Genomic and Structural Investigation of Mutations in Biotinidase (BTD) Gene Deficiency in Greater Middle Eastern Cohort: Insights from Molecular Dynamics Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Pathogenicity and Evolutionary Conservation Analysis
2.2. Variant Prioritization and In Silico Prediction Tools
2.3. Prediction of Deleterious nsSNPs
2.4. Prediction of Destabilizing nsSNPs
2.5. In Silico Structural Modeling and Molecular Dynamics Simulation
2.6. Modelled Structure of Biotinidase
2.7. Secondary Structure and Physicochemical Properties
2.8. Salt Bridge Analysis
2.9. Molecular Dynamics (MD) Simulation Studies
3. Results
3.1. Analysis of BTD Variants in Qatar Population
3.2. Conservation Analysis
3.3. Pathogenicity and Stability Prediction
3.4. Physicochemical Property Analysis
3.5. Secondary Structure Architecture and Catalytic Site Localization in BTD
3.6. Variant Prioritization for Structural Modeling and Molecular Dynamics Analysis
3.7. Quality Assessment of Native Protein and Single Mutants
3.8. Domain Organization and Mutant Localization in BTD
3.9. Structural Organization and Catalytic Site Topology of BTD
3.10. Structural Analysis of Missense Mutations in BTD
3.11. Impact of Missense Mutations on Salt Bridge Networks
3.12. Electrostatic Potential and Binding Affinity Analysis
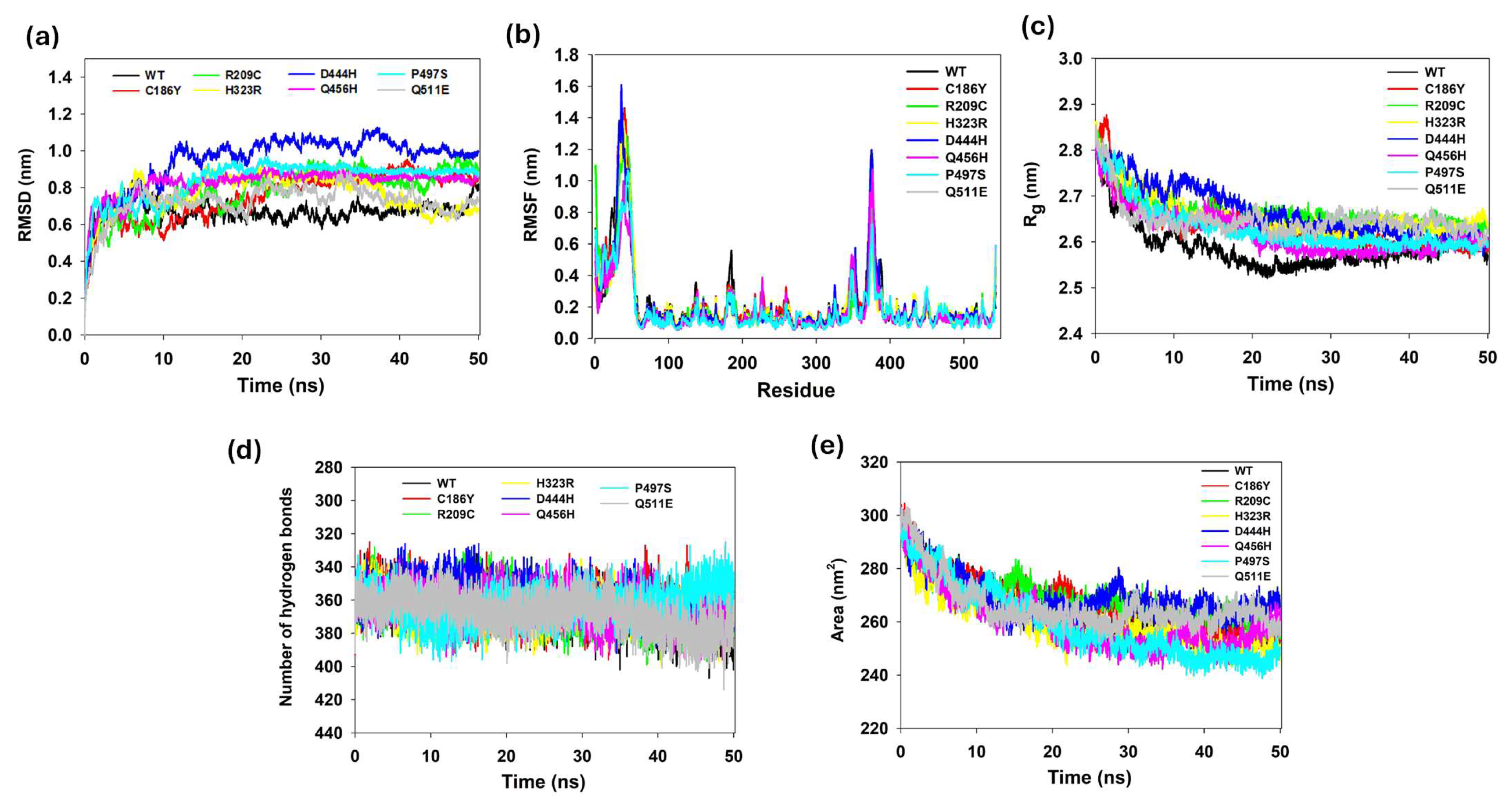
3.13. Protein Structure Conformational Flexibility and Stability Analyses
3.14. Hydrogen Bond Analysis of WT and Single Mutants
3.15. Solvent Accessibility and Surface Dynamics
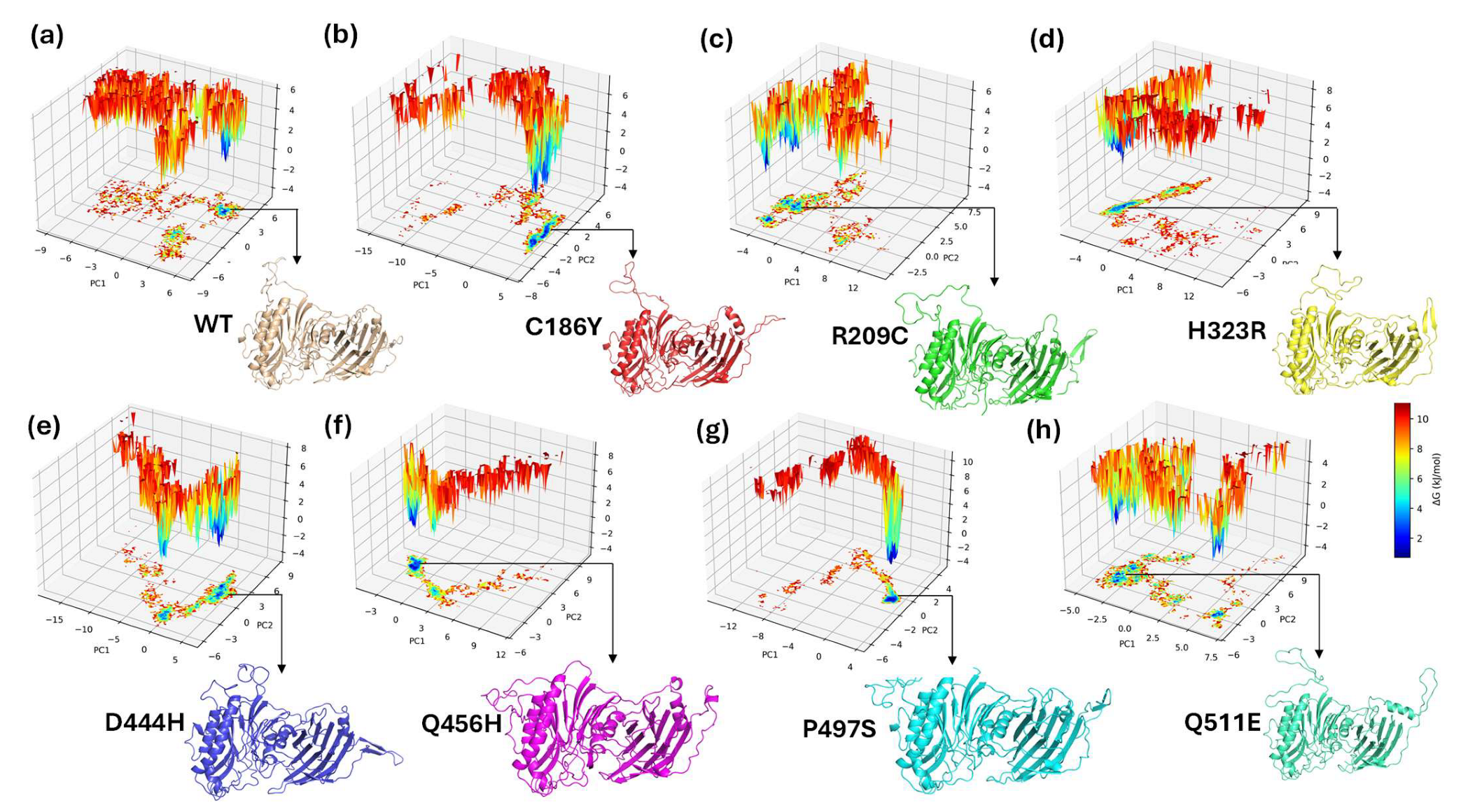
3.16. Principal Component Analysis (PCA) of Conformational Dynamics
3.17. Covariance Matrix Analysis of Residue Correlations in WT and Mutant BTDs
3.18. Free Energy Landscape of WT and Mutant BTDs
3.19. Clinical, Biochemical, and Genetic Findings of BTD Variants in Middle Eastern Countries
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BD | Biotinidase deficiency |
| BTD | Biotinidase |
| CADD | Combined annotation-dependent depletion |
| ESBRI | Electrostatic salt bridge |
| FEL | Free energy landscape |
| GRAVY | Grand average of hydropathy |
| LINCS | Linear constraint solver |
| MD | Molecular dynamics |
| NBS | Newborn screening |
| NMD | Nonsense-mediated decay |
| PCA | Principal component analysis |
| PDB | Protein Data Bank |
| PhD-SNP | Predictor of human deleterious SNPs |
| PME | Particle mesh Ewald |
| PolyPhen-2 | Polymorphism Phenotyping v2 |
| QGP | Qatar Genome Program |
| Rg | Radius of gyration |
| RMSD | Root mean square deviation |
| RMSF | Root mean square fluctuation |
| SASA | Solvent-accessible surface area |
| SIFT | Sorting Intolerant from Tolerant |
| SNAP | Screening for non-acceptable polymorphisms |
| SNP | Single-nucleotide polymorphism |
| SPDB | Swiss Protein Data Bank |
| SV | Structural variant |
References
- Wolf, B. Disorders of Biotin Metabolism. The Metabolic Basis of Inherited Disease; Academic Press: Cambridge, MA, USA, 1989; pp. 2083–2103. [Google Scholar]
- Wolf, B.; Grier, R.E.; Parker, W.D.; Goodman, S.I.; Allen, R.J. Deficient biotinidase activity in late-onset multiple carboxylase deficiency. N. Engl. J. Med. 1983, 308, 161. [Google Scholar] [PubMed]
- Knight, H.C.; Reynolds, T.R.; Meyers, G.A.; Pomponio, R.J.; Buck, G.A.; Wolf, B. Structure of the human biotinidase gene. Mamm. Genome 1998, 9, 327–330. [Google Scholar] [CrossRef] [PubMed]
- Pace, H.C.; Brenner, C. The nitrilase superfamily: Classification, structure and function. Genome Biol. 2001, 2, reviews0001-1. [Google Scholar] [CrossRef] [PubMed]
- Swango, K.; Hymes, J.; Brown, P.; Wolf, B. Amino acid homologies between human biotinidase and bacterial aliphatic amidases: Putative identification of the active site of biotinidase. Mol. Genet. Metab. 2000, 69, 111–115. [Google Scholar] [CrossRef] [PubMed]
