Authentication of Primary Murine Cell Lines by a Microfluidics-Based Lab-On-Chip System
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. Reagents, Plasmids and Media
2.3. Surface Marker Detection by FACS
2.4. Virus Production
2.5. Cell Lines
2.6. Differentiation of Immortalized HoxB8-FL Cells
2.7. DNA and RNA Isolation and Transcriptome Analysis
2.8. Short Tandem Repeat (STR)-Based Multiplex PCR
2.9. Fragment Length Analysis
3. Results
3.1. Differentiation and Characterization of Conditionally Immortalized HoxB8-FL Hematopoietic Progenitors
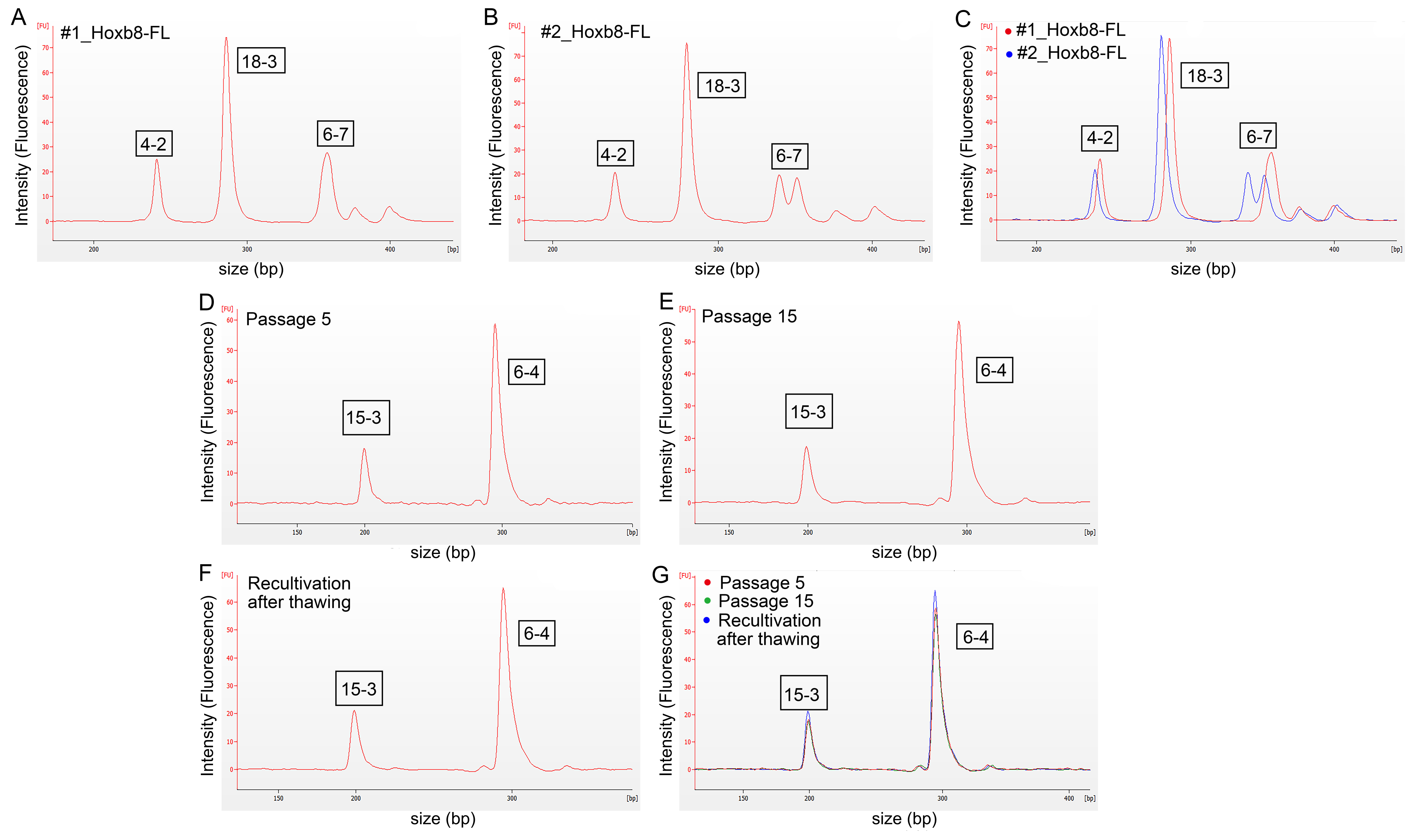
3.2. Authentication of HoxB8-FL Cell Lines by STR Profiling
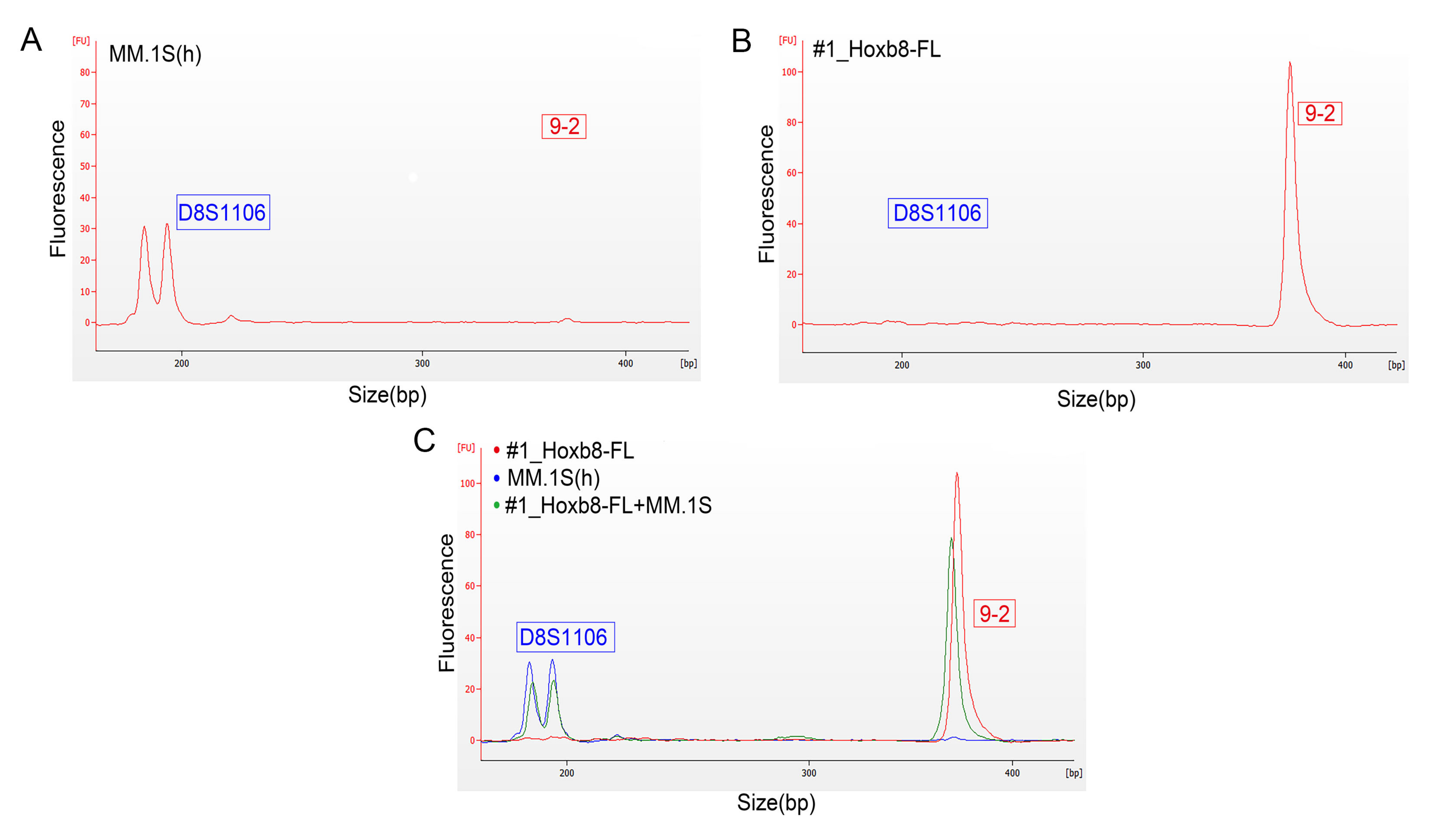
3.3. Validation of Microfluidics-Based STR Profiling in Primary Murine Pancreatic Cancer Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rieger, M.A.; Schroeder, T. Hematopoiesis. Cold Spring Harb. Perspect. Biol. 2012, 4. [Google Scholar] [CrossRef]
- Bhatlekar, S.; Fields, J.Z.; Boman, B.M. Role of HOX Genes in Stem Cell Differentiation and Cancer. Stem Cells Int. 2018, 2018, 3569493. [Google Scholar] [CrossRef]
- Redecke, V.; Wu, R.; Zhou, J.; Finkelstein, D.; Chaturvedi, V.; High, A.A.; Hacker, H. Hematopoietic progenitor cell lines with myeloid and lymphoid potential. Nat. Methods 2013, 10, 795–803. [Google Scholar] [CrossRef]
- Hammerschmidt, S.I.; Werth, K.; Rothe, M.; Galla, M.; Permanyer, M.; Patzer, G.E.; Bubke, A.; Frenk, D.N.; Selich, A.; Lange, L.; et al. CRISPR/Cas9 Immunoengineering of Hoxb8-Immortalized Progenitor Cells for Revealing CCR7-Mediated Dendritic Cell Signaling and Migration Mechanisms in vivo. Front. Immunol. 2018, 9, 1949. [Google Scholar] [CrossRef]
