Transcriptome Analysis of Acute Phase Liver Graft Injury in Liver Transplantation
Abstract
1. Introduction
2. Experimental Section
2.1. Clinical specimens
2.2. RNA sequencing (RNA-Seq) and data processing
2.3. Bioinformatics analyses
2.4. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
2.5. Statistical analyses
3. Results
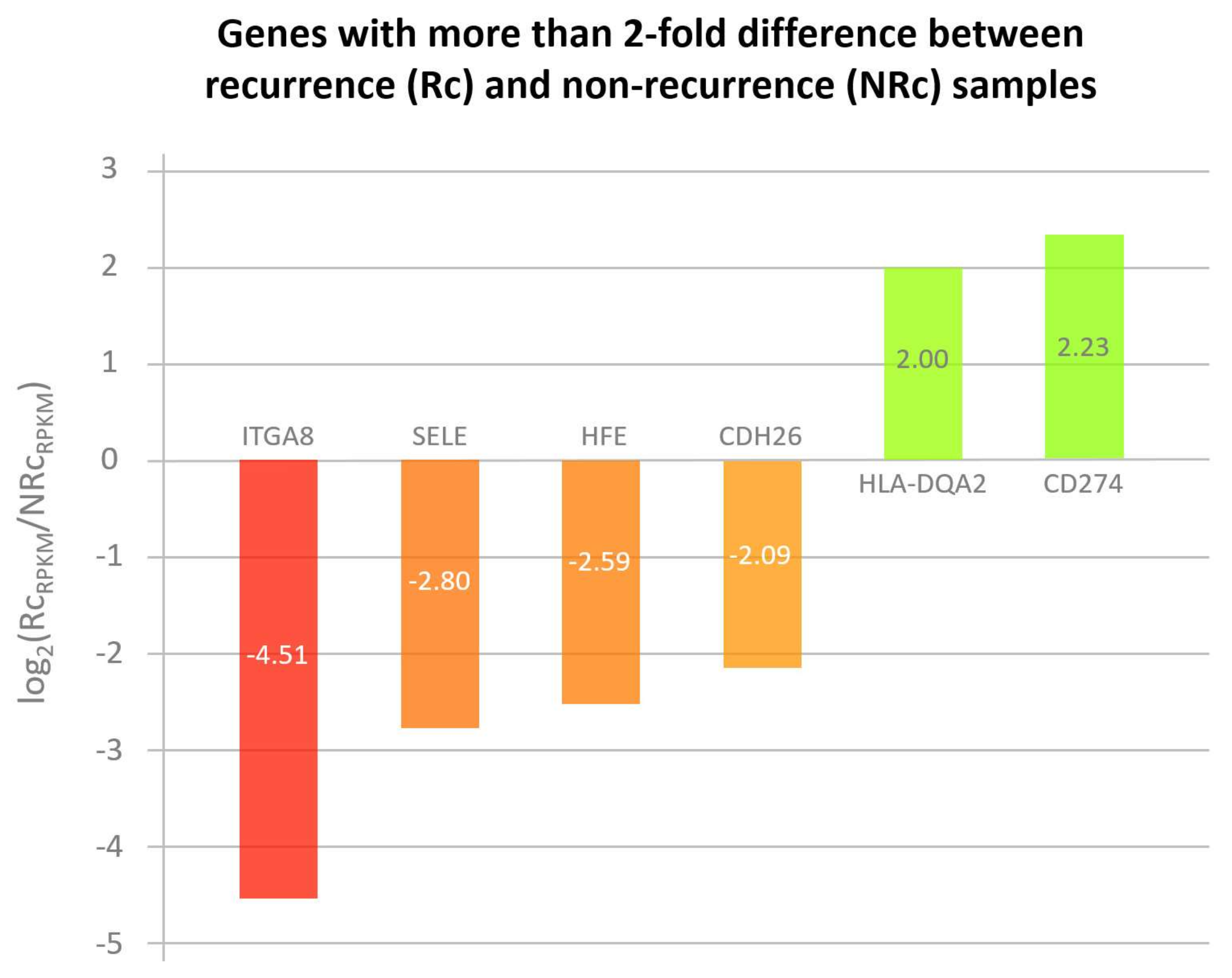
3.1. Cell adhesion molecules-related pathways in acute phase liver graft injury
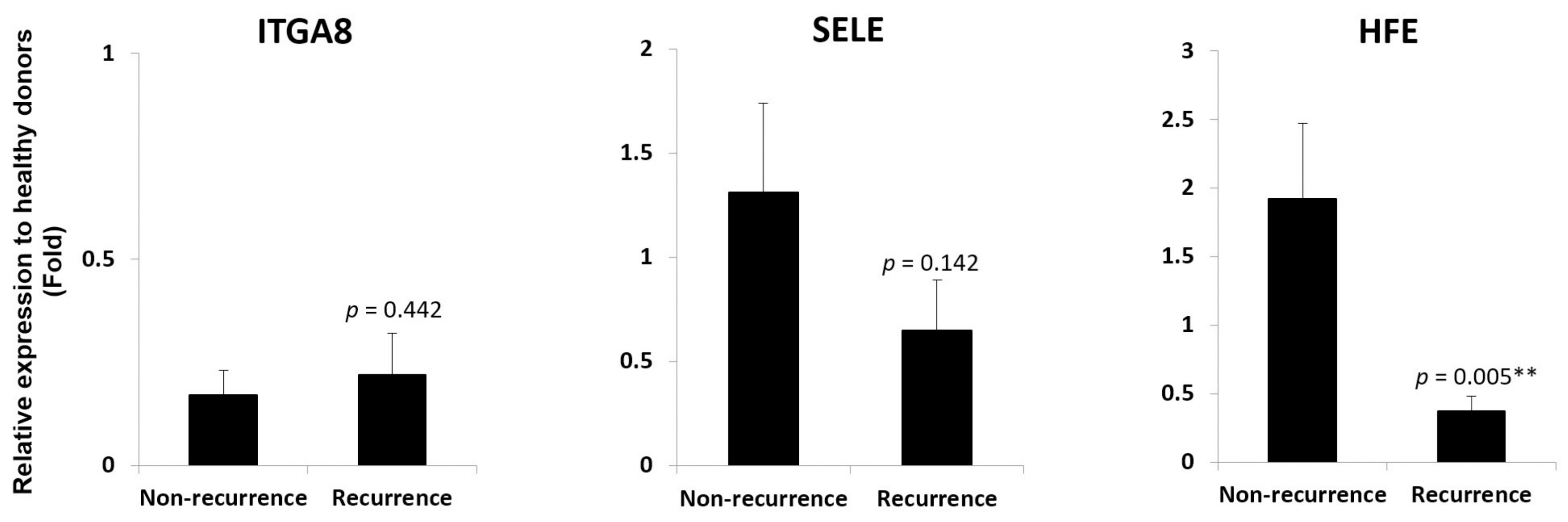
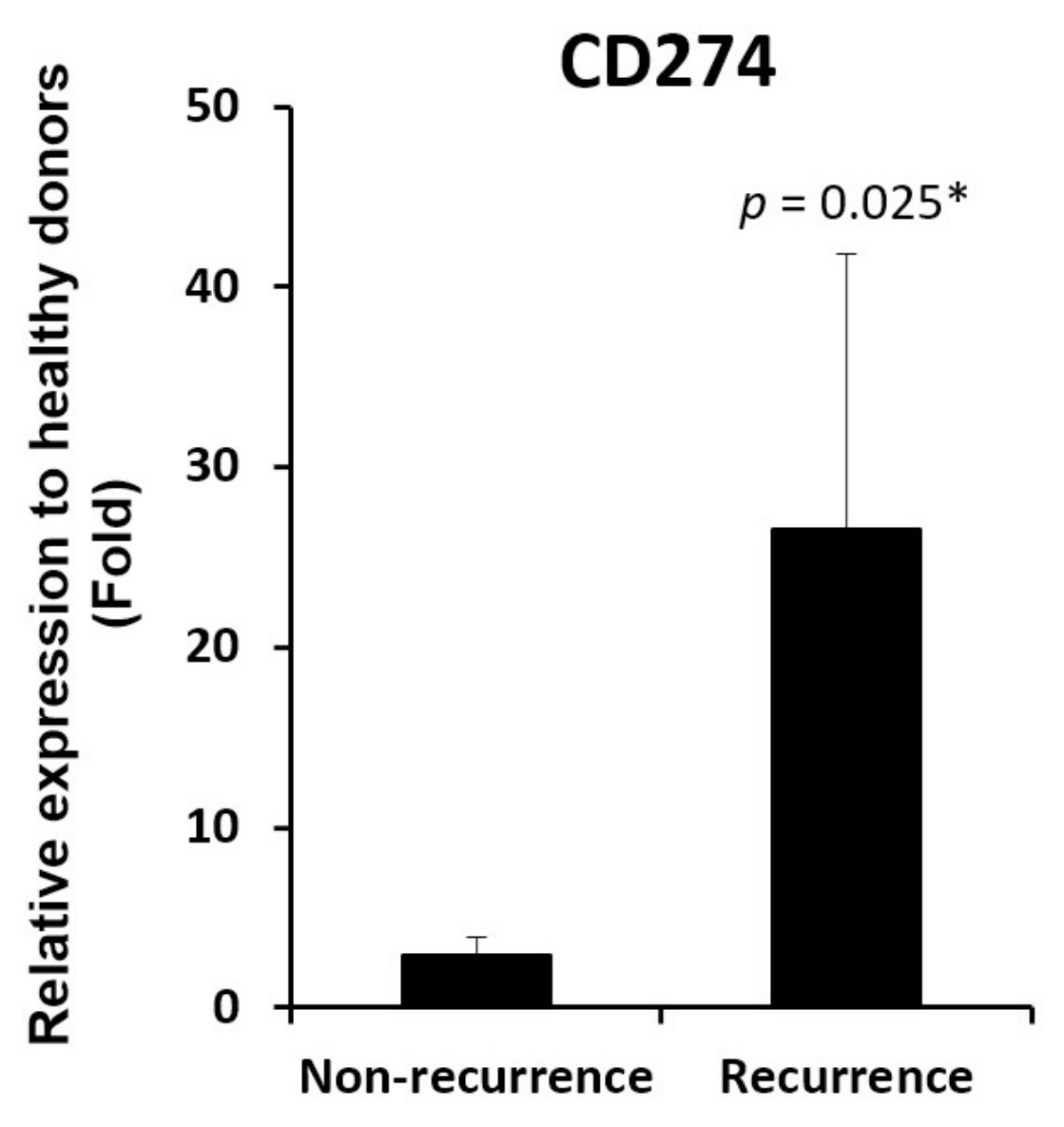
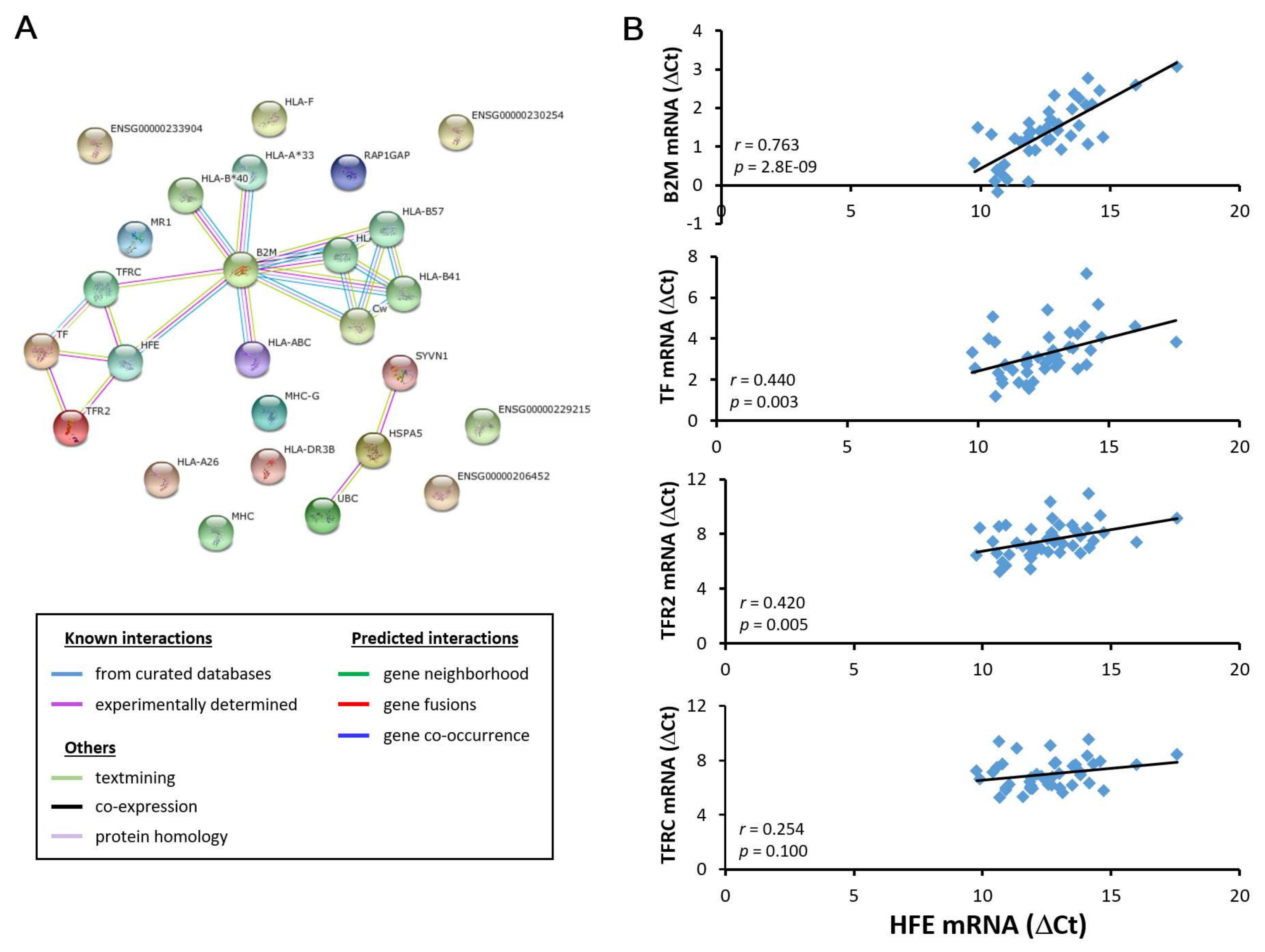
3.2. HFE and CD274 are two cell adhesion molecules with differential involvement in acute phase liver graft injury
3.3. HFE- and CD274-related molecules in acutely injured liver grafts
4. Discussion
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Lo, C.M. Liver transplantation in 2012: Transplantation for liver cancer—More with better results. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Busuttil, R.W.; Kupiec-Weglinski, J.W. Liver ischemia and reperfusion injury: New insights into mechanisms of innate-adaptive immune-mediated tissue inflammation. Am. J. Transplant. 2011, 11, 1563–1569. [Google Scholar] [CrossRef] [PubMed]
- Man, K.; Lo, C.M.; Xiao, J.W.; Ng, K.T.; Sun, B.S.; Ng, I.O.; Cheng, Q.; Sun, C.K.; Fan, S.T. The significance of acute phase small-for-size graft injury on tumor growth and invasiveness after liver transplantation. Ann. Surg. 2008, 247, 1049–1057. [Google Scholar] [CrossRef] [PubMed]