- Dobrowolski, S.F.; Angeletti, J.; Banas, R.A.; Naylor, E.W. Real time PCR assays to detect common mutations in the biotinidase gene and application of mutational analysis to newborn screening for biotinidase deficiency. Mol. Genet. Metab. 2003, 78, 100–107. [Google Scholar] [CrossRef]
- Sivri, H.S.K.; Genç, G.A.; Tokatlı, A.; Dursun, A.; Coşkun, T.; Aydın, H.İ.; Sennaroğlu, L.; Belgin, E.; Jensen, K.; Wolf, B. Hearing loss in biotinidase deficiency: Genotype-phenotype correlation. J. Pediatr. 2007, 150, 439–442. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase deficiency:“if you have to have an inherited metabolic disease, this is the one to have”. Genet. Med. 2012, 14, 565–575. [Google Scholar] [CrossRef]
- Wolf, B. Clinical issues and frequent questions about biotinidase deficiency. Mol. Genet. Metab. 2010, 100, 6–13. [Google Scholar] [CrossRef]
- Wolf, B. The neurology of biotinidase deficiency. Mol. Genet. Metab. 2011, 104, 27–34. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase Deficiency. In GeneReviews® [Internet]; Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 2023. [Google Scholar] [PubMed]
- Wolf, B. Biotinidase deficiency: New directions and practical concerns. Curr. Treat. Options Neurol. 2003, 5, 321–328. [Google Scholar] [CrossRef]
- Wolf, B.; Grier, R.; Secor McVoy, J.; Heard, G. Biotinidase deficiency: A novel vitamin recycling defect. J. Inherit. Metab. Dis. 1985, 8, 53–58. [Google Scholar] [CrossRef]
- Saudubray, J.-M.; Berghe, G.; Walter, J.H. Inborn Metabolic Diseases; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Cowan, T.M.; Blitzer, M.G.; Wolf, B.; Working Group of the American College of Medical Genetics (ACMG); Laboratory Quality Assurance Committee. Technical standards and guidelines for the diagnosis of biotinidase deficiency. Genet. Med. 2010, 12, 464–470. [Google Scholar] [CrossRef]
- Joshi, S.; Al Essa, M.; Archibald, A.; Ozand, P. Biotinidase deficiency: A treatable genetic disorder in the Saudi population. East. Mediterr. Health J. 1999, 5, 1213–1217. [Google Scholar] [CrossRef] [PubMed]
- Al-Shamsi, A.; Hertecant, J.L.; Al-Hamad, S.; Souid, A.-K.; Al-Jasmi, F. Mutation spectrum and birth prevalence of inborn errors of metabolism among Emiratis: A study from Tawam Hospital Metabolic Center, United Arab Emirates. Sultan Qaboos Univ. Med. J. 2014, 14, e42. [Google Scholar]
- Pomponio, R.; Coskun, T.; Demirkol, M.; Tokatli, A.; Ozalp, I.; Hüner, G.; Baykal, T.; Wolf, B. Novel mutations cause biotinidase deficiency in Turkish children. J. Inherit. Metab. Dis. 2000, 23, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, F.; Velayutham, D.; Alsharshani, M.; AlAlami, U.; AlDewik, M.; Abuarja, T.; Al Rifai, H.; Al-Dewik, N.I. Studying carrier frequency of spinal muscular atrophy in the State of Qatar and comparison to other ethnic groups: Pilot study. Mol. Genet. Genom. Med. 2023, 11, e2184. [Google Scholar] [CrossRef]
- Yavarna, T.; Al-Dewik, N.; Al-Mureikhi, M.; Ali, R.; Al-Mesaifri, F.; Mahmoud, L.; Shahbeck, N.; Lakhani, S.; AlMulla, M.; Nawaz, Z. High diagnostic yield of clinical exome sequencing in Middle Eastern patients with Mendelian disorders. Hum. Genet. 2015, 134, 967–980. [Google Scholar] [CrossRef]
- Ben-Omran, T.; Al Ghanim, K.; Yavarna, T.; El Akoum, M.; Samara, M.; Chandra, P.; Al-Dewik, N. Effects of consanguinity in a cohort of subjects with certain genetic disorders in Qatar. Mol. Genet. Genom. Med. 2020, 8, e1051. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Mohd, H.; Al-Mureikhi, M.; Ali, R.; Al-Mesaifri, F.; Mahmoud, L.; Shahbeck, N.; El-Akouri, K.; Almulla, M.; Al Sulaiman, R. Clinical exome sequencing in 509 Middle Eastern families with suspected Mendelian diseases: The Qatari experience. Am. J. Med. Genet. Part A 2019, 179, 927–935. [Google Scholar] [CrossRef]