- Zach, F.; Mueller, A.; Gessner, A. Production and Functional Characterization of Murine Osteoclasts Differentiated from ER-Hoxb8-Immortalized Myeloid Progenitor Cells. PLoS ONE 2015, 10, e0142211. [Google Scholar] [CrossRef]
- Wang, G.G.; Calvo, K.R.; Pasillas, M.P.; Sykes, D.B.; Hacker, H.; Kamps, M.P. Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8. Nat. Methods 2006, 3, 287–293. [Google Scholar] [CrossRef]
- Graham, M.L.; Prescott, M.J. The multifactorial role of the 3Rs in shifting the harm-benefit analysis in animal models of disease. Eur. J. Pharmacol. 2015, 759, 19–29. [Google Scholar] [CrossRef]
- Freedman, L.P.; Gibson, M.C.; Ethier, S.P.; Soule, H.R.; Neve, R.M.; Reid, Y.A. Reproducibility: Changing the policies and culture of cell line authentication. Nat. Methods 2015, 12, 493–497. [Google Scholar] [CrossRef]
- Marx, V. Cell-line authentication demystified. Nat. Methods 2014, 11, 483–488. [Google Scholar] [CrossRef]
- Masters, J.R.; Thomson, J.A.; Daly-Burns, B.; Reid, Y.A.; Dirks, W.G.; Packer, P.; Toji, L.H.; Ohno, T.; Tanabe, H.; Arlett, C.F.; et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl. Acad. Sci. USA 2001, 98, 8012–8017. [Google Scholar] [CrossRef]
- Almeida, J.L.; Hill, C.R.; Cole, K.D. Mouse cell line authentication. Cytotechnology 2014, 66, 133–147. [Google Scholar] [CrossRef]
- Butler, J.M. Genetics and genomics of core short tandem repeat loci used in human identity testing. J. Forensic Sci. 2006, 51, 253–265. [Google Scholar] [CrossRef]