- Man, K.; Shih, K.C.; Ng, K.T.; Xiao, J.W.; Guo, D.Y.; Sun, C.K.; Lim, Z.X.; Cheng, Q.; Liu, Y.; Fan, S.T.; et al. Molecular signature linked to acute phase injury and tumor invasiveness in small-for-size liver grafts. Ann. Surg. 2010, 251, 1154–1161. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.C.; Ng, K.T.; Shao, Y.; Geng, W.; Xiao, J.W.; Liu, H.; Li, C.X.; Liu, X.B.; Ma, Y.Y.; Yeung, W.H.; et al. Post-transplant endothelial progenitor cell mobilization via CXCL10/CXCR3 signaling promotes liver tumor growth. J. Hepatol. 2014, 60, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Ling, C.C.; Shao, Y.; Xu, A.; Li, X.C.; Ng, K.T.; Liu, X.B.; Ma, Y.Y.; Qi, X.; Liu, H.; et al. CXCL10/CXCR3 signaling mobilized-regulatory T cells promote liver tumor recurrence after transplantation. J. Hepatol. 2016, 65, 944–952. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Ng, K.T.; Shao, Y.; Li, C.X.; Geng, W.; Ling, C.C.; Ma, Y.Y.; Liu, X.B.; Liu, H.; Liu, J.; et al. The Clinical Significance and Potential Therapeutic Role of GPx3 in Tumor Recurrence after Liver Transplantation. Theranostics 2016, 6, 1934–1946. [Google Scholar] [CrossRef] [PubMed]