- Gallego-Villar, L.; Hannibal, L.; Häberle, J.; Thöny, B.; Ben-Omran, T.; Nasrallah, G.; Dewik, A.-N.; Kruger, W.; Blom, H. Cysteamine revisited: Repair of arginine to cysteine mutations. J. Inherit. Metab. Dis. 2017, 40, 555–567. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Ali, A.; Mahmoud, Y.; Shahbeck, N.; Ali, R.; Mahmoud, L.; Al-Mureikhi, M.; Al-Mesaifri, F.; Musa, S.; El-Akouri, K. Natural history, with clinical, biochemical, and molecular characterization of classical homocystinuria in the Qatari population. J. Inherit. Metab. Dis. 2019, 42, 818–830. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Al-Mureikhi, M.; Shahbeck, N.; Ali, R.; Al-Mesaifri, F.; Mahmoud, L.; Othman, A.; AlMulla, M.; Al Sulaiman, R.; Musa, S. Clinical genetics and genomic medicine in Qatar. Mol. Genet. Genom. Med. 2018, 6, 702. [Google Scholar] [CrossRef]
- Ismail, H.M.; Krishnamoorthy, N.; Al-Dewik, N.; Zayed, H.; Mohamed, N.A.; Giacomo, V.D.; Gupta, S.; Häberle, J.; Thöny, B.; Blom, H.J. In silico and in vivo models for Qatari-specific classical homocystinuria as basis for development of novel therapies. Hum. Mutat. 2019, 40, 230–240. [Google Scholar] [CrossRef]
- Gupta, S.; Gallego-Villar, L.; Wang, L.; Lee, H.O.; Nasrallah, G.; Al-Dewik, N.; Häberle, J.; Thöny, B.; Blom, H.J.; Ben-Omran, T. Analysis of the Qatari R336C cystathionine β-synthase protein in mice. J. Inherit. Metab. Dis. 2019, 42, 831–838. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.; Al-Dewik, N.; Mohammed, S.; Elfituri, M.; Agouba, S.; Musa, S.; Mahmoud, L.; Almulla, M.; El-Akouri, K.; Mohd, H. Expanding on the phenotypic spectrum of Woodhouse-Sakati syndrome due to founder pathogenic variant in DCAF17: Report of 58 additional patients from Qatar and literature review. Am. J. Med. Genet. Part A 2022, 188, 116–129. [Google Scholar] [CrossRef] [PubMed]
- Al-Sadeq, D.W.; Thanassoulas, A.; Islam, Z.; Kolatkar, P.; Al-Dewik, N.; Safieh-Garabedian, B.; Nasrallah, G.K.; Nomikos, M. Pyridoxine non-responsive p. R336C mutation alters the molecular properties of cystathionine beta-synthase leading to severe homocystinuria phenotype. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2022, 1866, 130148. [Google Scholar]
- Al-Sadeq, D.W.; Conter, C.; Thanassoulas, A.; Al-Dewik, N.; Safieh-Garabedian, B.; Martínez-Cruz, L.A.; Nasrallah, G.K.; Astegno, A.; Nomikos, M. Biochemical and structural impact of two novel missense mutations in cystathionine β-synthase gene associated with homocystinuria. Biochem. J. 2024, 481, 569–585. [Google Scholar] [CrossRef]
- Linga, B.G.; Mohammed, S.G.; Farrell, T.; Rifai, H.A.; Al-Dewik, N.; Qoronfleh, M.W. Genomic Newborn Screening for Pediatric Cancer Predisposition Syndromes: A Holistic Approach. Cancers 2024, 16, 2017. [Google Scholar] [CrossRef] [PubMed]
- Younes, S.; Elkahlout, R.; Kilani, H.; Okashah, S.; Al Sharshani, H.; Rezoug, Z.; Zayed, H.; Al-Dewik, N. Spectrum of Genetic Variants Associated with Maple Syrup Urine Disease in the Middle East and North Africa (MENA) Region: A Systematic Review; Research Square: Durham, NC, USA, 2024. [Google Scholar]
- Alkhidir, S.; El-Akouri, K.; Al-Dewik, N.; Khodjet-El-khil, H.; Okashah, S.; Islam, N.; Ben-Omran, T.; Al-Shafai, M. The genetic basis and the diagnostic yield of genetic testing related to nonsyndromic hearing loss in Qatar. Sci. Rep. 2024, 14, 420. [Google Scholar] [CrossRef]
- Nahas, L.D.; Datta, A.; Alsamman, A.M.; Adly, M.H.; Al-Dewik, N.; Sekaran, K.; Sasikumar, K.; Verma, K.; Doss, G.P.C.; Zayed, H. Genomic insights and advanced machine learning: Characterizing autism spectrum disorder biomarkers and genetic interactions. Metab Brain Dis. 2024, 39, 29–42. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Al-Jurf, R.; Styles, M.; Tahtamouni, S.; Alsharshani, D.; Alsharshani, M.; Ahmad, A.I.; Khattab, A.; Al Rifai, H.; Walid Qoronfleh, M. Overview and Introduction to Autism Spectrum Disorder (ASD). Adv. Neurobiol. 2020, 24, 3–42. [Google Scholar]
