Detailed Characterization of the Cooperative Binding of Piperine with Heat Shock Protein 70 by Molecular Biophysical Approaches
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Steady-State Fluorescence Spectroscopy
2.3. Time-Resolved Fluorescence
2.4. Circular Dichroism
2.5. Molecular Docking
2.6. Molecular Dynamics
3. Results and Discussions
3.1. Fluorescence Spectroscopy
3.2. Thermodynamic Parameters
3.3. Interaction Density Function (IDF)
3.4. Circular Dichroism
3.5. Molecular Modeling
3.5.1. Molecular Docking
3.5.2. Molecular Dynamics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dubrez, L.; Causse, S.; Bonan, N.B.; Dumetier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2019, 39, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M.; Westwood, I.M.; Osborne, J.D.; Matthews, T.P.; Cheeseman, M.D.; Rowlands, M.G.; Jeganathan, F.; Burke, R.; Lee, D.; Kadi, N. A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms. Sci. Rep. 2016, 6, 34701. [Google Scholar] [CrossRef] [PubMed]
- Asea, A.; Kraeft, S.-K.; Kurt-Jones, E.A.; Stevenson, M.A.; Chen, L.B.; Finberg, R.W.; Koo, G.C.; Calderwood, S.K. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat. Med. 2000, 6, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Asea, A.; Rehli, M.; Kabingu, E.; Boch, J.A.; Baré, O.; Auron, P.E.; Stevenson, M.A.; Calderwood, S.K. Novel signal transduction pathway utilized by extracellular HSP70 role of Toll-like receptor (TLR) 2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef] [PubMed]
- Somensi, N.; Brum, P.O.; de Miranda Ramos, V.; Gasparotto, J.; Zanotto-Filho, A.; Rostirolla, D.C.; da Silva Morrone, M.; Moreira, J.C.F.; Gelain, D.P. Extracellular HSP70 activates ERK1/2, NF-kB and pro-inflammatory gene transcription through binding with RAGE in A549 human lung cancer cells. Cell. Physiol. Biochem. 2017, 42, 2507–2522. [Google Scholar] [CrossRef]
- Pahl, H.L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene 1999, 18, 6853–6866. [Google Scholar] [CrossRef]
- Evans, C.G.; Chang, L.; Gestwicki, J.E. Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 2010, 53, 4585–4602. [Google Scholar] [CrossRef]
- de Oliveira, A.A.; Faustino, J.; de Lima, M.E.; Menezes, R.; Nunes, K.P. Unveiling the interplay between the TLR4/MD2 complex and HSP70 in the human cardiovascular system: A computational approach. Int. J. Mol. Sci. 2019, 20, 3121. [Google Scholar] [CrossRef]