- Asaoka, T.; Kato, T.; Marubashi, S.; Dono, K.; Hama, N.; Takahashi, H.; Kobayashi, S.; Takeda, Y.; Takemasa, I.; Nagano, H.; et al. Differential transcriptome patterns for acute cellular rejection in recipients with recurrent hepatitis C after liver transplantation. Liver Transplant. 2009, 15, 1738–1749. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.; Xu, A.; Cheng, Q.; Guo, D.Y.; Lim, Z.X.; Sun, C.K.; Fung, J.H.; Poon, R.T.; Fan, S.T.; Lo, C.M.; et al. Clinical relevance and therapeutic potential of angiopoietin-like protein 4 in hepatocellular carcinoma. Mol. Cancer 2014, 13, 196. [Google Scholar] [CrossRef] [PubMed]
- Ng, K.T.; Lo, C.M.; Guo, D.Y.; Qi, X.; Li, C.X.; Geng, W.; Liu, X.B.; Ling, C.C.; Ma, Y.Y.; Yeung, W.H.; et al. Identification of transmembrane protein 98 as a novel chemoresistance-conferring gene in hepatocellular carcinoma. Mol. Cancer Ther. 2014, 13, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Cescon, M.; Bertuzzo, V.R.; Ercolani, G.; Ravaioli, M.; Odaldi, F.; Pinna, A.D. Liver transplantation for hepatocellular carcinoma: Role of inflammatory and immunological state on recurrence and prognosis. World J. Gastroenterol. 2013, 19, 9174–9182. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Man, K.; Lo, C.M. The impact of liver graft injury on cancer recurrence posttransplantation. Transplantation 2017, 101, 2665–2670. [Google Scholar] [CrossRef] [PubMed]
