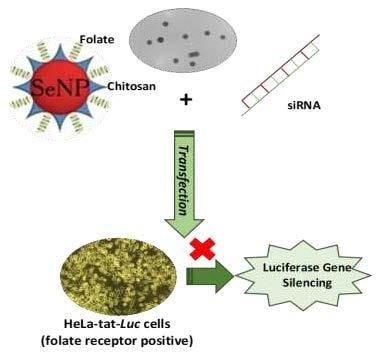
Polymerized Selenium Nanoparticles for Folate-Receptor-Targeted Delivery of Anti-Luc-siRNA: Potential for Gene Silencing
Abstract
1. Introduction
2. Experimental
2.1. Materials
2.2. Synthesis of SeNPs, SeCh, and SeChFA
2.3. Nanocomplex Formation
2.4. Nanoparticle and Nanocomplex Characterisation
2.5. siRNA Binding Studies
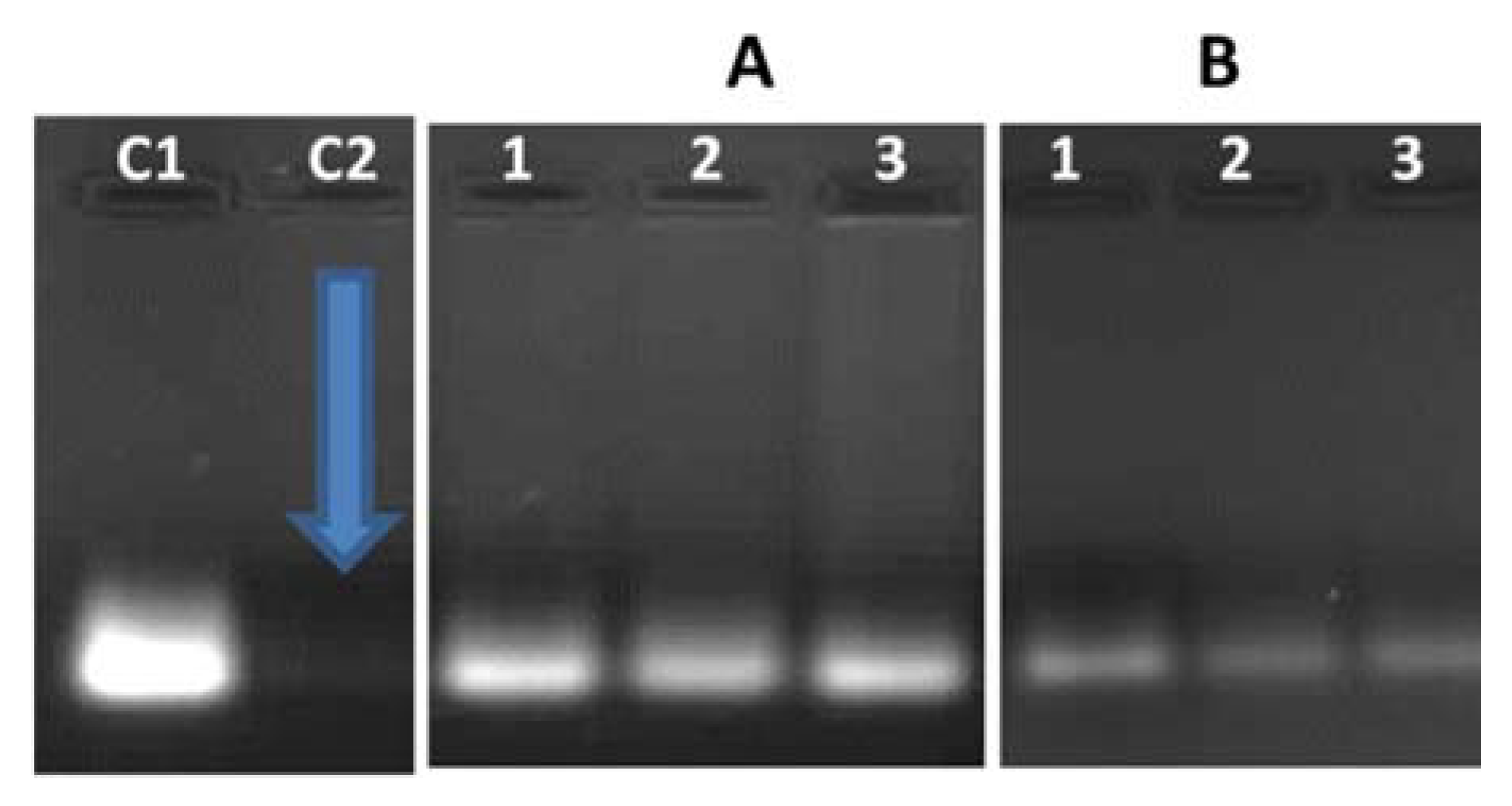
2.6. siRNA Protection Study
2.7. MTT Cell Viability Assay
2.8. Luciferase Gene Silencing In Vitro
2.9. Cellular Uptake study
2.10. Molecular Docking Studies
2.11. Statistical Analysis
3. Results
3.1. Synthesis and Characterisation of Nanoparticles and Nanocomplexes
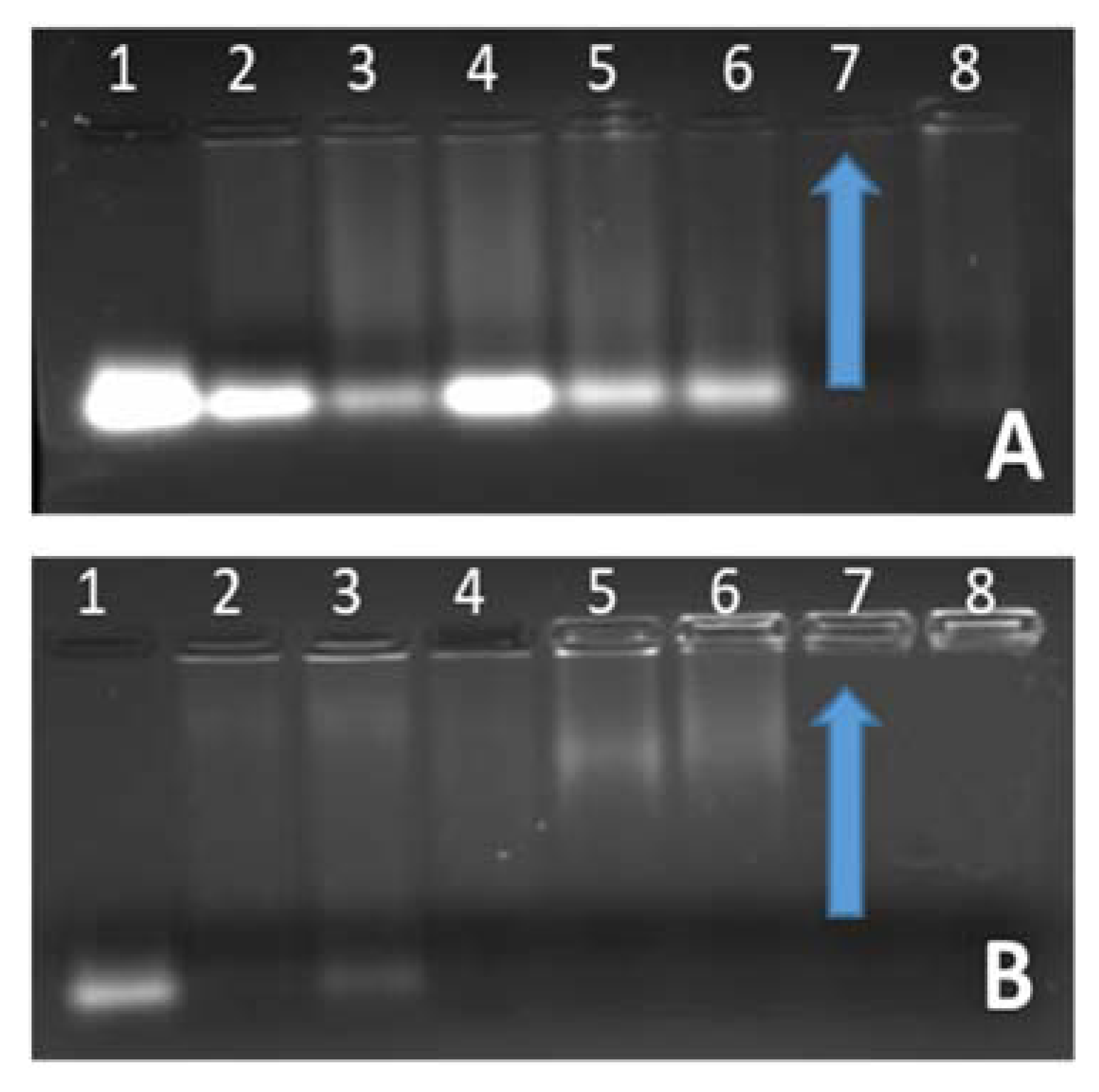
3.2. Nanoparticle: siRNA Interactions
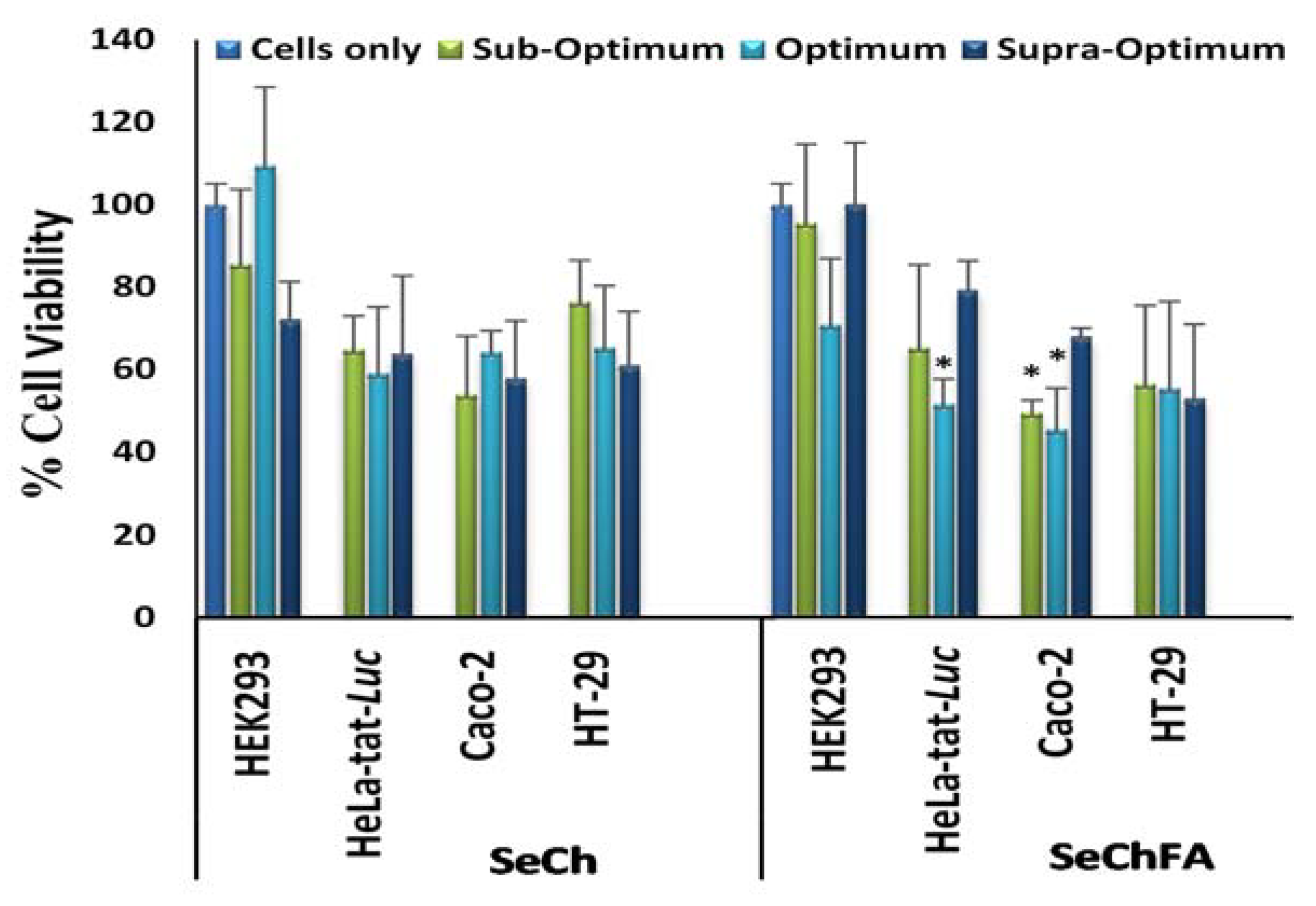
3.3. Cell Viability
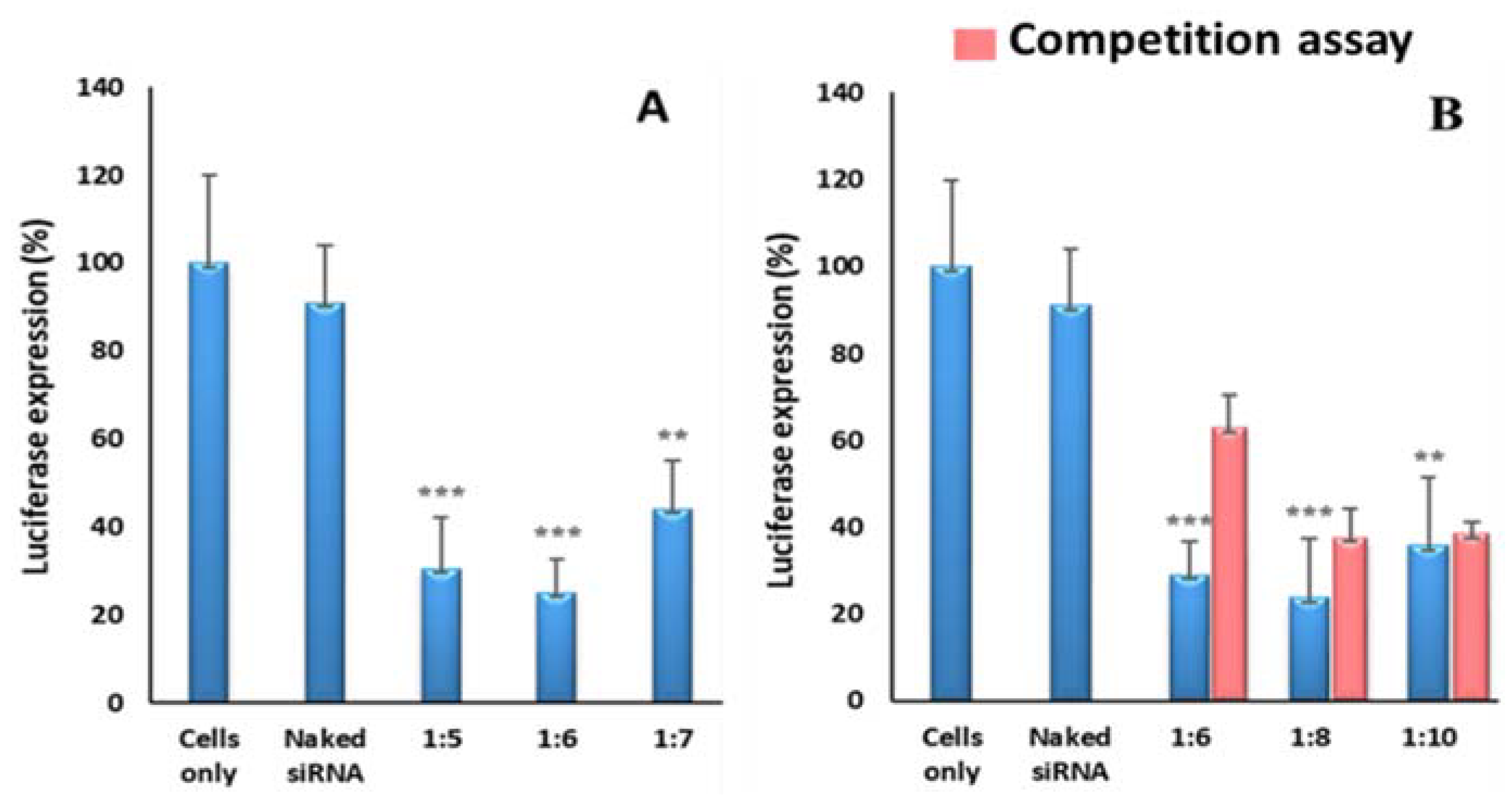
3.4. Gene Silencing
3.5. Molecular Docking Studies
3.6. Selenium Uptake
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kakde, D.; Jain, D.; Srivastava, V.; Kakde, R.; Patil, A. Cancer therapeutics-opportunities, challenges and advances in drug delivery. J. Appl. Pharm. Sci. 2011, 1, 1–10. [Google Scholar]
- Yang, T.; Li, B.; Qi, S.; Liu, Y.; Gai, Y.; Ye, P.; Yang, G.; Zhang, W.; Zhang, P.; He, X. Co-delivery of doxorubicin and Bmi1 siRNA by folate receptor targeted liposomes exhibits enhanced anti-tumor effects in vitro and in vivo. Theranostics 2014, 4, 1096–2011. [Google Scholar] [CrossRef] [PubMed]
- Teo, P.Y.; Cheng, W.; Hedrick, J.L.; Yang, Y.Y. Co-delivery of drugs and plasmid DNA for cancer therapy. Adv. Drug Deliv. Rev. 2016, 98, 41–63. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, F.; Ochiya, T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006, 97, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.H.; Kim, H.; Jang, B.; Cho, H.; Ryu, J.; Kim, B.; Park, Y.; Kim, J.; Lee, J.B.; Lee, H. Technological development of structural DNA/RNA-based RNAi systems and their applications. Adv. Drug Deliv. Rev. 2016, 104, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Shotorbani, S.S.; Baradaran, B. RNA interference and its role in cancer therapy. Adv. Pharm. Bull. 2014, 4, 313. [Google Scholar]
- Grimm, D.; Kay, M.A. RNAi and gene therapy: A mutual attraction. ASH Educ. Program Book 2007, 2007, 473–481. [Google Scholar] [CrossRef]
- Xu, C.-F.; Wang, J. Delivery systems for siRNA drug development in cancer therapy. Asian J. Pharm. Sci. 2015, 10, 1–12. [Google Scholar] [CrossRef]
- Pai, S.; Lin, Y.; Macaes, B.; Meneshian, A.; Hung, C.; Wu, T. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 13, 464–477. [Google Scholar] [CrossRef]
- Lee, S.H.; Kang, Y.Y.; Jang, H.-E.; Mok, H. Current preclinical small interfering RNA (siRNA)-based conjugate systems for RNA therapeutics. Adv. Drug Deliv. Rev. 2016, 104, 78–92. [Google Scholar] [CrossRef]
- Tabernero, J.; Shapiro, G.I.; Lorusso, P.M.; Cervantes, A.; Schwartz, G.K.; Weiss, G.J.; Paz-Ares, L.; Cho, D.C.; Infante, J.R.; Alsina, M. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013, 3, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.Y.; Silvestre, O.F.; Huang, X.; Min, K.H.; Howard, G.P.; Hida, N.; Jin, A.J.; Carvajal, N.; Lee, S.W.; Hong, J.-I. Versatile RNA interference nanoplatform for systemic delivery of RNAs. ACS Nano 2014, 8, 4559–4570. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-M.; Yoon, T.-J.; Cho, Y.-S. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. Biomed Res. Int. 2013, 2013, 782041. [Google Scholar] [CrossRef] [PubMed]
