Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target
Abstract
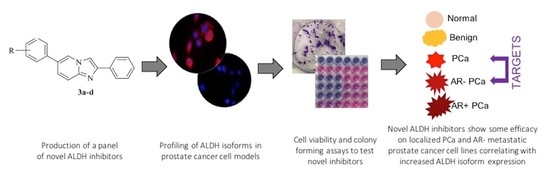
1. Introduction
2. Experimental Section
2.1. Chemistry
2.1.1. Materials and Methods
2.1.2. Synthesis of 6-Bromo-2-phenylimidazo [1,2-a]pyridine, 2
2.1.3. General Procedure for the Synthesis of 2,6-(Substituted)diphenylimidazo[1,2-a]pyridines, 3a–d
2.2. Biology
2.2.1. Materials and Methods
2.2.2. Protein Extraction
2.2.3. Protein Quantification
2.2.4. SDS-PAGE Gel Electrophoresis
2.2.5. Western Blot
2.2.6. Immunofluorescence
2.2.7. Cell Viability
2.2.8. Colony-Forming Assay
2.2.9. Real-Time Quantitative PCR
2.2.10. Quantification and Statistical Analysis
3. Results and Discussion
3.1. ALDH Expression Analysis
3.2. ALDH Inhibitors of the Imidazo[1,2-a]pyridine Series Have Anti-Proliferative Effects against Different Prostatic Tissue-Derived Cell Lines
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rawla, P. Epidemiology of prostate cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef] [PubMed]
- Center, M.M.; Jemal, A.; Lortet-Tieulent, J.; Ward, E.; Ferlay, J.; Brawley, O.; Bray, F. International variation in prostate cancer incidence and mortality rates. Eur. Urol. 2012, 61, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, S.; Ramamurthy, V.P.; Njar, V.C.O. Dissecting major signaling pathways in prostate cancer development and progression: Mechanisms and novel therapeutic targets. J. Steroid Biochem. Mol. Biol. 2017, 166, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Wu, Y.-M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castrate resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Harris, W.P.; Mostaghel, E.A.; Nelson, P.S.; Montgomery, B. Androgen deprivation therapy: Progress in understanding mechanisms of resistance and optimizing androgen depletion. Nat. Clin. Pract. Urol. 2009, 6, 76–85. [Google Scholar] [CrossRef]
- Drazer, M.W.; Stadler, W.M. The role of testosterone in the treatment of castration resistant prostate cancer. Cancer J. Sudbury Mass. 2016, 22, 330–333. [Google Scholar] [CrossRef][Green Version]
- Scott, L.J. Enzalutamide: A review in castration-resistant prostate cancer. Drugs 2018, 78, 1913–1924. [Google Scholar] [CrossRef]
- Hoy, S.M. Abiraterone acetate: A review of its use in patients with metastatic castration-resistant prostate cancer. Drugs 2013, 73, 2077–2091. [Google Scholar] [CrossRef]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Théodore, C.; James, N.D.; Turesson, I.; et al. for the TAX 327 Investigators. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef]
- Tsao, C.-K.; Cutting, E.; Martin, J.; Oh, W.K. The role of cabazitaxel in the treatment of metastatic castration-resistant prostate cancer. Ther. Adv. Urol. 2014, 6, 97–104. [Google Scholar] [CrossRef]
- Parker, C.; Heidenreich, A.; Nilsson, S.; Shore, N. Current approaches to incorporation of radium-223 in clinical practice. Prostate Cancer Prostatic Dis. 2018, 21, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Handy, C.E.; Antonarakis, E.S. Sipuleucel-T for the treatment of prostate cancer: Novel insights and future directions. Future Oncol. 2017, 14, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Frame, F.M.; Noble, A.R.; Klein, S.; Walker, H.F.; Suman, R.; Kasprowicz, R.; Mann, V.M.; Simms, M.S.; Maitland, N.J. Tumor heterogeneity and therapy resistance—Implications for future treatments of prostate cancer. J. Cancer Metastasis Treat. 2017, 3, 302–314. [Google Scholar] [CrossRef]
- Maitland, N.J.; Frame, F.M.; Rane, J.K.; Erb, H.H.; Packer, J.R.; Archer, L.K.; Pellacani, D. Resolution of cellular heterogeneity in human prostate cancers: Implications for diagnosis and treatment. Adv. Exp. Med. Biol. 2019, 1164, 207–224. [Google Scholar]
