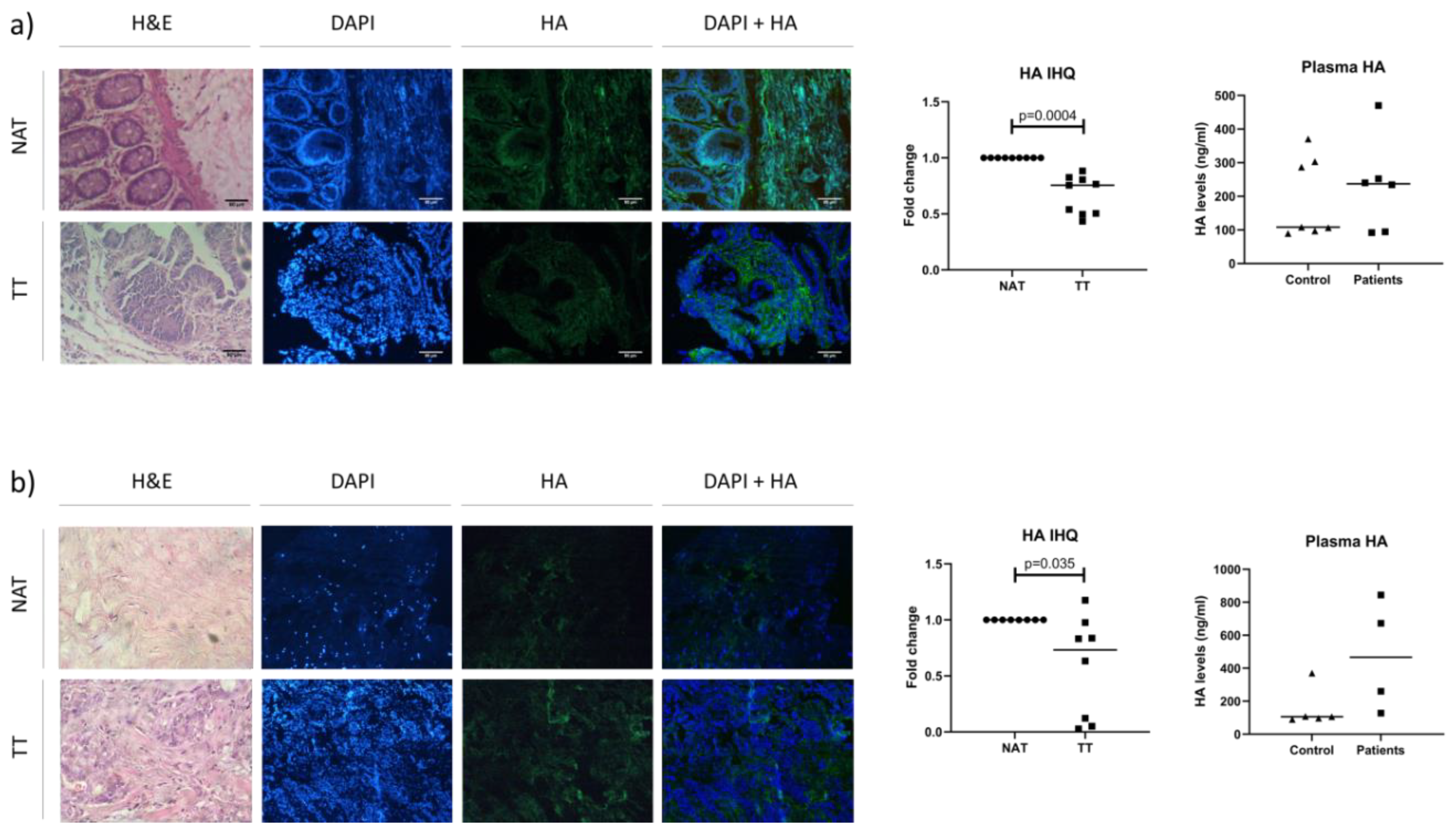
3.3. Analysis of HA Levels in Plasma and Tissue
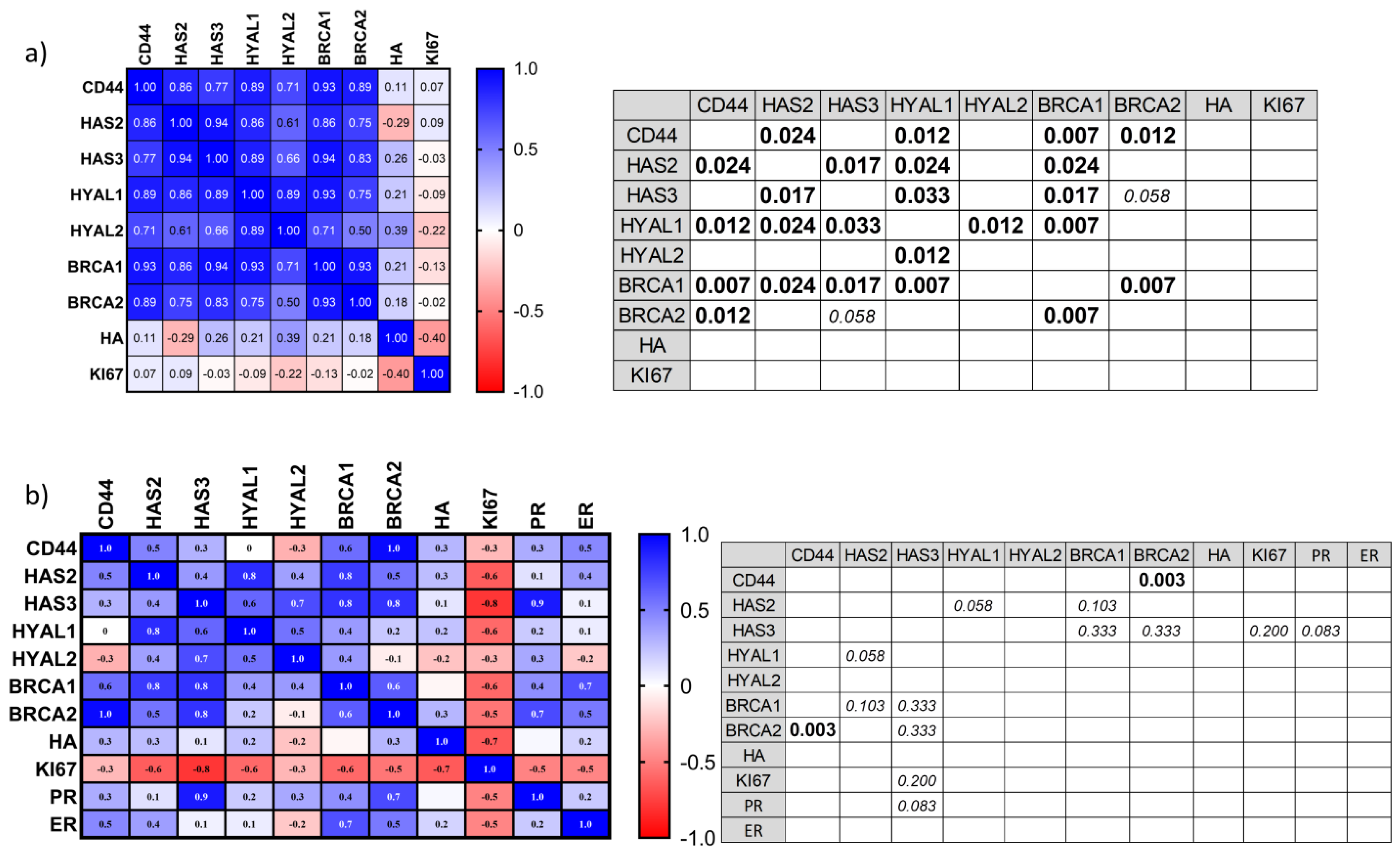
3.4. Correlation Analysis of Proposed Markers
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO. GLOBOCAN 2012: Estimated Incidence, Mortality and Prevalence Worldwide in 2012. 2012. Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx (accessed on 21 June 2019).