- Jaeschke, H. Cellular adhesion molecules: Regulation and functional significance in the pathogenesis of liver diseases. Am. J. Physiol. 1997, 273, G602–G611. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Mier, G.; Toledo-Pereyra, L.H.; Ward, P.A. Adhesion molecules in liver ischemia and reperfusion. J. Surg. Res. 2000, 94, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Viebahn, R.; Thoma, M.; Kinder, O.; Schenk, M.; Lauchart, W.; Becker, H.D. Analysis of intragraft adhesion molecules and their release in clinical liver transplantation: Impact of reperfusion injury. Transplant. Proc. 1998, 30, 4257–4259. [Google Scholar] [CrossRef]
- Li, C.X.; Ng, K.T.; Shao, Y.; Liu, X.B.; Ling, C.C.; Ma, Y.Y.; Geng, W.; Qi, X.; Cheng, Q.; Chung, S.K.; et al. The inhibition of aldose reductase attenuates hepatic ischemia-reperfusion injury through reducing inflammatory response. Ann. Surg. 2014, 260, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Li, C.X.; Lo, C.M.; Lian, Q.; Ng, K.T.; Liu, X.B.; Ma, Y.Y.; Qi, X.; Yeung, O.W.; Tergaonkar, V.; Yang, X.X.; et al. Repressor and activator protein accelerates hepatic ischemia reperfusion injury by promoting neutrophil inflammatory response. Oncotarget 2016, 7, 27711–27723. [Google Scholar] [CrossRef] [PubMed]
- Weston, C.; Connor, J. Evidence for the influence of the iron regulatory MHC class I molecule HFE on tumor progression in experimental models and clinical populations. Transl. Oncog. 2014, 6, 1–12. [Google Scholar]