- Young, S.W.S.; Stenzel, M.; Jia-Lin, Y. Nanoparticle-siRNA: A potential cancer therapy? Crit. Rev. Oncol. Hematol. 2016, 98, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, P.; Gajbhiye, V.; Jain, N.K. A review of nanocarriers for the delivery of small interfering RNA. Biomater 2012, 33, 7138–7150. [Google Scholar] [CrossRef]
- Rudzinski, W.E.; Aminabhavi, T.M. Chitosan as a carrier for targeted delivery of small interfering RNA. Int. J. Pharm. 2010, 399, 1–11. [Google Scholar] [CrossRef]
- Xue, W.; Dahlman, J.E.; Tammela, T.; Khan, O.F.; Sood, S.; Dave, A.; Cai, W.; Chirino, L.M.; Yang, G.R.; Bronson, R. Small RNA combination therapy for lung cancer. Proc. Natl. Acad. Sci. USA 2014, 111, E3553–E3561. [Google Scholar] [CrossRef]
- Tapiero, H.; Townsend, D.; Tew, K. The antioxidant role of selenium and seleno-compounds. Biomed. Pharmacother. 2003, 57, 134–144. [Google Scholar] [CrossRef]
- Abdulah, R.; Miyazaki, K.; Nakazawa, M.; Koyama, H. Chemical forms of selenium for cancer prevention. J. Trace Elem. Med. Biol. 2005, 19, 141–150. [Google Scholar] [CrossRef]
- Zeng, H.; Combs, G.F. Selenium as an anticancer nutrient: Roles in cell proliferation and tumor cell invasion. J. Nutr. Biochem. 2008, 19, 1–7. [Google Scholar] [CrossRef]
- Skalickova, S.; Milosavljevic, V.; Cihalova, K.; Horky, P.; Richtera, L.; Adam, V. Selenium nanoparticles as a nutritional supplement. Nutrition 2017, 33, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, Q.; Liu, Y.; Bhavsar, D.; Yang, L.; Ren, X.; Sun, D.; Zheng, W.; Liu, J.; Chen, L.-M. Multifunctional selenium nanoparticles: Chiral selectivity of delivering MDR-siRNA for reversal of multidrug resistance and real-time biofluorescence imaging. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Cao, C.; Liu, Y.; Yu, Q.; Zheng, C.; Sun, D.; Ren, X.; Liu, J. Multifunctional polyamidoamine-modified selenium nanoparticles dual-delivering siRNA and cisplatin to A549/DDP cells for reversal multidrug resistance. Acta Biomater. 2015, 11, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Zhao, M.; Hua, L.; Xu, T.; Wang, C.; Li, Y.; Zhu, B. Folate-targeted selenium nanoparticles deliver therapeutic siRNA to improve hepatocellular carcinoma therapy. RSC Adv. 2018, 8, 25932–25940. [Google Scholar] [CrossRef]
- Maiyo, F.; Singh, M. Selenium Nanoparticles: Potential in Cancer Gene and Drug Delivery. Nanomedicine 2017, 12, 1075–1089. [Google Scholar] [CrossRef]
- Chen, T.; Wong, Y.-S.; Zheng, W.; Bai, Y.; Huang, L. Selenium nanoparticles fabricated in Undaria pinnatifida polysaccharide solutions induce mitochondria-mediated apoptosis in A375 human melanoma cells. Colloids Surf. B Biointerfaces 2008, 67, 26–31. [Google Scholar] [CrossRef]
- Estevez, H.; Garcia-Lidon, J.C.; Luque-Garcia, J.L.; Camara, C. Effects of chitosan-stabilized selenium nanoparticles on cell proliferation, apoptosis and cell cycle pattern in HepG2 cells: Comparison with other selenospecies. Colloids Surf. B Biointerfaces 2014, 122, 184–193. [Google Scholar] [CrossRef]
- Maiyo, F.; Singh, M. Folate-Targeted mRNA Delivery Using Chitosan Functionalized Selenium Nanoparticles: Potential in Cancer Immunotherapy. Pharmaceuticals 2019, 12, 164. [Google Scholar] [CrossRef]
- Li, P.; Wang, Y.; Zeng, F.; Chen, L.; Peng, Z.; Kong, L.X. Synthesis and characterization of folate conjugated chitosan and cellular uptake of its nanoparticles in HT-29 cells. Carbohyd. Res. 2011, 346, 801–806. [Google Scholar] [CrossRef]