- Sierra, M.S.; Soerjomataram, I.; Antoni, S.; Laversanne, M.; Pineros, M.; De Vries, E.; Forman, D. Cancer patterns and trends in Central and South America. Cancer Epidemiol. 2016, 44, S23–S42. [Google Scholar] [CrossRef] [PubMed]
- Goss, P.E.; Lee, B.L. Epidemic rates of cancer incidence in Latin America. Nat. Rev. Clin. Oncol. 2013, 10, 304. [Google Scholar]
- Gress, D.M.; Edge, S.B.; Greene, F.L.; Washington, M.K.; Asare, E.A.; Brierley, J.D.; Byrd, D.R.; Compton, C.C.; Jessup, J.M.; Winchester, D.P.; et al. Principles of Cancer Staging; John Wiley & Sons: Hoboken, NJ, USA, 2017; pp. 3–30. [Google Scholar]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106, dju055. [Google Scholar] [CrossRef] [PubMed]
- Parise, C.A.; Caggiano, V. Risk of mortality of node-negative, ER/PR/HER2 breast cancer subtypes in T1, T2, and T3 tumors. Breast Cancer Res. Treat. 2017, 165, 743–750. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Pineda, E.; Adamo, B.; Galvan, P.; Fernandez, A.; Gaba, L.; Díez, M.; Viladot, M.; Arance, A.; Munoz, M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast 2015, 24, S26–S35. [Google Scholar] [CrossRef]
- Zhang, L.; Yu, Q.; Wu, X.C.; Hsieh, M.C.; Loch, M.; Chen, V.W.; Fontham, E.; Ferguson, T. Impact of chemotherapy relative dose intensity on cause-specific and overall survival for stage I-III breast cancer: ER+/PR+, HER2- vs. triple-negative. Breast Cancer Res. Treat. 2018, 169, 175–187. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, J.; Ding, M.; Xu, K.; Li, L.; Mao, L.; Zheng, J. Ki67 targeted strategies for cancer therapy. Clin. Transl. Oncol. 2018, 20, 570–575. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Bauer, J.; Rothley, M.; Schmaus, A.; Quagliata, L.; Ehret, M.; Biskup, M.; Orian-Rousseau, V.; Jackson, D.G.; Pettis, R.J.; Harvey, A.; et al. TGFβ counteracts LYVE-1-mediated induction of lymphangiogenesis by small hyaluronan oligosaccharides. J. Mol. Med. (Berl.) 2018, 96, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Gurski, L.A.; Xu, X.; Labrada, L.N.; Nguyen, N.T.; Xiao, L.; Van Golen, K.L.; Jia, X.; Farach-Carson, M.C. Hyaluronan (HA) interacting proteins RHAMM and hyaluronidase impact prostate cancer cell behavior and invadopodia formation in 3D HA-based hydrogels. PLoS ONE 2012, 7, e50075. [Google Scholar] [CrossRef] [PubMed]
- Franklin, O.; Billing, O.; Ohlund, D.; Berglund, A.; Herdenberg, C.; Wang, W.; Hellman, U.; Sund, M. Novel prognostic markers within the CD44-stromal ligand network in pancreatic cancer. J. Pathol. Clin. Res. 2019, 5, 130–141. [Google Scholar] [CrossRef] [PubMed]
- Toole, B.P. Hyaluronan: From extracellular glue to pericellular cue. Nat. Rev. Cancer 2004, 4, 528–539. [Google Scholar] [CrossRef]
- Lokeshwar, V.B.; Mirza, S.; Jordan, A. Targeting hyaluronic acid family for cancer chemoprevention and therapy. Adv. Cancer Res. 2014, 123, 35–65. [Google Scholar] [PubMed]
- Al Faraj, A.; Shaik, A.S.; Al Sayed, B.; Halwani, R.; Al Jammaz, I. Specific targeting and noninvasive imaging of breast cancer stem cells using single-walled carbon nanotubes as novel multimodality nanoprobes. Nanomedicine 2016, 11, 31–46. [Google Scholar] [CrossRef]