- Al-Dewik, N.; Alsharshani, M. New horizons for molecular genetics diagnostic and research in autism spectrum disorder. In Personalized Food Intervention and Therapy for Autism Spectrum Disorder Management; Springer: Berlin/Heidelberg, Germany, 2020; pp. 43–81. [Google Scholar]
- Styles, M.; Alsharshani, D.; Samara, M.; Alsharshani, M.; Khattab, A.; Qoronfleh, M.W.; Al-Dewik, N. Risk factors, diagnosis, prognosis and treatment of autism. Front. Biosci. Landmark 2020, 25, 1682–1717. [Google Scholar]
- Sarar, M.; Wafa, E.; Al-Aqeel, A.I.; Alhashem, A.M.; Ali, A.; Lujane, A.; Maher, A.; Fahad, A.; Horia, A.; Amer, A. Incidence of newborn screening disorders among 56632 infants in Central Saudi Arabia. Saudi Med. J. 2020, 41, 703–708. [Google Scholar]
- Canda, E.; Kalkan Uçar, S.; Çoker, M. Biotinidase Deficiency: Prevalence, Impact and Management Strategies. Pediatr. Health Med. Ther. 2020, 11, 127–133. [Google Scholar]
- CNeto, E.; Schulte, J.; Rubim, R.; Lewis, E.; DeMari, J.; Castilhos, C.; Brites, A.; Giugliani, R.; Jensen, K.; Wolf, B. Newborn screening for biotinidase deficiency in Brazil: Biochemical and molecular characterizations. Braz. J. Med. Biol. Res. 2004, 37, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Tadmouri, G.O.; Ali, M.T.A.; Ali, S.A.-H.; Khaja, N.A. CTGA: The database for genetic disorders in Arab populations. Nucleic Acids Res. 2006, 34, D602–D606. [Google Scholar] [CrossRef]
- Latif, M.; Hashmi, J.A.; Alayoubi, A.M.; Ayub, A.; Basit, S. Identification of Novel and Recurrent Variants in BTD, GBE1, AGL and ASL Genes in Families with Metabolic Disorders in Saudi Arabia. J. Clin. Med. 2024, 13, 1193. [Google Scholar] [CrossRef]
- Naseer, M.I.; Pushparaj, P.N.; Abdulkareem, A.A.; Muthaffar, O.Y. Whole-exome sequencing reveals a missense variant c. 1612C > T (p. Arg538Cys) in the BTD gene leading to neuromyelitis optica spectrum disorder in saudi families. Front. Pediatr. 2022, 9, 829251. [Google Scholar] [CrossRef]
- Wolf, B. Biotinidase deficiency should be considered in individuals exhibiting myelopathy with or without and vision loss. Mol. Genet. Metab. 2015, 116, 113–118. [Google Scholar] [CrossRef]
- Hymes, J.; Wolf, B. Biotinidase and its roles in biotin metabolism. Clin. Chim. Acta 1996, 255, 1–11. [Google Scholar] [CrossRef]
- Tankeu, A.T.; Van Winckel, G.; Elmers, J.; Jaccard, E.; Superti-Furga, A.; Wolf, B.; Tran, C. Biotinidase deficiency: What have we learned in forty years? Mol. Genet. Metab. 2023, 138, 107560. [Google Scholar] [CrossRef]
- Hymes, J.; Stanley, C.M.; Wolf, B. Mutations in BTD causing biotinidase deficiency. Hum. Mutat. 2001, 18, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Baykal, T.; Gokcay, G.; Gokdemir, Y.; Demir, F.; Seckin, Y.; Demirkol, M.; Jensen, K.; Wolf, B. Asymptomatic adults and older siblings with biotinidase deficiency ascertained by family studies of index cases. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2005, 28, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Couce, M.L.; Pérez-Cerdá, C.; Garcia Silva, M.T.; Martin-Hernandez, E.; Castiñeiras, D.; Pineda, M.; Navarrete, R.; Campistol, J.; Fraga, J.; Pérez, B. Clinical and genetic findings in patients with biotinidase deficiency detected through newborn screening or selective screening for hearing loss or inherited metabolic disease. Med. Clin. 2011, 137, 500–503. [Google Scholar] [CrossRef] [PubMed]
- Funghini, S.; Donati, M.; Pasquini, E.; Gasperini, S.; Ciani, F.; Morrone, A.; Zammarchi, E. Two new mutations in children affected by partial biotinidase deficiency ascertained by newborn screening. J. Inherit. Metab. Dis. 2002, 25, 328–330. [Google Scholar] [CrossRef]
- Pomponio, R.J.; Hymes, J.; Reynolds, T.R.; Meyers, G.A.; Fleischhauer, K.; Buck, G.A.; Wolf, B. Mutations in the human biotinidase gene that cause profound biotinidase deficiency in symptomatic children: Molecular, biochemical, and clinical analysis. Pediatr. Res. 1997, 42, 840–848. [Google Scholar] [CrossRef][Green Version]