- Cheeseman, M.D.; Westwood, I.M.; Barbeau, O.; Rowlands, M.; Dobson, S.; Jones, A.M.; Jeganathan, F.; Burke, R.; Kadi, N.; Workman, P. Exploiting protein conformational change to optimize adenosine-derived inhibitors of HSP70. J. Med. Chem. 2016, 59, 4625–4636. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; Le Duff, C.S.; Tang, B.; Cornelio, M.L.; Jones, A.M. Synthesis and Spectroscopic Analysis of Piperine-and Piperlongumine-Inspired Natural Product Scaffolds and Their Molecular Docking with IL-1β and NF-κB Proteins. Molecules 2020, 25, 2841. [Google Scholar] [CrossRef]
- Ying, X.; Chen, X.; Cheng, S.; Shen, Y.; Peng, L.; Xu, H. Piperine inhibits IL-β induced expression of inflammatory mediators in human osteoarthritis chondrocyte. Int. Immunopharmacol. 2013, 17, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Bang, J.S.; Choi, H.M.; Sur, B.-J.; Lim, S.-J.; Kim, J.Y.; Yang, H.-I.; Yoo, M.C.; Hahm, D.-H.; Kim, K.S. Anti-inflammatory and antiarthritic effects of piperine in human interleukin 1β-stimulated fibroblast-like synoviocytes and in rat arthritis models. Arthritis Res. Ther. 2009, 11, R49. [Google Scholar] [CrossRef] [PubMed]
- Ying, X.; Yu, K.; Chen, X.; Chen, H.; Hong, J.; Cheng, S.; Peng, L. Piperine inhibits LPS induced expression of inflammatory mediators in RAW 264.7 cells. Cell. Immunol. 2013, 285, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2004. [Google Scholar]