- Zinzi, L.; Contino, M.; Cantore, M.; Capparelli, E.; Leopoldo, M.; Colabufo, N.A. ABC transporters in CSCs membranes as a novel target for treating tumor relapse. Front. Pharmacol. 2014, 5, 163. [Google Scholar] [CrossRef]
- Boregowda, R.K.; Appaiah, H.N.; Siddaiah, M.; Kumarswamy, S.B.; Sunila, S.; Thimmaiah, K.N.; Mortha, K.; Toole, B.; d Banerjee, S. Expression of hyaluronan in human tumor progression. J. Carcinog. 2006, 5, 2. [Google Scholar] [CrossRef]
- Ghatak, S.; Misra, S.; Toole, B.P. Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phosphoinositide 3-kinase/Akt cell survival pathway. J. Biol. Chem. 2002, 277, 38013–38020. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Cancer Susceptibility and the Functions of BRCA1 and BRCA2. Cell 2002, 108, 171–182. [Google Scholar] [CrossRef]
- Tsibulak, I.; Wieser, V.; Degasper, C.; Shivalingaiah, G.; Wenzel, S.; Sprung, S.; Lax, S.F.; Marth, C.; Fiegl, H.; Zeimet, A.G. BRCA1 and BRCA2 mRNA-expression prove to be of clinical impact in ovarian cancer. Br. J. Cancer 2018, 119, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Lv, Y.; Gan, Z.; Zhang, Y.; Han, X.; Xu, Z. Identification of key genes involved in the metastasis of clear cell renal cell carcinoma. Oncol. Lett. 2019, 17, 4321–4328. [Google Scholar] [CrossRef] [PubMed]
- George, S.H.; Shaw, P. BRCA and Early Events in the Development of Serous Ovarian Cancer. Front. Oncol. 2014, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Graves, M.; Macdonald, G.; Cipollone, J.; Mueller, C.R.; Roskelley, C.D. Microenvironmental regulation of BRCA1 gene expression by c-Jun and Fra2 in premalignant human ovarian surface epithelial cells. Mol. Cancer Res. 2013, 11, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Phelan, C.M.; Iqbal, J.; Lynch, H.T.; Lubinski, J.; Gronwald, J.; Moller, P.; Ghadirian, P.; Foulkes, W.D.; Armel, S.; Eisen, A.; et al. Incidence of colorectal cancer in BRCA1 and BRCA2 mutation carriers: Results from a follow-up study. Br. J. Cancer 2014, 110, 530–534. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.V.; Katabathina, V.S.; Bowerson, M.L.; Mityul, M.I.; Shetty, A.S.; Elsayes, K.M.; Balachandran, A.; Bhosale, P.R.; McCullough, A.E.; Menias, C.O. BRCA-associated Cancers: Role of Imaging in Screening, Diagnosis, and Management. Radiographics 2017, 37, 1005–1023. [Google Scholar] [CrossRef]
- Maxwell, C.A.; Benıtez, J.; Gomez-Baldo, L.; Osorio, A.; Bonifaci, N.; Fernandez-Ramires, R.; Costes, S.V.; Guino, E.; Chen, H.; Evans, G.J.R.; et al. Interplay between BRCA1 and RHAMM Regulates Epithelial Apicobasal Polarization and May Influence Risk of Breast Cancer. PLoS Biol. 2011, 9, e1001199. [Google Scholar] [CrossRef]
- Spinelli, F.M.; Vitale, D.L.; Icardi, A.; Caon, I.; Brandone, A.; Giannoni, P.; Saturno, V.; Passi, A.; García, M.; Sevic, I.; et al. Hyaluronan preconditioning of monocytes/macrophages affects their angiogenic behavior and regulation of TSG-6 expression in a tumor type-specific manner. FEBS J. 2019, 286, 3433–3449. [Google Scholar] [CrossRef]
- Piccioni, F.; Fiore, E.; Bayo, J.; Atorrasagasti, C.; Peixoto, E.; Rizzo, M.; Malvicini, M.; Tirado-González, I.; García, M.G.; Alaniz, L.; et al. 4-Methylumbelliferone inhibits hepatocellular carcinoma growth by decreasing IL-6 production and angiogenesis. Glycobiology 2015, 25, 825–835. [Google Scholar] [CrossRef]