- Cattoretti, G.; Pasqualucci, L.; Ballon, G.; Tam, W.; Nandula, S.V.; Shen, Q.; Mo, T.; Murty, V.V.; Dalla-Favera, R. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 2005, 7, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Freitas, C.; Wittner, M.; Nguyen, J.; Rondeau, V.; Biajoux, V.; Aknin, M.L.; Gaudin, F.; Beaussant-Cohen, S.; Bertrand, Y.; Bellanne-Chantelot, C.; et al. Lymphoid differentiation of hematopoietic stem cells requires efficient Cxcr4 desensitization. J. Exp. Med. 2017, 214, 2023–2040. [Google Scholar] [CrossRef] [PubMed]
- Amend, S.R.; Valkenburg, K.C.; Pienta, K.J. Murine Hind Limb Long Bone Dissection and Bone Marrow Isolation. J. Vis. Exp. 2016. [Google Scholar] [CrossRef] [PubMed]
- Dobrovolny, P.L.; Bess, D. Optimized PCR-based detection of mycoplasma. J. Vis. Exp. 2011. [Google Scholar] [CrossRef] [PubMed]
- von Burstin, J.; Eser, S.; Paul, M.C.; Seidler, B.; Brandl, M.; Messer, M.; von Werder, A.; Schmidt, A.; Mages, J.; Pagel, P.; et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009, 137, 361–371.e5. [Google Scholar] [CrossRef] [PubMed]
- Bamopoulos, S.A.; Batcha, A.M.N.; Jurinovic, V.; Rothenberg-Thurley, M.; Janke, H.; Ksienzyk, B.; Philippou-Massier, J.; Graf, A.; Krebs, S.; Blum, H.; et al. Clinical presentation and differential splicing of SRSF2, U2AF1 and SF3B1 mutations in patients with acute myeloid leukemia. Leukemia 2020. [Google Scholar] [CrossRef]
- Agilent DNA 1000; Agilent Technologies, Inc.: Santa Clara, CA, USA, 2016; Volume G2938-90014, p. 24.