- Song, H.; Su, C.; Cui, W.; Zhu, B.; Liu, L.; Chen, Z.; Zhao, L. Folic acid-chitosan conjugated nanoparticles for improving tumor-targeted drug delivery. BioMed Res. Int. 2013, 2013, 723158. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Despande, N.; Addess, K.J.; Bluhm, W.F.; Merino-Ott, J.C.; Townsend-Merino, W.; Zhang, Q.; Knezevich, C.; Xie, L.; Chen, L.; Feng, Z. The RCSB Protein Data Bank: A redesigned query system and relational database based on the mmCIF schema. Nucleic Acid Res. 2005, 33, D233–D237. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Fuhrmann, J.; Rurainski, A.; Lenhof, H.P.; Neumann, D. A new Lamarckian genetic algorithm for flexible ligand-receptor docking. J. Comput. Chem. 2010, 31, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Ingole, A.R.; Thakare, S.R.; Khati, N.; Wankhade, A.V.; Burghate, D. Green synthesis of selenium nanoparticles under ambient condition. Chalcogenide Lett. 2010, 7, 485–489. [Google Scholar]
- Ramamurthy, C.; Sampath, K.; Arunkumar, P.; Kumar, M.S.; Sujatha, V.; Premkumar, K.; Thirunavukkarasu, C. Green synthesis and characterization of selenium nanoparticles and its augmented cytotoxicity with doxorubicin on cancer cells. Bioprocess Biosyst. Eng. 2013, 36, 1131–1139. [Google Scholar] [CrossRef]
- Prasad, K.S.; Selvaraj, K. Biogenic synthesis of selenium nanoparticles and their effect on As (III)-induced toxicity on human lymphocytes. Biol. Trace Elem. Res. 2014, 157, 275–283. [Google Scholar] [CrossRef]
- Esfandiarpour-Boroujeni, S.; Bagheri-Khoulenjani, S.; Mirzadesh, H. Modeling and optimization of degree of folate grafted on chitosan and carboxymethyl-chitosan. Progress Biomater. 2016, 5, 1–8. [Google Scholar] [CrossRef]