- Onitilo, A.A.; Engel, J.M.; Greenlee, R.T.; Mukesh, B.N. Breast cancer subtypes based on ER/PR and Her2 expression: Comparison of clinicopathologic features and survival. Clin. Med. Res. 2009, 7, 4–13. [Google Scholar] [CrossRef]
- Kast, K.; Link, T.; Friedrich, K.; Petzold, A.; Niedostatek, A.; Schoffer, O.; Werner, C.; Klug, S.J.; Werner, A.; Gatzweiler, A.; et al. Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res. Treat. 2015, 150, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Schlfiter, C.; Duchrow, M.; Wohlenberg, C.; Becker, M.H.G.; Key, G.; Flad, H.-D.; Gerdes, J. The Cell Proliferation-associated antigen of antibody Ki-67: A very large, ubiquitous nuclear protein with numerous repeated elements, representing a new kind of cell cycle-maintaining proteins. J. Cell Biol. 1993, 123, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Nielsen, T.O.; A’hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in breast cancer: Recommendations from the International Ki67 in breast cancer working group. JNCI J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.-W.; Zhu, M.-G.; Zhang, Z.-Q.; Ye, F.-J.; Huang, W.-H.; Luo, X.-Z. Increased expression of Ki-67 is a poor prognostic marker for colorectal cancer patients: A meta analysis. BMC Cancer 2019, 19, 123. [Google Scholar] [CrossRef] [PubMed]
- Penault-Llorca, F.; Radosevic-Robin, N. Ki67 assessment in breast cancer: An update. Pathology 2017, 49, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Li, L.T.; Jiang, G.; Chen, Q.; Zheng, J.N. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol. Med. Rep. 2015, 11, 1566–1572. [Google Scholar] [CrossRef] [PubMed]
- Yerushalmi, R.; Woods, R.; Ravdin, P.M.; Hayes, M.M.; Gelmon, K.A. Ki67 in breast cancer: Prognostic and predictive potential. Lancet Oncol. 2010, 11, 174–183. [Google Scholar] [CrossRef]
- Itano, N.; Kimata, K. Mammalian hyaluronan synthases. IUBMB Life 2002, 54, 195–199. [Google Scholar] [CrossRef]
- Simpson, M.A.; Wilson, C.M.; Furcht, L.T.; Spicer, A.P.; Oegema, T.R.; Mccarthy, J.B. Manipulation of hyaluronan synthase expression in prostate adenocarcinoma cells alters pericellular matrix retention and adhesion to bone marrow endothelial cells. J. Biol. Chem. 2002, 277, 10050–10057. [Google Scholar] [CrossRef]
- Spinelli, F.; Vitale, D.; Sevic, I.; Alaniz, L. Hyaluronan in the tumor microenvironment. In Tumor Microenvironment; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Auvinen, P.; Rilla, K.; Tumelius, R.; Tammi, M.; Sironen, R.; Soini, Y.; Kosma, V.M.; Mannermaa, A.; Viikari, J.; Tammi, R. Hyaluronan synthases (HAS1-3) in stromal and malignant cells correlate with breast cancer grade and predict patient survival. Breast Cancer Res. Treat. 2014, 143, 277–286. [Google Scholar] [CrossRef]
- Rilla, K.; Oikari, S.; Jokela, T.A.; Hyttinen, J.M.; Karna, R.; Tammi, R.H.; Tammi, M.I. Hyaluronan synthase 1 (HAS1) requires higher cellular UDP-GlcNAc concentration than HAS2 and HAS3. J. Biol. Chem. 2013, 288, 5973–5983. [Google Scholar] [CrossRef] [PubMed]
- Tammi, M.I.; Oikari, S.; Pasonen-Seppanen, S.; Rilla, K.; Auvinen, P.; Tammi, R.H. Activated hyaluronan metabolism in the tumor matrix—Causes and consequences. Matrix Biol. 2019, 78–79, 147–164. [Google Scholar] [CrossRef]