- Carlyle, J.R.; Michie, A.M.; Furlonger, C.; Nakano, T.; Lenardo, M.J.; Paige, C.J.; Zuniga-Pflucker, J.C. Identification of a novel developmental stage marking lineage commitment of progenitor thymocytes. J. Exp. Med. 1997, 186, 173–182. [Google Scholar] [CrossRef]
- Schmitt, T.M.; Zuniga-Pflucker, J.C. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity 2002, 17, 749–756. [Google Scholar] [CrossRef]
- Barallon, R.; Bauer, S.R.; Butler, J.; Capes-Davis, A.; Dirks, W.G.; Elmore, E.; Furtado, M.; Kline, M.C.; Kohara, A.; Los, G.V.; et al. Recommendation of short tandem repeat profiling for authenticating human cell lines, stem cells, and tissues. In Vitro Cell Dev. Biol. Anim. 2010, 46, 727–732. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Fillmore, H.L.; Vouri, M.; Pilkington, G.J. Brain tumor cell line authentication, an efficient alternative to capillary electrophoresis by using a microfluidics-based system. Neuro Oncol. 2014, 16, 265–273. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.; Engleitner, T.; Maresch, R.; Zukowska, M.; Lange, S.; Kaltenbacher, T.; Konukiewitz, B.; Ollinger, R.; Zwiebel, M.; Strong, A.; et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018, 554, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Almeida, J.L.; Cole, K.D.; Plant, A.L. Standards for Cell Line Authentication and Beyond. PLoS Biol. 2016, 14, e1002476. [Google Scholar] [CrossRef]
- Aboud, M.J.; Gassmann, M.; McCord, B.R. The development of mini pentameric STR loci for rapid analysis of forensic DNA samples on a microfluidic system. Electrophoresis 2010, 31, 2672–2679. [Google Scholar] [CrossRef]
- Cardoso, S.; Valverde, L.; Odriozola, A.; Elcoroaristizabal, X.; de Pancorbo, M.M. Quality standards in Biobanking: Authentication by genetic profiling of blood spots from donor’s original sample. Eur. J. Hum. Genet. 2010, 18, 848–851. [Google Scholar] [CrossRef]
- Yuille, M.; van Ommen, G.J.; Brechot, C.; Cambon-Thomsen, A.; Dagher, G.; Landegren, U.; Litton, J.E.; Pasterk, M.; Peltonen, L.; Taussig, M.; et al. Biobanking for Europe. Brief. Bioinform. 2008, 9, 14–24. [Google Scholar] [CrossRef]

| Medium | Concentration | Media/Additive | Catalog No. | Storage |
|---|---|---|---|---|
| RP-10 Medium | RPMI1640 | Gibco: 21875-034 | 4 °C | |
| 10% | FBS (not heat inactivated) | Gibco: 10270-106 | −20 °C | |
| 0.10% | Mercapto Ethanol | Gibco: 31350-010 50 mM | 4 °C | |
| 1% | Antibiotic Antimycotic | Gibco: 15240-062 | −20 °C | |
| Progenitor Outgrowth Medium | RPMI1640 | Gibco: 21875-034 | 4 °C | |
| 10% | FBS (not heat inactivated) | Gibco: 10270-106 | −20 °C | |
| 0.10% | Mercapto Ethanol | Gibco: 31350-010 50 mM | 4 °C | |
| 1% | Antibiotic Antimycotic | Gibco: 15240-062 | −20 °C | |
| 1 µM | β-estradiol | Sigma: E-2758 (stock 100 mM) | −20 °C | |
| 5% | FLT3L-SNT | from B16 melanoma cell line | −20 °C | |
| B-Cell Differentiation Medium | RPMI1640 | Gibco: 21875-034 | 4 °C | |