- Norrgard, K.J.; Pomponio, R.J.; Hymes, J.; Wolf, B. Mutations causing profound biotinidase deficiency in children ascertained by newborn screening in the United States occur at different frequencies than in symptomatic children. Pediatr. Res. 1999, 46, 20–27. [Google Scholar] [CrossRef]
- Laszlo, A.; Schuler, E.; Sallay, E.; Endreffy, E.; Somogyi, C.; Varkonyi, A.; Havass, Z.; Jansen, K.; Wolf, B. Neonatal screening for biotinidase deficiency in Hungary: Clinical, biochemical and molecular studies. J. Inherit. Metab. Dis. 2003, 26, 693–698. [Google Scholar] [CrossRef]
- Milánkovics, I.; Kámory, E.; Csókay, B.; Fodor, F.; Somogyi, C.; Schuler, Á. Mutations causing biotinidase deficiency in children ascertained by newborn screening in Western Hungary. Mol. Genet. Metab. 2007, 90, 345–348. [Google Scholar] [CrossRef]
- Swango, K.L.; Demirkol, M.; Hüner, G.; Pronicka, E.; Sykut-Cegielska, J.; Schulze, A.; Wolf, B. Partial biotinidase deficiency is usually due to the D444H mutation in the biotinidase gene. Hum. Genet. 1998, 102, 571–575. [Google Scholar] [CrossRef]
- Iqbal, F.; Vilaseca, M.A.; Jalan, A.; Mühl, A.; Couce, M.L.; Duat, A.; Delgado, M.P.; Bosch, J.; Puche, A.; Campistol, J. The identification of novel mutations in the biotinidase gene using denaturing high pressure liquid chromatography (dHPLC). Mol. Genet. Metab. 2010, 100, 42–45. [Google Scholar] [CrossRef]
- Procter, M.; Wolf, B.; Crockett, D.K.; Mao, R. The biotinidase gene variants registry: A paradigm public database. G3: Genes Genomes Genet. 2013, 3, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-L.; Li, Y.; Zhou, S.-F. A bioinformatics approach for the phenotype prediction of nonsynonymous single nucleotide polymorphisms in human cytochromes P450. Drug Metab. Dispos. 2009, 37, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Valastyan, J.S.; Lindquist, S. Mechanisms of protein-folding diseases at a glance. Dis. Models Mech. 2014, 7, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, P.; Rajasekaran, R. Detailed computational analysis revealed mutation V210I on PrP induced conformational conversion on β2–α2 loop and α2–α3. Mol. Biosyst. 2016, 12, 3223–3233. [Google Scholar] [CrossRef]
- Thirumal Kumar, D.; Eldous, H.G.; Mahgoub, Z.A.; George Priya Doss, C.; Zayed, H. Computational modelling approaches as a potential platform to understand the molecular genetics association between Parkinson’s and Gaucher diseases. Metab. Brain Dis. 2018, 33, 1835–1847. [Google Scholar] [CrossRef]
- Chasman, D.; Adams, R.M. Predicting the functional consequences of non-synonymous single nucleotide polymorphisms: Structure-based assessment of amino acid variation. J. Mol. Biol. 2001, 307, 683–706. [Google Scholar] [CrossRef]
- Sunyaev, S.; Hanke, J.; Aydin, A.; Wirkner, U.; Zastrow, I.; Reich, J.; Bork, P. Prediction of nonsynonymous single nucleotide polymorphisms in human disease-associated genes. J. Mol. Med. 1999, 77, 754–760. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. 2001, 11, 863–874. [Google Scholar] [CrossRef]
- Bromberg, Y.; Rost, B. SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 2007, 35, 3823–3835. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Rossi, I.; Casadio, R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinform. 2008, 9, S6. [Google Scholar] [CrossRef]
- Acharya, V.; Nagarajaram, H.A. Hansa: An automated method for discriminating disease and neutral human nsSNPs. Hum. Mutat. 2012, 33, 332–337. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.; Edwards, K.J.; Day, I.N.; Gaunt, T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Zhernakova, A.; Van Diemen, C.C.; Wijmenga, C. Detecting shared pathogenesis from the shared genetics of immune-related diseases. Nat. Rev. Genet. 2009, 10, 43–55. [Google Scholar] [CrossRef]