- Nykopp, T.K.; Pasonen-Seppanen, S.; Tammi, M.I.; Tammi, R.H.; Kosma, V.M.; Anttila, M.; Sironen, R. Decreased hyaluronidase 1 expression is associated with early disease recurrence in human endometrial cancer. Gynecol. Oncol. 2015, 137, 152–159. [Google Scholar] [CrossRef] [PubMed]
- Nykopp, T.K.; Rilla, K.; Sironen, R.; Tammi, M.I.; Tammi, R.H.; Hämäläinen, K.; Heikkinen, A.M.; Komulainen, M.; Kosma, V.M.; Anttila, M. Expression of hyaluronan synthases (HAS1-3) and hyaluronidases (HYAL1-2) in serous ovarian carcinomas: Inverse correlation between HYAL1 and hyaluronan content. BMC Cancer 2009, 9, 143. [Google Scholar] [CrossRef]
- Tan, J.X.; Wang, X.Y.; Li, H.Y.; Su, X.L.; Wang, L.; Ran, L.; Zheng, K.; Ren, G.S. HYAL1 overexpression is correlated with the malignant behavior of human breast cancer. Int. J. Cancer 2011, 128, 1303–1315. [Google Scholar] [CrossRef] [PubMed]
- Fede, C.; Stecco, C.; Angelini, A.; Fan, C.; Belluzzi, E.; Pozzuoli, A.; Ruggieri, P.; De Caro, R. Variations in contents of hyaluronan in the peritumoral micro-environment of human chondrosarcoma. J. Orthop. Res. 2019, 37, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Stern, R. Devising a pathway for hyaluronan catabolism: Are we there yet? Glycobiology 2003, 13, 105R–115R. [Google Scholar] [CrossRef]
- Oikari, S.; Kettunen, T.; Tiainen, S.; Hayrinen, J.; Masarwah, A.; Sudah, M.; Sutela, A.; Vanninen, R.; Tammi, M.; Auvinen, P. UDP-sugar accumulation drives hyaluronan synthesis in breast cancer. Matrix Biol. 2018, 67, 63–74. [Google Scholar] [CrossRef]
- Twarock, S.; Reichert, C.; Peters, U.; Gorski, D.J.; Röck, K.; Fischer, J.W. Hyperglycaemia and aberrated insulin signalling stimulate tumour progression via induction of the extracellular matrix component hyaluronan. Int. J. Cancer 2017, 141, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Tavianatou, A.-G.; Piperigkou, Z.; Barbera, C.; Beninatto, R.; Masola, V.; Caon, I.; Onisto, M.; Franchi, M.; Galesso, D.; Karamanos, N.K. Molecular size-dependent specificity of hyaluronan on functional properties, morphology and matrix composition of mammary cancer cells. Matrix Biol. Plus 2019, 3, 100008. [Google Scholar] [CrossRef]
- Josefsson, A.; Adamo, H.; Hammarsten, P.; Granfors, T.; Stattin, P.; Egevad, L.; Laurent, A.E.; Wikström, P.; Bergh, A. Prostate cancer increases hyaluronan in surrounding nonmalignant stroma, and this response is associated with tumor growth and an unfavorable outcome. Am. J. Pathol. 2011, 179, 1961–1968. [Google Scholar] [CrossRef] [PubMed]
- Cowman, M.K.; Lee, H.G.; Schwertfeger, K.L.; Mccarthy, J.B.; Turley, E.A. The content and size of hyaluronan in biological fluids and tissues. Front. Immunol. 2015, 6, 261. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Xue, C.; Yu, Y.; Chen, J.; Chen, X.; Ren, F.; Ren, Z.; Cui, G.; Sun, R. CD44 is overexpressed and correlated with tumor progression in gallbladder cancer. Cancer Manag. Res. 2018, 10, 3857–3865. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.; Mcbride, A.; Yun, S.; Bhattacharjee, S.; Slack, M.; Martin, J.R.; Jeter, J.; Abraham, I. BRCA1 and BRCA2 gene mutations and colorectal cancer risk: Systematic review and meta-analysis. J. Natl. Cancer Inst. 2018, 110, 1178–1189. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, I.; Min, H.; Bang, Y.J.; Kim, W. Clinical significance of BRCA1 and BRCA2 mRNA and protein expression in patients with sporadic gastric cancer. Oncol. Lett. 2019, 17, 4383–4392. [Google Scholar] [CrossRef]