- Brissot, P.; Bardou-Jacquet, E.; Jouanolle, A.M.; Loreal, O. Iron disorders of genetic origin: A changing world. Trends. Mol. Med. 2011, 17, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Kew, M.C. Hepatic iron overload and hepatocellular carcinoma. Liver Cancer 2014, 3, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kowdley, K.V.; Brandhagen, D.J.; Gish, R.G.; Bass, N.M.; Weinstein, J.; Schilsky, M.L.; Fontana, R.J.; McCashland, T.; Cotler, S.J.; Bacon, B.R.; et al. National Hemochromatosis Transplant Registry. Survival after liver transplantation in patients with hepatic iron overload: The national hemochromatosis transplant registry. Gastroenterology 2005, 129, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Brandhagen, D.J.; Alvarez, W.; Therneau, T.M.; Kruckeberg, K.E.; Thibodeau, S.N.; Ludwig, J.; Porayko, M.K. Iron overload in cirrhosis-HFE genotypes and outcome after liver transplantation. Hepatology 2000, 31, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Barton, J.C.; Edwards, C.Q.; Acton, R.T. HFE gene: Structure, function, mutations, and associated iron abnormalities. Gene 2015, 574, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Gianchecchi, E.; Delfino, D.V.; Fierabracci, A. Recent insights into the role of the PD-1/PD-L1 pathway in immunological tolerance and autoimmunity. Autoimmun. Rev. 2013, 12, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Carreno, B.M.; Collins, M. The B7 family of ligands and its receptors: New pathways for costimulation and inhibition of immune responses. Annu. Rev. Immunol. 