- Lindner, M.; Abdoh, G.; Fang-Hoffmann, J.; Shabeck, N.; Al Sayrafi, M.; Al Janahi, M.; Ho, S.; Abdelrahman, M.; Ben-Omran, T.; Bener, A. Implementation of extended neonatal screening and a metabolic unit in the State of Qatar: Developing and optimizing strategies in cooperation with the Neonatal Screening Center in Heidelberg. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2007, 30, 522–529. [Google Scholar] [CrossRef]
- Ben-Rebeh, I.; Hertecant, J.L.; Al-Jasmi, F.A.; Aburawi, H.E.; Al-Yahyaee, S.A.; Al-Gazali, L.; Ali, B.R. Identification of mutations underlying 20 inborn errors of metabolism in the United Arab Emirates population. Genet. Test. Mol. Biomark. 2012, 16, 366–371. [Google Scholar] [CrossRef]
- Varghese, S.E.; Otol, E.; Mohammad, R.H.; Al Olama, F.S.; Elbadawi, S.A.M. The importance of early detection of genetic diseases. Dubai Med. J. 2021, 4, 133–141. [Google Scholar] [CrossRef]
- Al-Jasmi, F.A.; Al-Shamsi, A.; Hertecant, J.L.; Al-Hamad, S.M.; Souid, A.-K. Inborn errors of metabolism in the United Arab Emirates: Disorders detected by newborn screening (2011–2014). JIMD Rep. 2016, 28, 127–135. [Google Scholar] [PubMed]
- Saleh, S.; Beyyumi, E.; Al Kaabi, A.; Hertecant, J.; Barakat, D.; Al Dhaheri, N.S.; Al-Gazali, L.; Al Shamsi, A. Spectrum of neuro-genetic disorders in the United Arab Emirates national population. Clin. Genet. 2021, 100, 573–600. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.O. Juvenile progressive optic atrophy as the presenting feature of biotinidase deficiency, a treatable metabolic disorder. J. Am. Assoc. Pediatr. Ophthalmol. Strabismus 2021, 25, 248–250. [Google Scholar] [CrossRef]
- Dabbagh, O.; Brismar, J.; Gascon, G.; Ozand, P. The clinical spectrum of biotin-treatable encephalopathies in Saudi Arabia. Brain Dev. 1994, 16, 72–80. [Google Scholar] [CrossRef]
- Moammar, H.; Cheriyan, G.; Mathew, R.; Al-Sannaa, N. Incidence and patterns of inborn errors of metabolism in the Eastern Province of Saudi Arabia, 1983–2008. Ann. Saudi Med. 2010, 30, 271–277. [Google Scholar] [CrossRef] [PubMed]
- McCandless, M.S.; Scaglia, F. Program for SIMD annual meeting March 31–April 3, 2012, The Westin Charlotte, Charlotte, NC. Mol. Genet. Metab. 2012, 105, 273–366. [Google Scholar] [CrossRef]
- Sweetman, L.; Nyhan, W.; Sakati, N.; Ohlsson, A.; Mange, M.; Boychuk, R.; Kaye, R. Organic aciduria in neonatal multiple carboxylase deficiency. J. Inherit. Metab. Dis. 1982, 5, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.; Daly, M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- Consortium, G.P. A global reference for human genetic variation. Nature 2015, 526, 68. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef] [PubMed]
- Glaser, F.; Pupko, T.; Paz, I.; Bell, R.E.; Bechor-Shental, D.; Martz, E.; Ben-Tal, N. ConSurf: Identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics 2003, 19, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Altman, R.B.; Bromberg, Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013, 14, S2. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Grishin, N.V. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics 2001, 17, 700–712. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, B.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct. Funct. Bioinform. 2006, 62, 1125–1132. [Google Scholar] [CrossRef]
- Montanucci, L.; Capriotti, E.; Frank, Y.; Ben-Tal, N.; Fariselli, P. DDGun: An untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinform. 2019, 20, 335. [Google Scholar] [CrossRef]
- Mi, H.; Guo, N.; Kejariwal, A.; Thomas, P.D. PANTHER version 6: Protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007, 35, D247–D252. [Google Scholar] [CrossRef]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Johnson, A.D.; Handsaker, R.E.; Pulit, S.L.; Nizzari, M.M.; O’Donnell, C.J.; De Bakker, P.I. SNAP: A web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008, 24, 2938–2939. [Google Scholar] [CrossRef]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2022. Available online: http://www.pymol.org/ (accessed on 22 June 2024).