- Sreerama, N.; Woody, R.W. Estimation of protein secondary structure from circular dichroism spectra: Comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000, 287, 252–260. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; de Lima, M.F.; Cornélio, M.L. Experimental Approaches and Computational Modeling of Rat Serum Albumin and Its Interaction with Piperine. Int. J. Mol. Sci. 2019, 20, 2856. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Morris, G.M.; Goodsell, D.S.; Pique, M.E.; Lindstrom, W.; Huey, R.; Forli, S.; Hart, W.E.; Halliday, S.; Belew, R.; Olson, A.J. User Guide AutoDock Version 4.2. Automated Docking of Flexible Ligands to Flexible Receptors; AutoGrid: Redwood City, CA, USA, 2010. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: The GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. 2004, 25, 1656–1676. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Wu, Y.; Tepper, H.L.; Voth, G.A. Flexible simple point-charge water model with improved liquid-state properties. J. Chem. Phys. 2006, 124, 24503. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Batcho, P.F.; Case, D.A.; Schlick, T. Optimized particle-mesh Ewald/multiple-time step integration for molecular dynamics simulations. J. Chem. Phys. 2001, 115, 4003–4018. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Roux, B. The calculation of the potential of mean force using computer simulations. Comput. Phys. Commun. 1995, 91, 275–282. [Google Scholar] [CrossRef]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992, 13, 1011–1021. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Consortium, O.S.D.D.; Lynn, A. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Povinelli, A.P.R.; Zazeri, G.; de Freitas Lima, M.; Cornélio, M.L. Details of the cooperative binding of piperlongumine with rat serum albumin obtained by spectroscopic and computational analyses. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef]
- Soares, S.; Mateus, N.; De Freitas, V. Interaction of different polyphenols with bovine serum albumin (BSA) and human salivary α-amylase (HSA) by fluorescence quenching. J. Agric. Food Chem. 2007, 55, 6726–6735. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Lohman, T.M.; Bujalowski, W. Thermodynamic methods for model-independent determination of equilibrium binding isotherms for protein-DNA interactions: Spectroscopic approaches to monitor binding. Methods Enzymol. 1991, 208, 258–290. [Google Scholar] [PubMed]