2002, 20, 29–53. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, M.L.; Buhler, L.; Gibbons, C.; Tian, J.; Rodriguez-Barbosa, J.I. PD-1/PD-L1, PD-1/PD-L2, and other co-inhibitory signaling pathways in transplantation. Transpl. Int. 2008, 21, 1015–1028. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, N.P.; Wu, H.; Ng, K.T.P.; Luo, R.; Lam, T.-W.; Lo, C.-M.; Man, K. Transcriptome Analysis of Acute Phase Liver Graft Injury in Liver Transplantation. Biomedicines 2018, 6, 41. https://doi.org/10.3390/biomedicines6020041
Lee NP, Wu H, Ng KTP, Luo R, Lam T-W, Lo C-M, Man K. Transcriptome Analysis of Acute Phase Liver Graft Injury in Liver Transplantation. Biomedicines. 2018; 6(2):41. https://doi.org/10.3390/biomedicines6020041
Chicago/Turabian StyleLee, Nikki P., Haiyang Wu, Kevin T.P. Ng, Ruibang Luo, Tak-Wah Lam, Chung-Mau Lo, and Kwan Man. 2018. "Transcriptome Analysis of Acute Phase Liver Graft Injury in Liver Transplantation" Biomedicines 6, no. 2: 41. https://doi.org/10.3390/biomedicines6020041
APA StyleLee, N. P., Wu, H., Ng, K. T. P., Luo, R., Lam, T.-W., Lo, C.-M., & Man, K. (2018). Transcriptome Analysis of Acute Phase Liver Graft Injury in Liver Transplantation. Biomedicines, 6(2), 41. https://doi.org/10.3390/biomedicines6020041