| 10% | FBS (not heat inactivated) | Gibco: 10270-106 | −20 °C | |
| 0.10% | Mercapto Ethanol | Gibco: 31350-010 50 mM | 4 °C | |
| 1% | Antibiotic Antimycotic | Gibco: 15240-062 | −20 °C | |
| 1 µM | β-estradiol | Sigma: E-2758 (stock 100 mM) | −20 °C | |
| 5 ng/mL | FLT3L | R & D Systems: 427-FL | −20 °C | |
| 25 ng/mL | SCF | R & D Systems: 455-MC | −20 °C | |
| 7 ng/mL | IL-7 | R & D Systems: 407-ML | −20 °C | |
| BBMM (Basal Bone Marrow Medium) | IMDM | Gibco: 12440-053 | 4 °C | |
| 30% | FBS (not heat inactivated) | Gibco: 10270-106 | −20 °C | |
| 0.20% | Mercapto Ethanol | Gibco: 31350-010 50 mM | 4 °C | |
| 0.50% | Antibiotic Antimycotic | Gibco: 15240-062 | −20 °C | |
| 1% | Glutamine | Corning: 25-005-CV | −20 °C | |
| 0.50% | BSA | Sigma-Aldrich: B6917 | 4 °C | |
| HF2 Buffer | ddH2O | Ampuwa: 250166 | RT | |
| 30% | FBS (not heat inactivated) | Gibco: 10270-106 | −20 °C | |
| 0.50% | Antibiotic Antimycotic | Gibco: 15240-062 | −20 °C | |
| 1% | HEPES | Gibco: 15630-049 | 4 °C |
| Cell Line | 18-3 | 4-2 | 6-7 | 9-2 | 15-3 | 6-4 | 12-1 | 5-5 | X-1 |
|---|---|---|---|---|---|---|---|---|---|
| #1_HoxB8FL (wild-type) | 16 | 19 | 19 | 17 | 22 | 19 | 15 | 16 | 23 |
| #2_HoxB8FL (Iμ-HA-Bcl6) | 15 | 18 | 15, 18 | 15 | 20 | 18 | 16 | 17 | 24 |
| #3_HoxB8FL (Cxcr4WHIM) | 16 | 19 | 19 | 15 | 22 | 19 | 16 | 17 | 24 |
| Cell Line | 18-3 | 4-2 | 6-7 | 9-2 | 15-3 | 6-4 | 12-1 | 5-5 | X-1 |
|---|---|---|---|---|---|---|---|---|---|
| mPDAC06 | 17, 18 | 18 | 20 | 16 | 20, 22 | 17 | 16, 20 | 14, 16 | 20, 24 |
| mPDAC09 | 17, 19 | 17, 19 | 20, 23 | 18, 22 | 20 | 18 | 16 | 16, 17 | 18, 25 |
| mPDAC95 | 17 | 19, 19 | 17 | 18, 20 | 21 | 19 | 16 | 16, 17 | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, Y.; Singh, N.; Bamopoulos, S.; Gjerga, E.; Schmalbrock, L.K.; Balabanian, K.; Schick, M.; Keller, U.; Wirth, M. Authentication of Primary Murine Cell Lines by a Microfluidics-Based Lab-On-Chip System. Biomedicines 2020, 8, 590. https://doi.org/10.3390/biomedicines8120590
Hong Y, Singh N, Bamopoulos S, Gjerga E, Schmalbrock LK, Balabanian K, Schick M, Keller U, Wirth M. Authentication of Primary Murine Cell Lines by a Microfluidics-Based Lab-On-Chip System. Biomedicines. 2020; 8(12):590. https://doi.org/10.3390/biomedicines8120590
Chicago/Turabian StyleHong, Yingfen, Nikita Singh, Stefanos Bamopoulos, Enio Gjerga, Laura K. Schmalbrock, Karl Balabanian, Markus Schick, Ulrich Keller, and Matthias Wirth. 2020. "Authentication of Primary Murine Cell Lines by a Microfluidics-Based Lab-On-Chip System" Biomedicines 8, no. 12: 590. https://doi.org/10.3390/biomedicines8120590
APA StyleHong, Y., Singh, N., Bamopoulos, S., Gjerga, E., Schmalbrock, L. K., Balabanian, K., Schick, M., Keller, U., & Wirth, M. (2020). Authentication of Primary Murine Cell Lines by a Microfluidics-Based Lab-On-Chip System. Biomedicines, 8(12), 590. https://doi.org/10.3390/biomedicines8120590