- Shityakov, S.; Förster, C. In silica predictive model to determine vector-mediated transport properties for the blood-brain barrier choline transporter. Adv. Appl. Bioinform. Chem. 2014, 7, 23–36. [Google Scholar]
- Maney, V.; Singh, M. An in vitro assessment of Chitosan/ Bimetallic PtAu nanocomposites as delivery vehicles for Doxorubicin. Nanomedicine 2017, 12, 2625–2640. [Google Scholar] [CrossRef] [PubMed]
- Akinyelu, J.; Singh, M. Chitosan stabilized Gold-Folate-Poly(lactide-co-glycolide) Nanoplexes Facilitate Efficient Gene Delivery in Hepatic and Breast Cancer Cells. J. Nanosci. Nanotechnol. 2018, 18, 4478–4486. [Google Scholar] [CrossRef] [PubMed]
- Akinyelu, J.; Singh, M. Folate-tagged chitosan functionalized gold nanoparticles for enhanced delivery of 5-fluorouracil to cancer cells. Appl. Nanosci. 2019, 9, 7–17. [Google Scholar] [CrossRef]
- Daniels, A.N.; Singh, M. Sterically stabilised siRNA: Gold nanocomplexes enhance c-MYC silencing in a breast cancer cell model. Nanomedicine 2019, 14, 1387–1401. [Google Scholar] [CrossRef]
- Jin, L.; Zeng, X.; Liu, M.; Deng, Y.; He, N. Current progress in gene delivery technology based on chemical methods and nano-carriers. Theranostics 2014, 4, 240–255. [Google Scholar] [CrossRef]
- Lin, J.; Li, Y.; Li, Y.; Wu, H.; Yu, F.; Zhou, S.; Xie, L.; Luo, F.; Lin, C.; Hou, Z. Drug/dye-loaded, multifunctional PEG–chitosan–iron oxide nanocomposites for methotraxate synergistically self-targeted cancer therapy and dual model imaging. ACS Appl. Mater. Interfaces 2015, 7, 11908–11920. [Google Scholar] [CrossRef]
- Gorle, S.; Ariatti, M.; Singh, M. Novel serum-tolerant lipoplexes target the folate receptor efficiently. Eur. J. Pharm. Sci. 2014, 59, 83–93. [Google Scholar] [CrossRef]