- Atkinson, R.L.; Yang, W.T.; Rosen, D.G.; Landis, M.D.; Wong, H.; Lewis, M.T.; Creighton, C.J.; Sexton, K.R.; Hilsenbeck, S.G.; Sahin, A.A.; et al. Cancer stem cell markers are enriched in normal tissue adjacent to triple negative breast cancer and inversely correlated with DNA repair deficiency. Breast Cancer Res. 2013, 15, R77. [Google Scholar] [CrossRef]
- Wang, X.; Stolla, M.; Ring, B.Z.; Yang, Q.; Laughlin, T.S.; Rothberg, P.G.; Skinner, K.; Hicks, D.G. p53 alteration in morphologically normal/benign breast tissue in patients with triple-negative high-grade breast carcinomas: Breast p53 signature? Hum. Pathol. 2016, 55, 196–201. [Google Scholar] [CrossRef]
| Patient Characteristics | Breast Cancer | Colorectal Cancer |
|---|---|---|
| Number of patients | 8 | 9 |
| Average age ± SD, years | 61.3 ± 12.4 | 67.4 ± 10.3 |
| Gender, Male/Female | 0/8 | 6/3 |
| Prior treatment history: | ||
| Chemotherapy | 1 | 0 |
| Radiotherapy | 2 | 0 |
| Tumor size | ||
| T1 | 1 | 0 |
| T2 | 6 | 0 |
| T3 | 1 | 4 |
| T4 | 0 | 2 |
| Unknown | 0 | 3 |
| Lymph node status | ||
| N0 | 2 | 3 |
| N1 | 1 | 3 |
| N2 | 2 | 0 |
| N3 | 1 | 0 |
| Unknown | 2 | 3 |
| Metastasis | 0 | 0 |
| Primer | Primer Direction | Sequence |
|---|---|---|
| GAPDH | Forward | 5′-GGGGCTGCCCAGAACATCAT-3′ |
| Reverse | 5′-GCCTGCTTCACCACCTTCTTG-3′ | |
| HAS2 | Forward | 5′-TACACAGCCTTCAGAGCACTG-3′ |
| Reverse | 5′-ATGAGGCTGGGTCAAGCATAG-3′ | |
| HAS3 | Forward | 5′-TGCACCATCGAGATGCTTCG-3′ |
| Reverse | 5′-CCATGAGTCGTACTTGTTGAGG-3′ | |
| HYAL1 | Forward | 5′-GGCTATGAGGAAACTGAGTCAC-3′ |
| Reverse | 5′-TAGGAGTGCAAGGGCTGTAC-3′ | |
| HYAL2 | Forward | 5′-ATCTCTACCATTGGCGAGAGTG-3′ |
| Reverse | 5′-ATCTTTGAGGTACTGGCAGGTC-3′ | |
| CD44 | Forward | 5′-GTGATGGCACCCGCTATG-3′ |
| Reverse | 5′-ACTGTCTTCGTCTGGGATGG-3′ | |
| BRCA1 | Forward | 5′-GGCTATCCTCTCAGAGTGACATTT-3′ |
| Reverse | 5′-GCTTTATCAGGTTATGTTGCATGG-3′ | |
| BRCA2 | Forward | 5′-CCAAGTGGTCCACCCCAAC-3′ |
| Reverse | 5′-CACAATTAGGAGAAGACATCAGAAGC-3′ |
| Patient | Sex | Age at Sample Acquisition (yr) | Histopathologic Diagnosis | TNM Stage | Chemotherapy | Radiotherapy | KI67 | HA | CD44 | HAS2 | HAS3 | HYAL1 | HYAL2 | BRCA1 | BRCA2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 58 | Adenocarcinoma of caecum | pT4pN1b | NO | NO | 62% | ↓ | ↑ | ↓ | ↓ | ↓↓ | ↑ | ↓↓ | ↓ |
| 2 | M | 58 | Villous adenoma with high grade dysplasia | NA | NO | NO | 43% | ↓ | ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | ↓ |
| 3 | M | 67 | Adenocarcinoma of colon | NA | NO | NO | 63% | ↓ | ↓ | ↓ | ↓ | ↓↓ | ↓ | ↓↓ | ↓ |
| 4 | M | 64 | Adenocarcinoma of colon | pT3pN1 | NO | NO | 42% | ↓ | ↑ | ↑ | ↑↑↑ | ↑ | ↑ | ↑↑↑ | ↑↑↑ |
| 5 | M | 65 | Adenocarcinoma of colon | pT4pN0 | NO | NO | 44% | ↓ | ↑ | ↑ | NA | ↓ | ↑ | ↑↑ | ↑↑↑ |
| 6 | F | 74 | Adenocarcinoma of colon | pT3pN0 | NO | NO | 44% | ↓ | ↑↑ | ↑ | ↑↑ | ↑↑ | ↑ | ↑↑↑ | ↑↑↑ |
| 7 | M | 57 | Adenocarcinoma of colon | NA | NO | NO | 7% | ↓ | ↓↓ | ↓↓↓ | ↓↓ | ↓↓↓ | ↓ | ↓↓↓ | ↓↓ |