- Scatchard, G. The attractions of proteins for small molecules and ions. Ann. N. Y. Acad. Sci. 1949, 51, 660–672. [Google Scholar] [CrossRef]
- Povinelli, A.P.R.; Zazeri, G.; Cornélio, M.L. Molecular Mechanism of Flavonoids Using Fluorescence Spectroscopy and Computational Tools. In Flavonoids-A Coloring Model For Cheering Up Life; IntechOpen: Rijeka, Croatia, 2019. [Google Scholar]
- Bordbar, A.K.; Saboury, A.A.; Moosavi-Movahedi, A.A. The shapes of Scatchard plots for systems with two sets of binding sites. Biochem. Educ. 1996, 24, 172–175. [Google Scholar] [CrossRef]
- Barcroft, J.; Hill, A.V. The nature of oxyhaemoglobin, with a note on its molecular weight. J. Physiol. 1910, 39, 411. [Google Scholar] [CrossRef]
- Zazeri, G.; Povinelli, A.P.R.; de Freitas Lima, M.; Cornélio, M.L. The Cytokine IL-1β and Piperine Complex Surveyed by Experimental and Computational Molecular Biophysics. Biomolecules 2020, 10, 1337. [Google Scholar] [CrossRef]
- Borges, J.C.; Ramos, C.H.I. Spectroscopic and thermodynamic measurements of nucleotide-induced changes in the human 70-kDa heat shock cognate protein. Arch. Biochem. Biophys. 2006, 452, 46–54. [Google Scholar] [CrossRef]
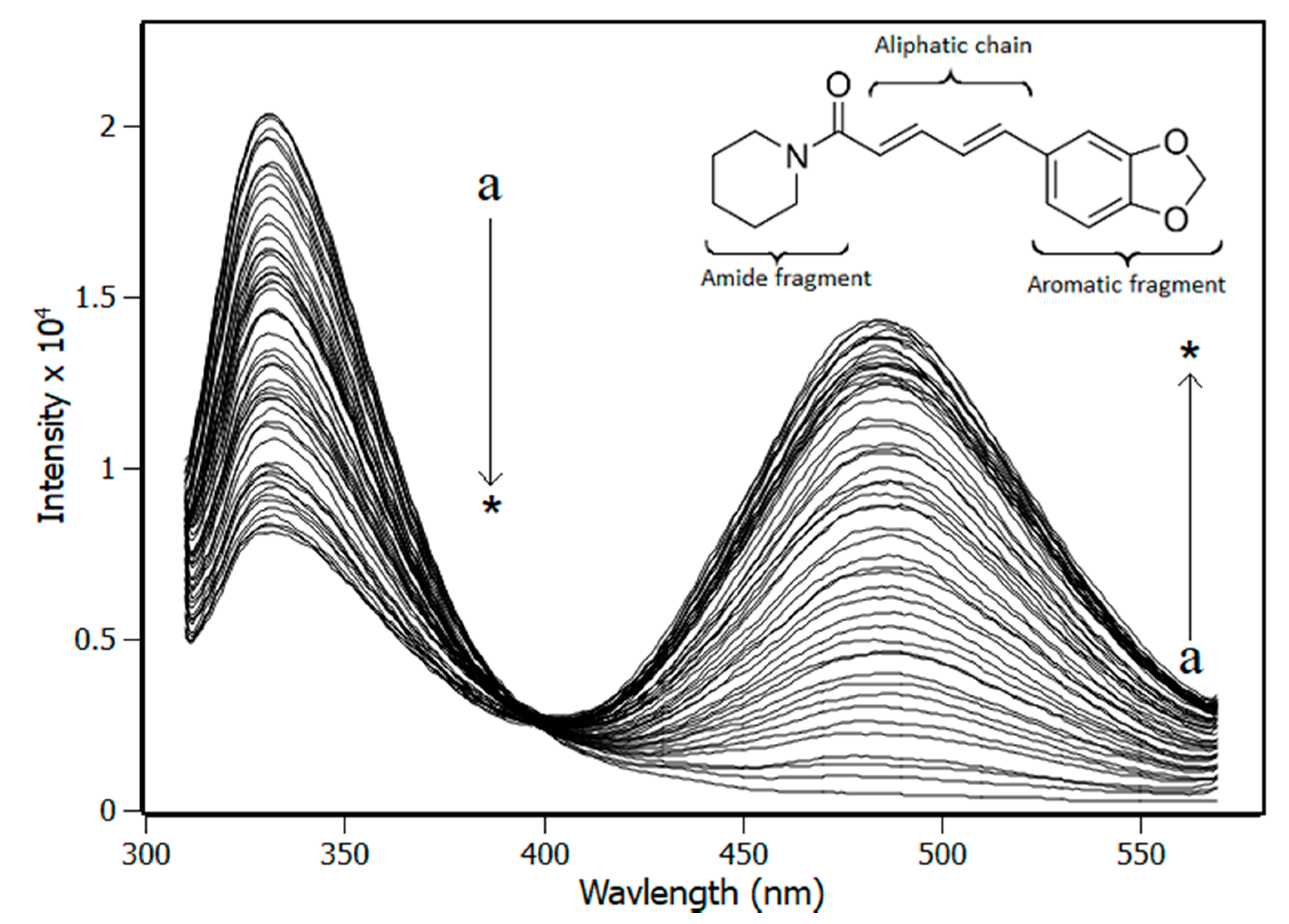