- McKenna, D.J.; Errington, R.; Pors, K. Current challenges and opportunities in treating hypoxic prostate tumors. J. Cancer Metastasis Treat. 2018, 4, 1–13. [Google Scholar] [CrossRef][Green Version]
- Carnero, A.; Garcia-Mayea, Y.; Mir, C.; Lorente, J.; Rubio, I.T.; Leonart, M.E. The cancer stem-cell signaling network and resistance to therapy. Cancer Treat. Rev. 2016, 49, 25–36. [Google Scholar] [CrossRef]
- Wang, T.; Shigdar, S.; Gantier, M.P.; Hou, Y.; Wang, L.; Li, Y.; Al Shamaileh, H.; Yin, W.; Zhou, S.-F.; Zhao, X.; et al. Cancer stem cell targeted therapy: Progress amid controversies. Oncotarget 2015, 6, 44191–44206. [Google Scholar] [CrossRef]
- Maitland, N.J.; Collins, A.T. Prostate cancer stem cells: A new target for therapy. J. Clin. Oncol. 2008, 26, 2862–2870. [Google Scholar] [CrossRef]
- Patrawala, L.; Calhoun, T.; Schneider-Broussard, R.; Li, H.; Bhatia, B.; Tang, S.; Reilly, J.G.; Chandra, D.; Zhou, J.; Claypool, K.; et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene 2006, 25, 1696–1708. [Google Scholar] [CrossRef]
- Collins, A.T.; Habib, F.K.; Maitland, N.J.; Neal, D.E. Identification and isolation of human prostate epithelial stem cells based on alpha(2)beta(1)-integrin expression. J. Cell Sci. 2001, 114, 3865–3872. [Google Scholar]
- Richardson, G.D.; Robson, C.N.; Lang, S.H.; Neal, D.E.; Maitland, N.J.; Collins, A.T. CD133, a novel marker for human prostatic epithelial stem cells. J. Cell Sci. 2004, 117, 3539–3545. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.I.M.; Sadiq, M.; Frame, F.M.; Maitland, N.J.; Pors, K. Expression and regulation of aldehyde dehydrogenases in prostate cancer. J. Cancer Metastasis Treat. 2018, 4, 1–17. [Google Scholar]
- Vassalli, G. Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 13, 3904645. [Google Scholar] [CrossRef] [PubMed]
- Cojoc, M.; Peitzsch, C.; Kurth, I.; Trautmann, F.; Kunz-Schughart, L.A.; Telegeev, G.D.; Stakhovsky, E.A.; Walker, J.R.; Simin, K.; Lyle, S.; et al. Aldehyde dehydrogenase is regulated by β-catenin/TCF and promotes radioresistance in prostate cancer progenitor cells. Cancer Res. 2015, 75, 1482–1494. [Google Scholar] [CrossRef]
- Pors, K.; Moreb, J.S. Aldehyde dehydrogenases in cancer: An opportunity for biomarker and drug development? Drug Discov. Today 2014, 19, 1953–1963. [Google Scholar] [CrossRef]
- Le Magnen, C.; Bubendorf, L.; Rentsch, C.A.; Mengus, C.; Gsponer, J.; Zellweger, T.; Rieken, M.; Thalmann, G.N.; Cecchini, M.G.; Germann, M.; et al. Characterization and clinical relevance of ALDHbright populations in prostate cancer. Clin. Cancer Res. 2013, 19, 5361–5371. [Google Scholar] [CrossRef]
- Kim, H.; Lapointe, J.; Kaygusuz, G.; Ong, D.E.; Li, C.; van de Rijn, M.; Brooks, J.D.; Pollack, J.R. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005, 65, 8118–8124. [Google Scholar] [CrossRef]
- van den Hoogen, C.; van der Horst, G.; Cheung, H.; Buijs, J.T.; Lippitt, J.M.; Guzmán-Ramírez, N.; Hamdy, F.C.; Eaton, C.L.; Thalmann, G.N.; Cecchini, M.G.; et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2020, 70, 5163–5173. [Google Scholar] [CrossRef]
- Li, T.; Su, Y.; Mei, Y.; Leng, Q.; Leng, B.; Liu, Z.; Stass, S.A.; Jiang, F. ALDH1A1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab. Investig. 2010, 90, 234–244. [Google Scholar] [CrossRef]
- Quattrini, L.; Gelardi, E.L.M.; Coviello, V.; Sartini, S.; Ferraris, D.M.; Mori, M.; Nakano, I.; Garavaglia, S.; La Motta, C. Imidazo[1,2-a]pyridine derivatives as aldehyde dehydrogenase inhibitors: Novel chemotypes to target glioblastoma stem cells. J. Med. Chem. 2020, 14, 4603–4616. [Google Scholar] [CrossRef]
- Quattrini, L.; Gelardi, E.L.M.; Petrarolo, G.; Colombo, G.; Ferraris, D.M.; Picarazzi, F.; Rizzi, M.; Garavaglia, S.; La Motta, C. Progress in the field of aldehyde dehydrogenase inhibitors: Novel imidazo[1,2-a]pyridines against the 1A family. ACS Med. Chem. Lett. 2020, 11, 963–970. [Google Scholar] [CrossRef] [PubMed]