- Makarov, V. Computer programs for eukaryotic gene prediction. Brief. Bioinform. 2002, 3, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.U.; Zoete, V.; Michielin, O.; Guex, N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinform. 2012, 13, 173. [Google Scholar] [CrossRef] [PubMed]
- Colovos, C.; Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993, 2, 1511–1519. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Laskowski, R.; MacArthur, M.; Thornton, J. PROCHECK: Validation of Protein-Structure Coordinates; Wiley: Oxford, UK, 2006. [Google Scholar]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007, 35, W407–W410. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Chistyakov, V.V.; Thornton, J.M. PDBsum more: New summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005, 33, D266–D268. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.e.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server; Springer: Berlin/Heidelberg, Germany, 2005. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Costantini, S.; Colonna, G.; Facchiano, A.M. ESBRI: A web server for evaluating salt bridges in proteins. Bioinformation 2008, 3, 137. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, H.J.; Postma, J.V.; Van Gunsteren, W.F.; DiNola, A.; Haak, J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Monks, S. SigmaPlot 8.0. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2002, 3, 141–145. [Google Scholar] [CrossRef]
- Mikati, M.A.; Zalloua, P.; Karam, P.; Habbal, M.-Z.; Rahi, A.C. Novel mutation causing partial biotinidase deficiency in a Syrian boy with infantile spasms and retardation. J. Child Neurol. 2006, 21, 978–981. [Google Scholar] [CrossRef]
- Sarafoglou, K.; Bentler, K.; Gaviglio, A.; Redlinger-Grosse, K.; Anderson, C.; McCann, M.; Bloom, B.; Babovic-Vuksanovic, D.; Gavrilov, D.; Berry, S. High incidence of profound biotinidase deficiency detected in newborn screening blood spots in the Somalian population in Minnesota. J. Inherit. Metab. Dis. Off. J. Soc. Study Inborn Errors Metab. 2009, 32, 169–173. [Google Scholar] [CrossRef]
- Li, H.; Spencer, L.; Nahhas, F.; Miller, J.; Fribley, A.; Feldman, G.; Conway, R.; Wolf, B. Novel mutations causing biotinidase deficiency in individuals identified by newborn screening in Michigan including an unique intronic mutation that alters mRNA expression of the biotinidase gene. Mol. Genet. Metab. 2014, 112, 242–246. [Google Scholar] [CrossRef]
- Al-Dewik, N.; Abuarja, T.; Younes, S.; Nasrallah, G.; Alsharshani, M.; Ibrahim, F.E.; Samara, M.; Farrell, T.; Abdulrouf, P.V.; Qoronfleh, M.W. Precision medicine activities and opportunities for shaping maternal and neonatal health in Qatar. Pers. Med. 2024, 21, 313–333. [Google Scholar] [CrossRef]
- Procter, M.; Wolf, B.; Mao, R. Forty-eight novel mutations causing biotinidase deficiency. Mol. Genet. Metab. 2016, 117, 369–372. [Google Scholar] [CrossRef]
- Jezela-Stanek, A.; Suchoń, L.; Sobczyńska-Tomaszewska, A.; Czerska, K.; Kuśmierska, K.; Taybert, J.; Ołtarzewski, M.; Sykut-Cegielska, J. Molecular Background and Disease Prevalence of Biotinidase Deficiency in a Polish Population—Data Based on the National Newborn Screening Programme. Genes 2022, 13, 802. [Google Scholar] [CrossRef]
- Wolf, B.; Jensen, K.P.; Barshop, B.; Blitzer, M.; Carlson, M.; Goudie, D.R.; Gokcay, G.H.; Demirkol, M.; Baykal, T.; Demir, F. Biotinidase deficiency: Novel mutations and their biochemical and clinical correlates. Hum. Mutat. 2005, 25, 413. [Google Scholar] [CrossRef]
- Pindolia, K.; Jordan, M.; Wolf, B. Analysis of mutations causing biotinidase deficiency a. Hum. Mutat. 2010, 31, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Wolf, B.; Jensen, K. Evolutionary conservation of biotinidase: Implications for the enzyme’s structure and subcellular localization. Mol. Genet. Metab. 2005, 86, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Sun, Y.; Zhao, Y.; Xiong, W.; Zhong, M.; Zhang, Y.; Zhao, Q.; Bao, Z.; Cheng, J.; Lu, Y. Two novel BTD mutations causing profound biotinidase deficiency in a Chinese patient. Mol. Genet. Genom. Med. 2021, 9, e1591. [Google Scholar] [CrossRef] [PubMed]
- Göksoy, E. Evaluation of newborn screening for biotinidase deficiency from southeastern region of Türkiye. Trends Pediatr. 2023, 4, 247–252. [Google Scholar] [CrossRef]
- Al-Dewik, N.I.; Younes, S.N.; Essa, M.M.; Pathak, S.; Qoronfleh, M.W. Making biomarkers relevant to healthcare innovation and precision medicine. Processes 2022, 10, 1107. [Google Scholar] [CrossRef]
- Al-Dewik, N.I.; Qoronfleh, M.W. Genomics and precision medicine: Molecular diagnostics innovations shaping the future of healthcare in Qatar. Adv. Public Health 2019, 2019, 3807032. [Google Scholar] [CrossRef]
- Zhai, K.; Yousef, M.S.; Mohammed, S.; Al-Dewik, N.I.; Qoronfleh, M.W. Optimizing clinical workflow using precision medicine and advanced data analytics. Processes 2023, 11, 939. [Google Scholar] [CrossRef]