- Gaspar, V.M.; Costa, E.C.; Queiroz, J.A.; Pichon, C.; Sousa, F.; Correia, I.J. Folate-targeted multifunctional amino acid-chitosan nanoparticles for improved cancer therapy. Pharm. Res. 2015, 32, 562–577. [Google Scholar] [CrossRef]
- Mbatha, L.S.; Maiyo, F.C.; Singh, M. Dendrimer Functionalized Folate-Targeted Gold Nanoparticles for Luciferase Gene Silencing in vitro: A Proof of Principle Study. Acta Pharm. 2019, 69, 49–61. [Google Scholar] [CrossRef]
- Yoo, H.S.; Park, T.G. Folate-receptor-targeted delivery of doxorubicin nano-aggregates stabilized by doxorubicin–PEG–folate conjugate. J. Control Release 2004, 100, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Mary, T.A.; Shanthi, K.; Vimala, K.; Soundarapandian, K. PEG functionalized selenium nanoparticles as a carrier of crocin to achieve anticancer synergism. RSC Adv. 2016, 6, 22936–22949. [Google Scholar] [CrossRef]
- Ostolska, I.; Wisniewska, M. Application of the zeta potential measurements to explanation of colloidal Cr2O3stability mechanism in the presence of the ionic polyamino acids. Colloid Polym. Sci. 2014, 292, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Honary, S.; Zahir, F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 1). Trop. J. Pharm. Res. 2013, 12, 255–264. [Google Scholar]
- Daniels, A.; Singh, M.; Ariatti, M. Pegylated and Non-Pegylated siRNA lipoplexes formulated with cholesteryl cytofectins promote efficient Luciferase knockdown in HeLa tat luc cells. Nucleosides Nucleotides Nucleic Acids 2013, 32, 206–220. [Google Scholar] [CrossRef]