| 8 | F | 76 | Adenocarcinoma of colon | pT3pN1 | NO | NO | 49% | ↓ | NA | NA | NA | NA | NA | NA | NA |
| 9 | M | 88 | Adenocarcinoma of colon | pT3pN0 | NO | NO | 9% | ↓ | NA | NA | NA | NA | NA | NA | NA |
| 6M/3F | 67.4 ± 10.3 | ||||||||||||||
| Patient | Sex | Age at sample Acquisition (yr) | Histopathologic Diagnosis | TNM Stage | Chemotherapy | Radiotherapy | RE | RP | HER2 | KI67 | HA | CD44 | HAS2 | HAS3 | HYAL1 | HYAL2 | BRCA1 | BRCA2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 53 | Invasive carcinoma of no special type (NST) | pT1 pNo | NO | NO | 98% intense | 10% | NEG | 15% | ↑ | ↑↑ | ↑↑ | ↓ | ↓ | ↓↓ | ↑↑ | ↑↑↑ |
| 2 | F | 63 | Invasive carcinoma of no special type (NST) | T2pN2a | NO | NO | 30% intense | NEG | POS 3+ | 40% | ↓ | ↓↓ | ↓ | ↓↓ | ↓ | ↓ | ↓↓ | ↓ |
| 3 | F | 61 | Invasive carcinoma of no special type (NST) | pT2pNo | NO | NO | 95% intense | 70% | NEG | 40% | ↓↓ | ↑ | ↓ | ↑ | ↓↓ | ↓ | ↑ | ↑ |
| 4 | F | 75 | Invasive carcinoma of no special type (NST) | pT2pNx | NO | * | 90% moderate | 90% | NEG | 10% | ↓ | ↑↑ | ↑↑↑ | ↑↑↑ | ↓ | ↑↑↑ | ↑↑↑ | ↑↑↑ |
| 5 | F | 35 | Invasive carcinoma of no special type (NST) | pT3pN3a | NO | NO | 70% intense | 30% | NEG | 30% | ↓ | ↓ | ↓ | NA | ↓↓ | ↓ | ↑ | ↑ |
| 6 | F | 69 | Invasive carcinoma of no special type (NST) | T2Nx | * | * | 95% intense | 10% intense | NEG | 35% | ↓↓ | ↓ | ↑↑ | NA | ↓ | ↑ | ↑↑↑ | ↑ |
| 7 | F | 69 | Invasive carcinoma of no special type (NST) | pT2N2a | NO | NO | 35% moderate | 30% moderate | NEG | 70% | ↓ | ↓ | NA | NA | ↓↓ | ↓ | ↑ | ↑ |
| 8 | F | 65 | Invasive carcinoma of no special type (NST) | T2pN1 | NO | NO | 95% intense | 95% intense | NEG | 10% | ↓ | ↓ | NA | ↑ | ↓ | ↓ | NA | NA |
| 61.3 ±12.4 | ||||||||||||||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sevic, I.; Spinelli, F.M.; Vitale, D.L.; Icardi, A.; Romano, L.; Brandone, A.; Giannoni, P.; Cristina, C.; Bolontrade, M.F.; Alaniz, L. Hyaluronan Metabolism is Associated with DNA Repair Genes in Breast and Colorectal Cancer. Screening of Potential Progression Markers Using qPCR. Biomedicines 2020, 8, 183. https://doi.org/10.3390/biomedicines8070183
Sevic I, Spinelli FM, Vitale DL, Icardi A, Romano L, Brandone A, Giannoni P, Cristina C, Bolontrade MF, Alaniz L. Hyaluronan Metabolism is Associated with DNA Repair Genes in Breast and Colorectal Cancer. Screening of Potential Progression Markers Using qPCR. Biomedicines. 2020; 8(7):183. https://doi.org/10.3390/biomedicines8070183
Chicago/Turabian StyleSevic, Ina, Fiorella Mercedes Spinelli, Daiana Lujan Vitale, Antonella Icardi, Lucia Romano, Alejandra Brandone, Paula Giannoni, Carolina Cristina, Marcela Fabiana Bolontrade, and Laura Alaniz. 2020. "Hyaluronan Metabolism is Associated with DNA Repair Genes in Breast and Colorectal Cancer. Screening of Potential Progression Markers Using qPCR" Biomedicines 8, no. 7: 183. https://doi.org/10.3390/biomedicines8070183
APA StyleSevic, I., Spinelli, F. M., Vitale, D. L., Icardi, A., Romano, L., Brandone, A., Giannoni, P., Cristina, C., Bolontrade, M. F., & Alaniz, L. (2020). Hyaluronan Metabolism is Associated with DNA Repair Genes in Breast and Colorectal Cancer. Screening of Potential Progression Markers Using qPCR. Biomedicines, 8(7), 183. https://doi.org/10.3390/biomedicines8070183