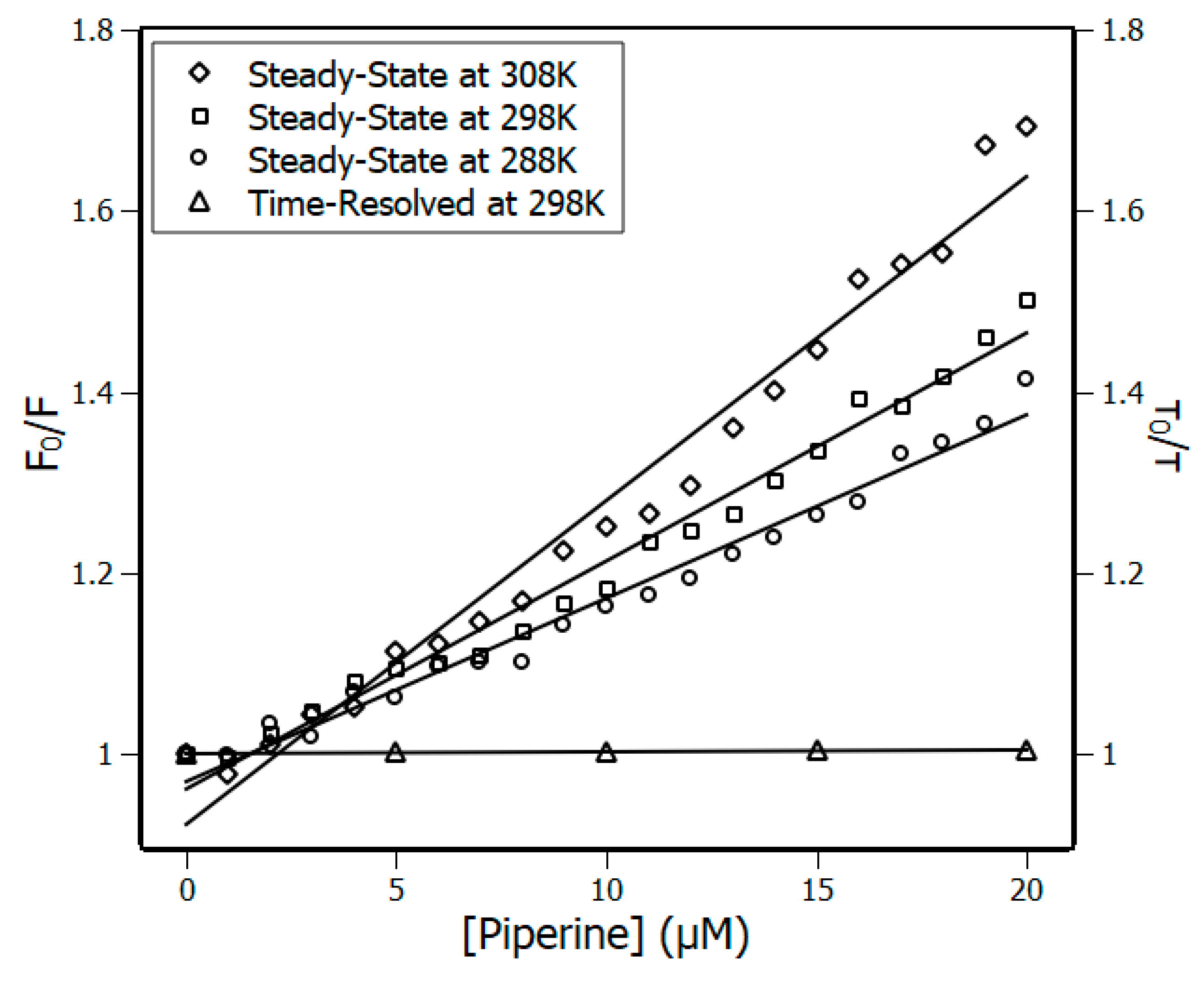
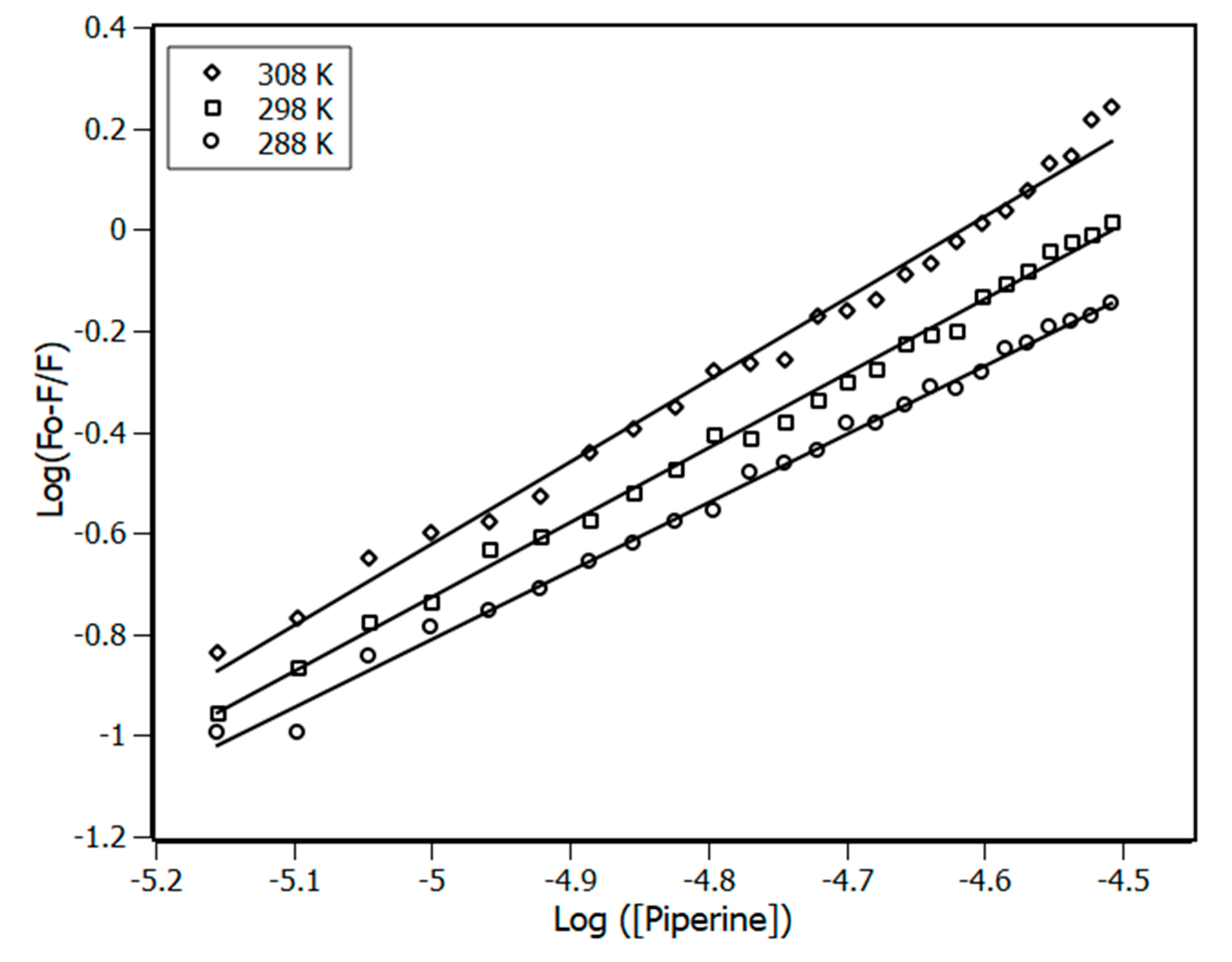

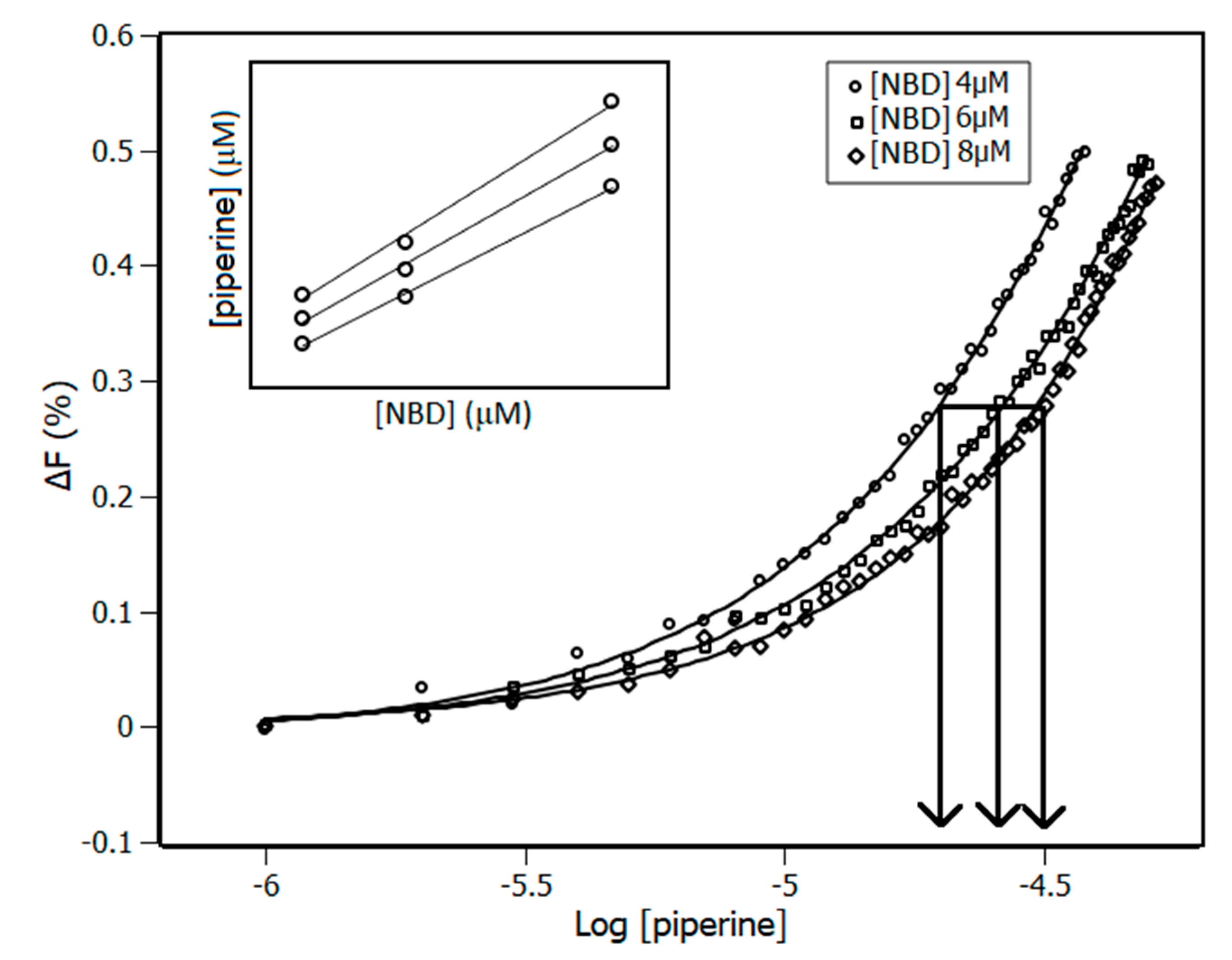
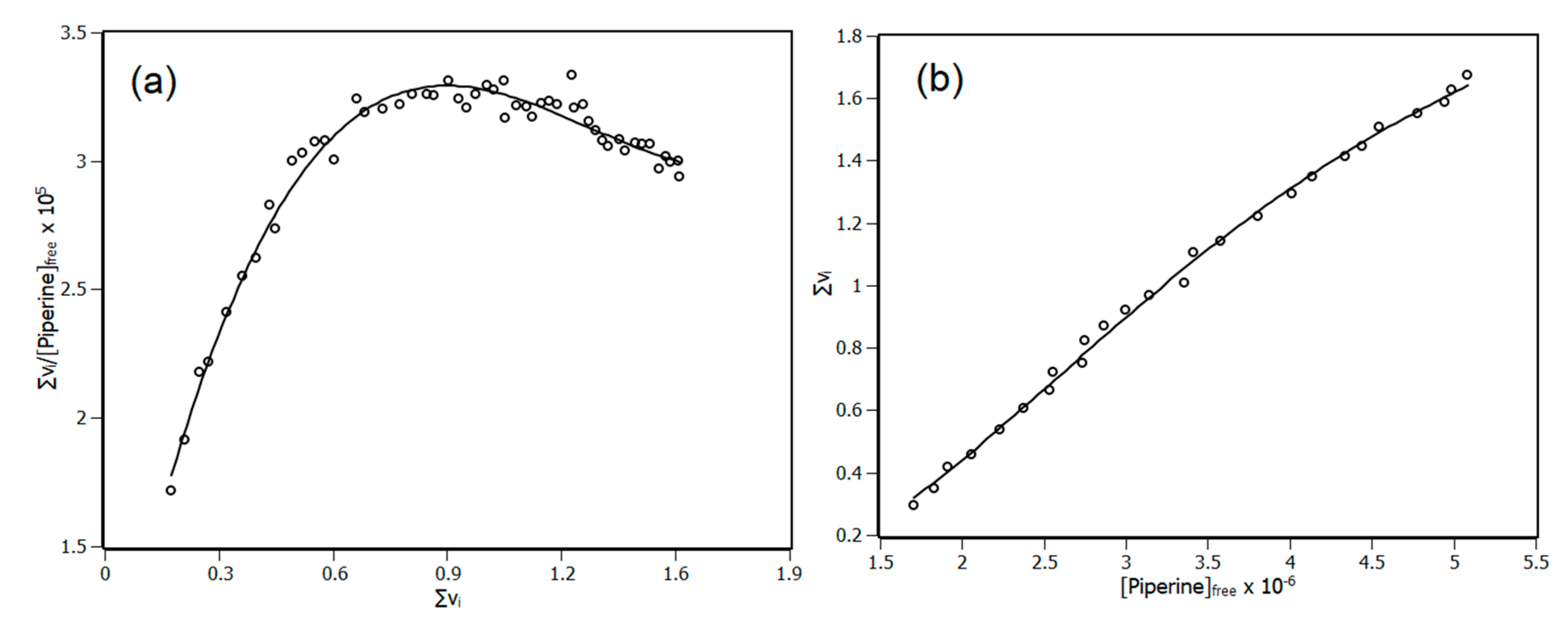
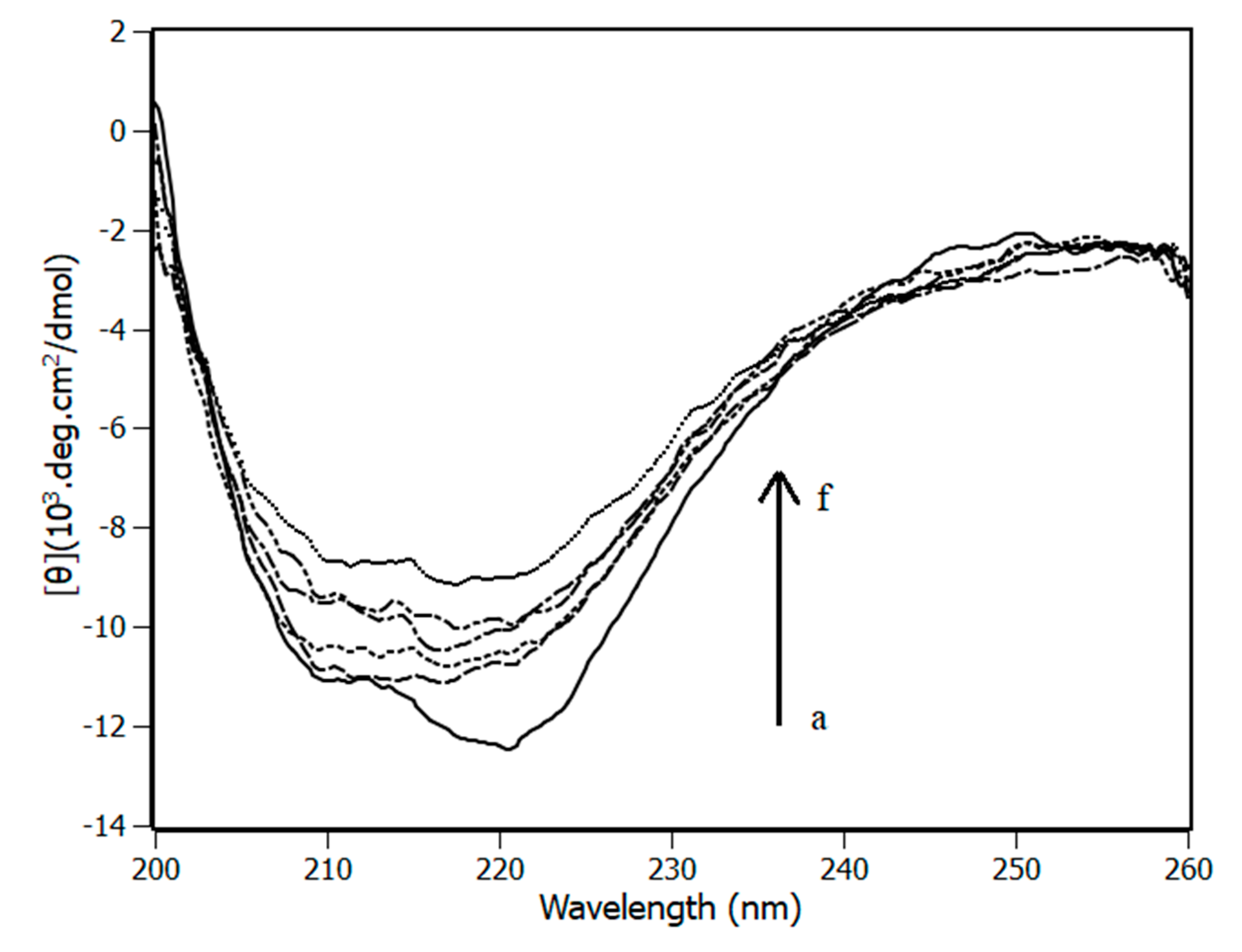
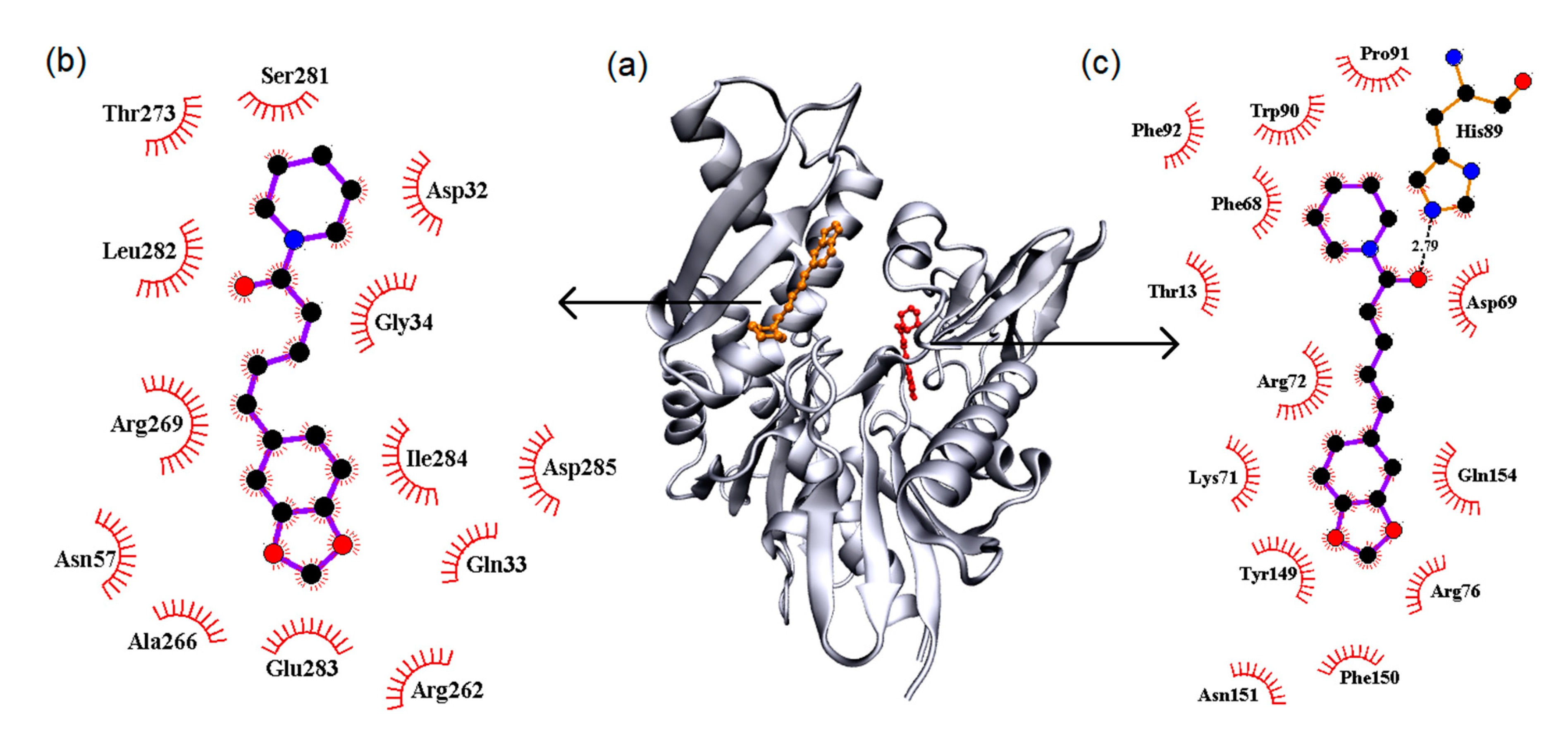
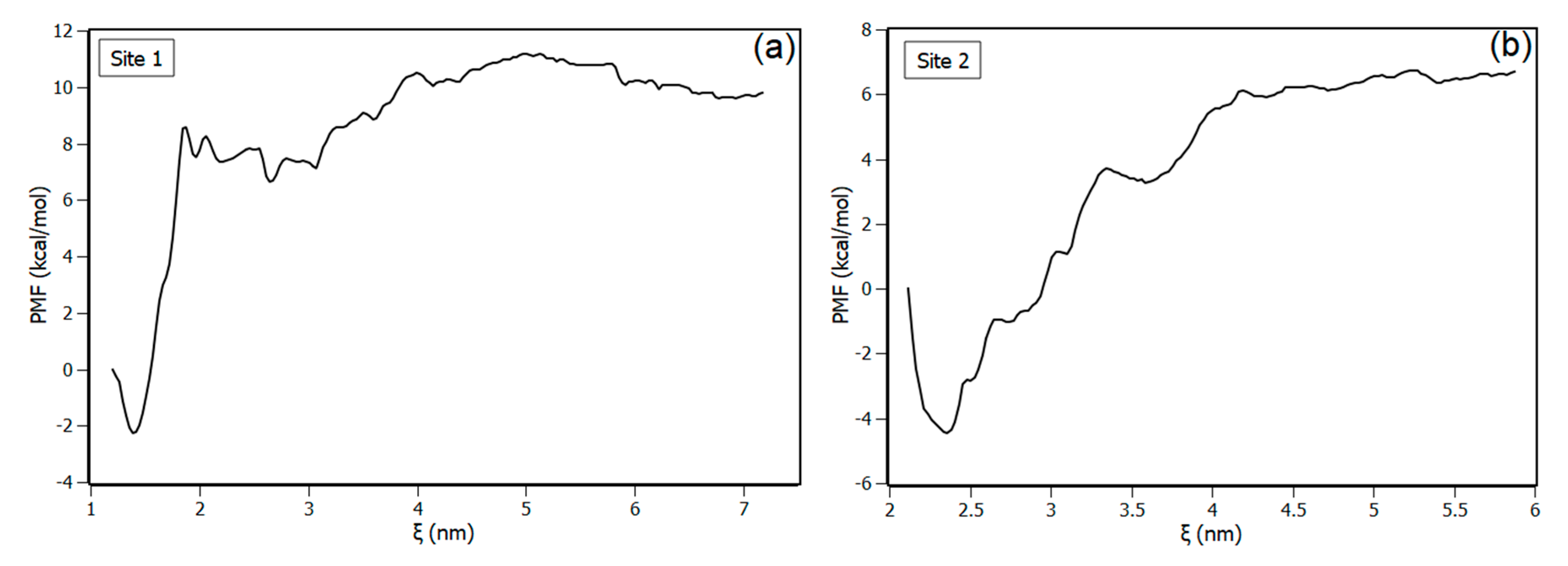
| Temperature (K) | Stern-Volmer (KSV) ×104 M−1 | Bimolecular (Kq) ×1012 M−1·s−1 | Binding (Ka) ×105 M−1 |
|---|---|---|---|
| 288 | 2.02 ± 0.07 | 9.18 ± 0.01 | 8.82 ± 0.07 |
| 298 | 2.52 ± 0.08 | 11.45 ± 0.01 | 44.60 ± 0.10 |
| 308 | 3.58 ± 0.1 | 16.27 ± 0.01 | 301.37 ± 1.1 |
| T (K) | ∆G (kJ/mol) | ∆H (kJ/mol) | T.∆S (kJ/mol) |
|---|---|---|---|
| 288 | −32.65 ± 1.31 | 130.13 ± 8.69 | 162.78 ± 8.4 |
| 298 | −38.3 ± 1.94 | 130.13 ± 8.69 | 168.43 ± 8.69 |
| 308 | −43.95 ± 2.06 | 130.13 ± 8.69 | 174.01 ± 8.98 |
| Energies (kJ/mol) | Site 1 | Site 2 | Both Sites Occupied | |
|---|---|---|---|---|
| Site 1 | Site 2 | |||
| Binding free energy ∆G | −35.22 ± 2.45 | −34.59 ± 2.60 | −49.92 ± 3.01 | −46.15 ± 2.03 |
| van der Waals | −97.63 ± 1.86 | −119.61 ± 2.25 | −73.74 ± 1.98 | −96.12 ± 1.45 |
| Electrostatic | −11.76 ± 0.85 | −25.23 ± 0.95 | −2.80 ± 1.73 | −12.31 ± 1.17 |
| Polar solvation | 86.80 ± 2.89 | 124.32 ± 3.52 | 36.25 ± 3.34 | 73.88 ± 2.1 |