- Lopez-Heras, I.; Sanchez-Diaz, R.; Anunciação, D.S.; Madrid, Y.; Luque-Garcia, J.L.; Camara, C. Effect of Chitosan-stabilized Selenium nanoparticles on cell cycle arrest and invasiveness in hepatocarcinoma cells revealed by quantitative proteomics. J. Nanomed. Nanotechnol. 2014, 5, 5. [Google Scholar] [CrossRef]
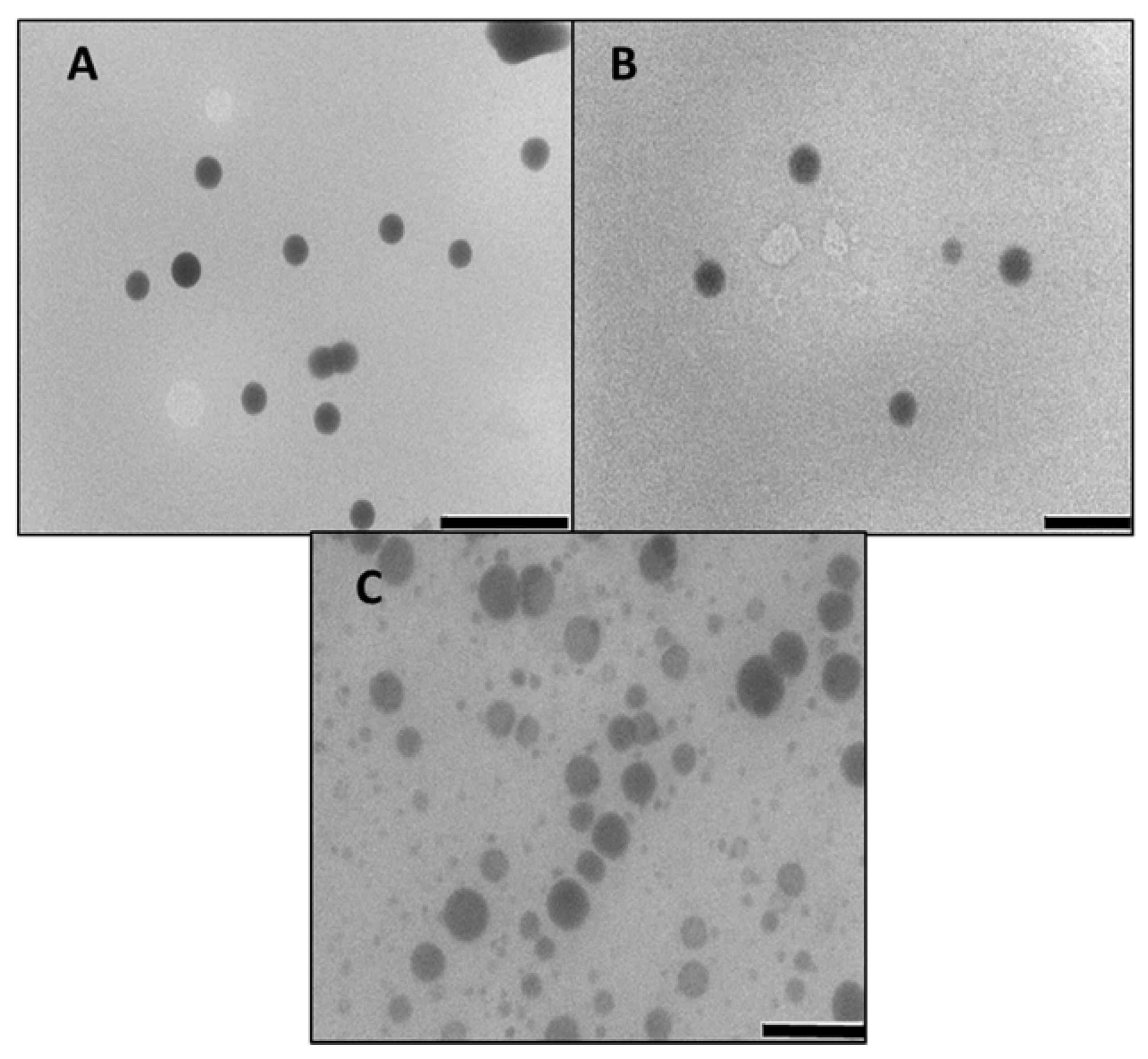
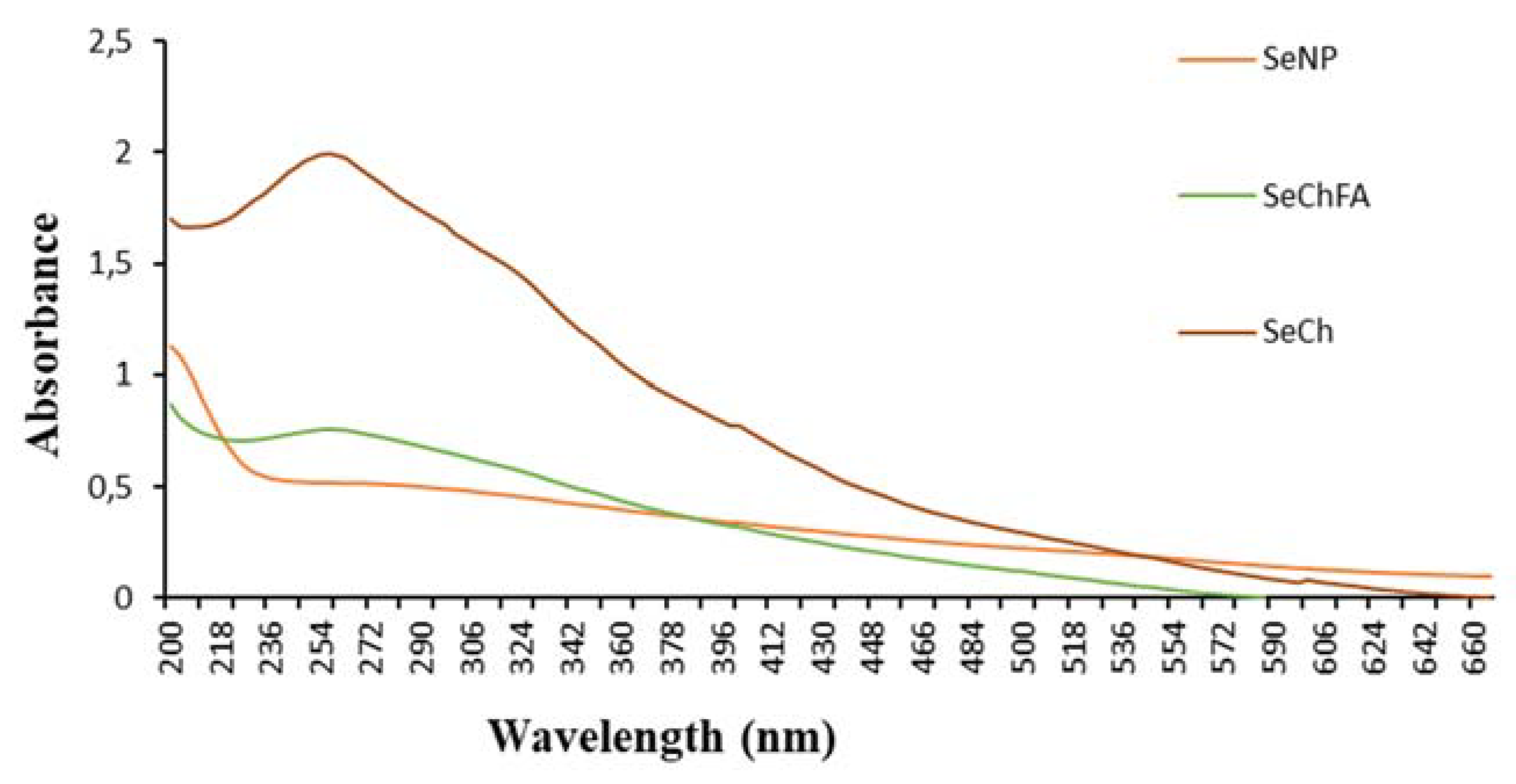
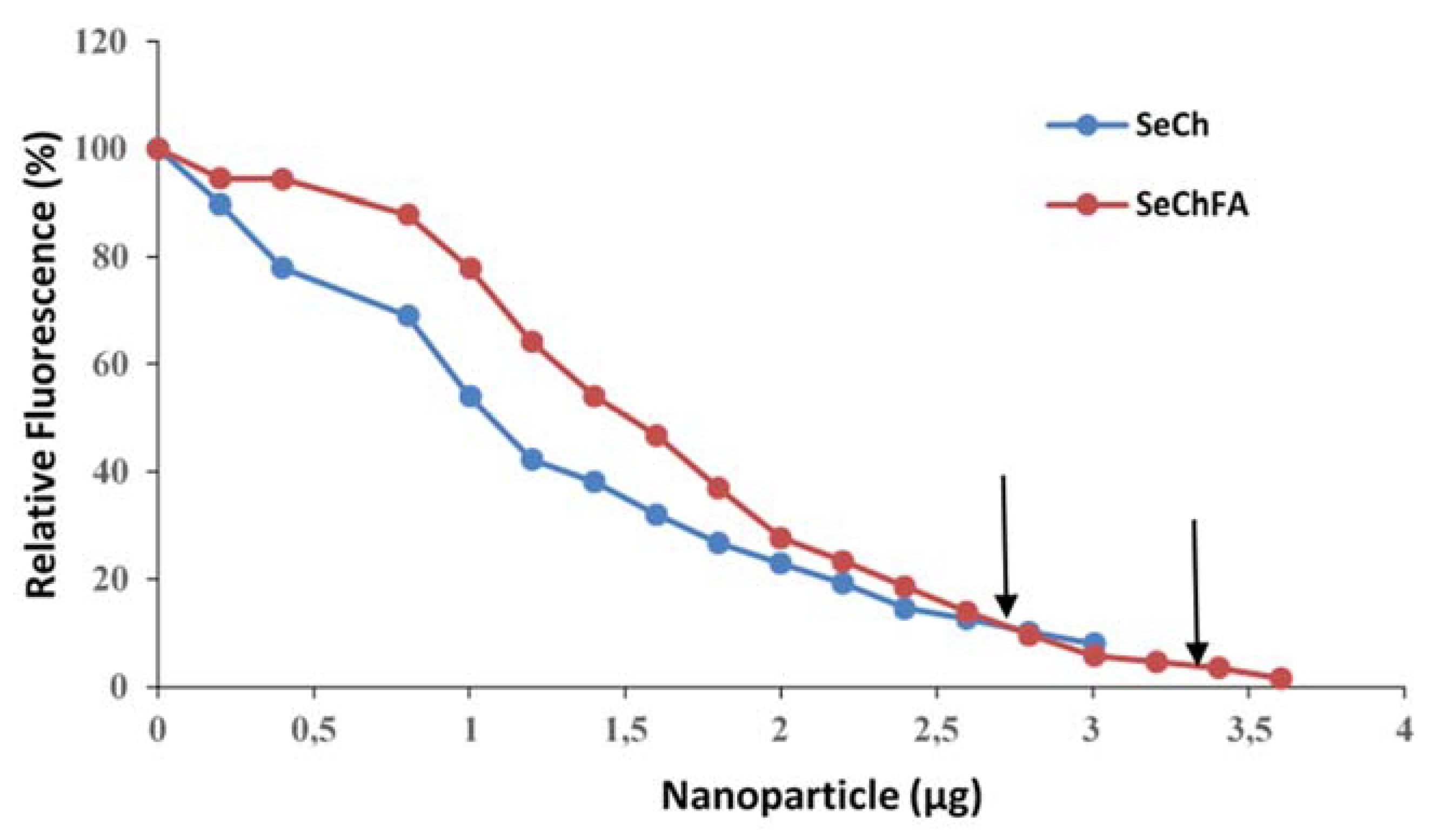
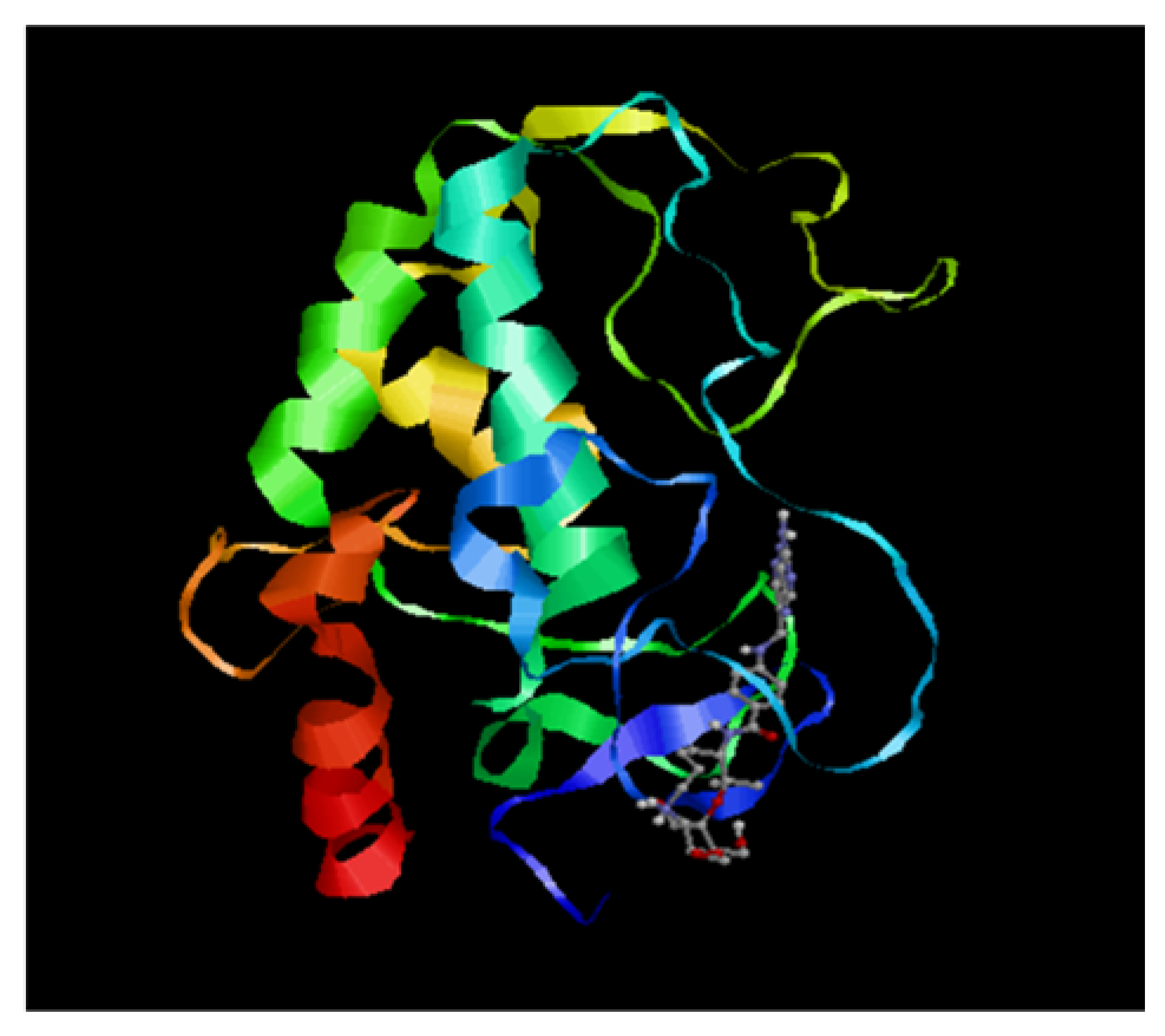
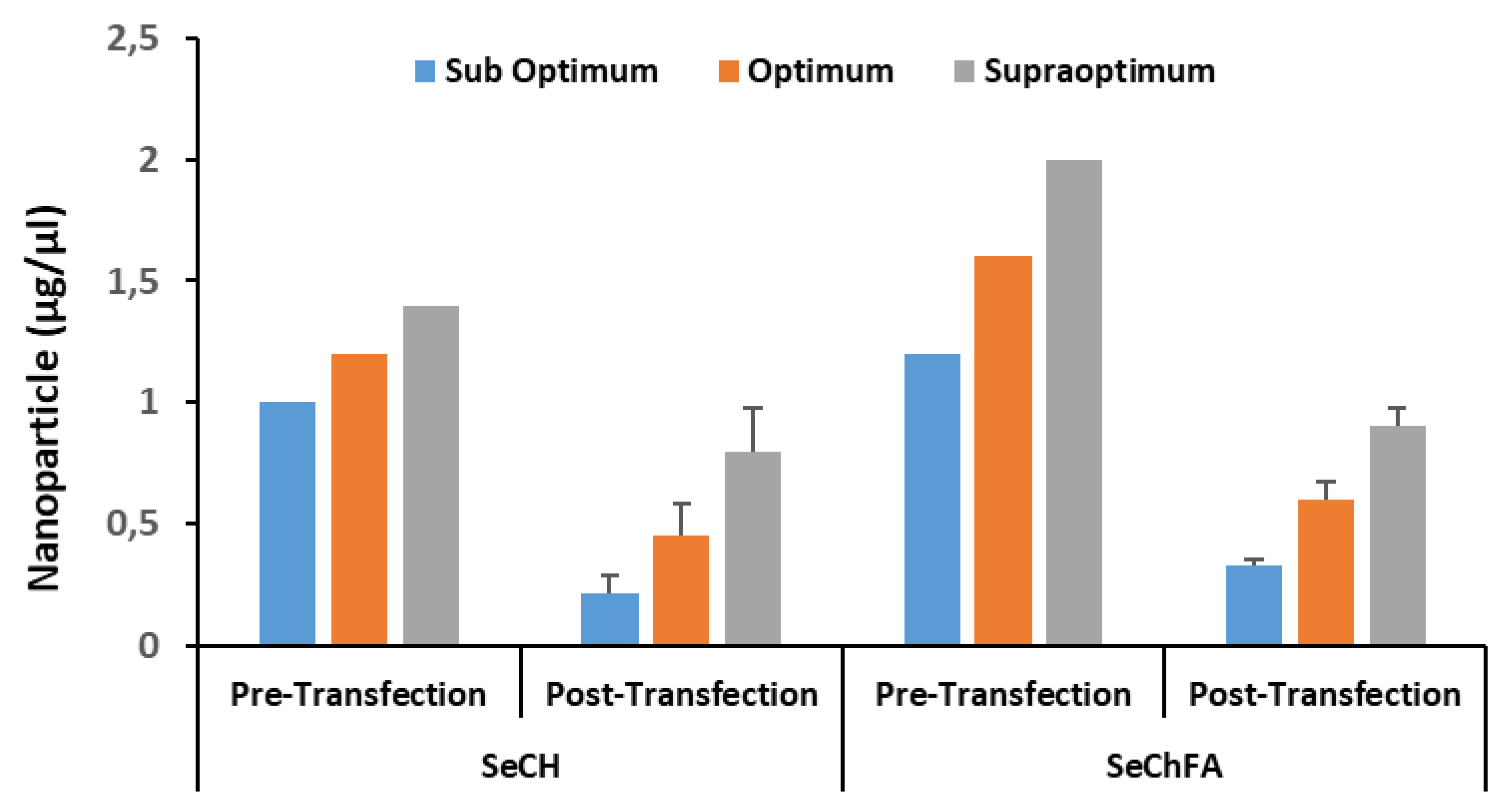
| Nanoparticle | Size (nm) | ζ Potential (mV) | End-Point Ratios (Optimum Binding) | ||
|---|---|---|---|---|---|
| w/w | Size | ζ Potential | |||
| SeNP | 85.3 ± 8 | −14.8 ± −3.6 | n/a | n/a | n/a |
| SeCh | 59.6 ± 0.1 | 21.0 ± 0.2 | 1:6 | 57.1 ± 0.3 | 31.8 ± 1.2 |
| SeChFA | 75.6 ± 1.4 | 9.0 ± 0.3 | 1:8 | 63.3 ± 0.7 | 19.3 ± 0.8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maiyo, F.; Singh, M. Polymerized Selenium Nanoparticles for Folate-Receptor-Targeted Delivery of Anti-Luc-siRNA: Potential for Gene Silencing. Biomedicines 2020, 8, 76. https://doi.org/10.3390/biomedicines8040076
Maiyo F, Singh M. Polymerized Selenium Nanoparticles for Folate-Receptor-Targeted Delivery of Anti-Luc-siRNA: Potential for Gene Silencing. Biomedicines. 2020; 8(4):76. https://doi.org/10.3390/biomedicines8040076
Chicago/Turabian StyleMaiyo, Fiona, and Moganavelli Singh. 2020. "Polymerized Selenium Nanoparticles for Folate-Receptor-Targeted Delivery of Anti-Luc-siRNA: Potential for Gene Silencing" Biomedicines 8, no. 4: 76. https://doi.org/10.3390/biomedicines8040076
APA StyleMaiyo, F., & Singh, M. (2020). Polymerized Selenium Nanoparticles for Folate-Receptor-Targeted Delivery of Anti-Luc-siRNA: Potential for Gene Silencing. Biomedicines, 8(4), 76. https://doi.org/10.3390/biomedicines8040076