- Ulukaya, E.; Frame, F.M.; Cevatemre, B.; Pellacani, D.; Walker, H.; Mann, V.M.; Simms, M.S.; Stower, M.J.; Yilmaz, V.T.; Maitland, N.J. Differential cytotoxic activity of a novel palladium based compound on prostate cell lines, primary prostate epithelial cells and prostate stem cells. PLoS ONE 2013, 8, e64278. [Google Scholar] [CrossRef]
- Frame, F.M.; Pellacani, D.; Collins, A.T.; Maitland, N.J. Harvesting human prostate tissue material and culturing primary prostate epithelial cells. Methods Mol. Biol. 2016, 1443, 181–201. [Google Scholar] [PubMed]
- Frame, F.M.; Pellacani, D.; Collins, A.T.; Simms, M.S.; Mann, V.M.; Jones, G.D.D.; Meuth, M.; Bristow, R.G.; Maitland, N.J. HDAC inhibitor confers radiosensitivity to prostate stem-like cells. Br. J. Cancer 2013, 109, 3023–3033. [Google Scholar] [CrossRef] [PubMed]
- Pippione, A.C.; Boschi, D.; Pors, K.; Oliaro-Bosso, S.; Lolli, M.L. Androgen-AR axis in primary and metastatic prostate cancer: Chasing steroidogenic enzymes for therapeutic intervention. J. Cancer Metastasis Treat. 2017, 3, 328–361. [Google Scholar] [CrossRef]
- Pors, K. Topic: How does the prostate cancer microenvironment affect the metastatic process and/or treatment outcome? J. Cancer Metastasis Treat. 2018, 4, 11. [Google Scholar]
- Di Zazzo, E.; Galasso, G.; Giovannelli, P.; Di Donato, M.; Di Santi, A.; Cernera, G.; Rossi, V.; Abbondanza, C.; Moncharmont, B.; Agostino Sinisi, A.; et al. Prostate cancer stem cells: The role of androgen and estrogen receptors. Oncotarget 2016, 7, 193–208. [Google Scholar] [CrossRef]
- Clark, D.W.; Palle, K. Aldehyde dehydrogenases in cancer stem cells: Potential as therapeutic targets. Ann. Transl. Med. 2016, 4, 518. [Google Scholar] [CrossRef]




| Company and Code | Antibody | Concentration |
|---|---|---|
| Cell Signaling Tech, D9J7R | ALDH1A1 1° Ab | 1:500 |
| Gene Tex, GTX110784 | ALDH1A3 1° Ab | 1:750 |
| Abcam, AB9485 | GAPDH 1° Ab | 1:10,000 |
| Cell Signaling Tech, 7074S | HRP-linked 2° Antibody | 1:10,000 |
| Company and Code | Antibody | Concentration |
|---|---|---|
| Cell Signaling Tech, D9J7R | ALDH1A1 1° Ab | 1:400 |
| Gene Tex, GTX110784 | ALDH1A3 1° Ab | 1:400 |
| Abcam, AB175471 | Goat Anti-Rabbit IgG H&L (Alexa Fluor® 568) 2° Antibody | 1:10,000 |
| EC50 (nM a) | ||||||
|---|---|---|---|---|---|---|
| N | R | P4E6 | PC3 | LNCaP | PNT2-C2 | BPH1 |
| 3a | H | 60.72 | 239.4 | 314.8 | 422.6 | 113.0 |
| 3b | 4-F | 4.038 | 70.92 | 240.0 | 217.0 | 41.47 |
| 3c | 4-Cl | 33.39 | 321.9 | 323.1 | 324.7 | 416.0 |
| 3d | 3-CN | 439.6 | n.t. b | 7180.0 | 1718.0 | 351.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quattrini, L.; Sadiq, M.; Petrarolo, G.; Maitland, N.J.; Frame, F.M.; Pors, K.; La Motta, C. Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target. Biomedicines 2020, 8, 569. https://doi.org/10.3390/biomedicines8120569
Quattrini L, Sadiq M, Petrarolo G, Maitland NJ, Frame FM, Pors K, La Motta C. Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target. Biomedicines. 2020; 8(12):569. https://doi.org/10.3390/biomedicines8120569
Chicago/Turabian StyleQuattrini, Luca, Maria Sadiq, Giovanni Petrarolo, Norman J. Maitland, Fiona M. Frame, Klaus Pors, and Concettina La Motta. 2020. "Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target" Biomedicines 8, no. 12: 569. https://doi.org/10.3390/biomedicines8120569
APA StyleQuattrini, L., Sadiq, M., Petrarolo, G., Maitland, N. J., Frame, F. M., Pors, K., & La Motta, C. (2020). Aldehyde Dehydrogenases and Prostate Cancer: Shedding Light on Isoform Distribution to Reveal Druggable Target. Biomedicines, 8(12), 569. https://doi.org/10.3390/biomedicines8120569