| SASA | −12.64 ± 0.24 | −14.07 ± 0.23 | −9.64 ± 0.25 | −11.59 ± 0.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zazeri, G.; Povinelli, A.P.R.; Lima, M.d.F.; Cornélio, M.L. Detailed Characterization of the Cooperative Binding of Piperine with Heat Shock Protein 70 by Molecular Biophysical Approaches. Biomedicines 2020, 8, 629. https://doi.org/10.3390/biomedicines8120629
Zazeri G, Povinelli APR, Lima MdF, Cornélio ML. Detailed Characterization of the Cooperative Binding of Piperine with Heat Shock Protein 70 by Molecular Biophysical Approaches. Biomedicines. 2020; 8(12):629. https://doi.org/10.3390/biomedicines8120629
Chicago/Turabian StyleZazeri, Gabriel, Ana Paula Ribeiro Povinelli, Marcelo de Freitas Lima, and Marinônio Lopes Cornélio. 2020. "Detailed Characterization of the Cooperative Binding of Piperine with Heat Shock Protein 70 by Molecular Biophysical Approaches" Biomedicines 8, no. 12: 629. https://doi.org/10.3390/biomedicines8120629
APA StyleZazeri, G., Povinelli, A. P. R., Lima, M. d. F., & Cornélio, M. L. (2020). Detailed Characterization of the Cooperative Binding of Piperine with Heat Shock Protein 70 by Molecular Biophysical Approaches. Biomedicines, 8(12), 629. https://doi.org/10.3390/biomedicines8120629