- Elkahlout, R.; Mohammed, S.G.A.A.; Najjar, A.; Farrell, T.; Rifai, H.A.; Al-Dewik, N.; Qoronfleh, M.W. Application of Proteomics in Maternal and Neonatal Health: Advancements and Future Directions. Proteom. Clin. Appl. 2025, 19, e70004. [Google Scholar] [CrossRef]
- Mohammed, S.G.A.A.; Qoronfleh, M.W.; Acar, A.; Al-Dewik, N.I. Holistic precision wellness: Paving the way for next-generation precision medicine (ngPM) with AI, biomedical informatics, and clinical medicine. FASEB Bioadv. 2025, 7, e70005. [Google Scholar] [CrossRef]







| Genomic Coordinates | RS ID | Nucleotide Change | Amino Acid Change | Classification (ClinVar) | Genotype | Genotype (GT) n (%) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| * QGP (n = 14,669) | & IP (n = 2504) | ||||||||||
| Homo | Hetero | WT | Homo | Hetero | WT | ||||||
| 3:15644413 | rs397514369 | BTD: c.557G>A | C186Y | P/LP | AA/AG/GG | 0 (0%) | 14 (0.09%) | 14,655 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15644481 | rs369102875 | BTD: c.565C>T | R209C | P | TT/TC/CC | 1 (0.006%) | 7 (0.04%) | 14,661 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15644824 | rs397507176 | BTD: c.968A>G | H323R | LP | GG/GA/AA | 2 (0.01%) | 49 (0.33%) | 14,618 (99.44%) | 0 (0%) | 24 (0.95%) | 2480 (99.04%) |
| 3:15645186 | rs13078881 | BTD: c.1330G>C | D444H | P | CC/CG/GG | 22 (0.14%) | 608 (4.14%) | 14,039 (95.7%) | 4 (0.15%) | 85 (3.39%) | 2415 (96.44%) |
| 3:15645224 | rs80338685 | BTD: c.1368A>C | Q456H | P | CC/CA/AA | 0 (0%) | 10 (0.06%) | 14,659 (99.9%) | 0 (0%) | 1 (0.03%) | 2503 (99.9%) |
| 3:15645345 | rs138818907 | BTD: c.1489C>T | P497S | P | TT/TC/CC | 0 (0%) | 6 (0.04%) | 14,663 (99.9%) | 0 (0%) | 1 (0.03%) | 2503 (99.9%) |
| 3:15645387 | rs397514427 | BTD: c.1531C>G | Q511E | LP | GG/GC/CC | 0 (0%) | 2 (0.01%) | 14,667 (99.9%) | 0 (0%) | 0 (0%) | 2504 (99.9%) |
| 3:15645288 | rs181396238 | BTD: c.G>A | A478T | P | AA/AG/GG | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 3 (0.11%) | 2501 (99.88%) |
| 3:15645292 | rs142249642 | BTD: c.C>T | T479M | P/LP | TT/TC/CC | 0 (0%) | 6 (0.04%) | 14,663 (99.9%) | 0 (0%) | 3 (0.11%) | 2501 (99.8%) |
| 3:15645451 | rs104893688 | BTD: c.C>T | T532M | P/LP | TT/TC/CC | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 1 (0.03%) | 2503 (99.9%) |
| 3:15645484 | rs1050035768 | BTD: c.A>G | D543G | LP | GG/GA/AA | 0 (0%) | 2 (0.01%) | 14,667 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15635478 | rs765906887 | BTD: c.CGG>C | G34fs | P | CC/CG/GG | 0 (0%) | 9 (0.06%) | 14,660 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15635550 | rs397514339 | BTD: c.T>G | Y57Ter | P | GG/GT/TT | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15644322 | rs397514362 | BTD: c.C>T | Q156Ter | P | TT/TC/CC | 0 (0%) | 2 (0.01%) | 14,667 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15644345 | rs397514365 | BTD: c.CAG>C | R164fs | LP | CC/GC/GG | 0 (0%) | 3 (0.02%) | 14,666 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15641920 | rs976185636 | BTD: c.A>G | I108V | P ¥ | GG/GA/AA | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15645295 | rs558477960 | BTD: c.G>A | G480E | P ¥ | AA/AG/GG | 0 (0%) | 20 (0.13%) | 14,649 (99.8%) | 0 (0%) | 1 (0.03%) | 2503 (99.9%) |
| 3:15645459 | rs771537277 | BTD: c.C>G | L535V | P ¥ | GG/GC/CC | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15645647 | rs530872564 | BTD: c.G>A | - | P ¥ | AA/AG/GG | 0 (0%) | 1 (0.006%) | 14,668 (99.9%) | 0 (0%) | 2 (0.07%) | 2502 (99.9%) |
| 3:15635478 | rs765906887 | BTD:c.CGG>TGG,C | - | P | TGG:TGG/CGG:TGG/CGG:CGG | 0 (0%) | 9 (0.06%) | 14,660 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| 3:15644345 | rs397514365 | BTD:c.CAG>C | - | LP | C:C/CAG:C/CAG:CAG | 0 (0%) | 3 (0.02%) | 14,666 (99.9%) | 0 (0%) | 0 (0%) | 2504 (100%) |
| RS ID | Mutation | # PANTHER Score | & PhD-SNP Score | $ SIFT Score | ^ SNAP Score | % Meta-SNP Score | RI * |
|---|---|---|---|---|---|---|---|
| rs397514369 | C186Y | Disease 0.918 | Disease 0.862 | Disease 0.010 | Disease 0.735 | Disease 0.799 | 6 |
| rs369102875 | R209C | Disease 0.979 | Disease 0.867 | Disease 0.000 | Disease 0.720 | Disease 0.819 | 6 |
| rs397507176 | H323R | Disease 0.511 | Disease 0.318 | Neutral 0.270 | Disease 0.695 | Neutral 0.315 | 4 |
| rs13078881 | D444H | Disease 0.692 | Disease 0.793 | Disease 0.010 | Disease 0.825 | Disease 0.811 | 6 |
| rs80338685 | Q456H | Disease 0.820 | Disease 0.755 | Disease 0.000 | Disease 0.810 | Disease 0.825 | 6 |
| rs138818907 | P497S | Disease 0.779 | Disease 0.643 | Disease 0.000 | Disease 0.720 | Disease 0.697 | 4 |
| rs397514427 | Q511E | Neutral 0.194 | Neutral 0.170 | Disease 0.800 | Neutral 0.125 | Neutral 0.156 | 7 |
| rs976185636 | I108V | Neutral 0.331 | Neutral 0.485 | Disease 0.000 | Disease 0.560 | Neutral 0.425 | 2 |
| rs181396238 | A478T | Disease 0.606 | Neutral 0.410 | Disease 0.000 | Disease 0.640 | Disease 0.535 | 1 |
| rs142249642 | T479M | Neutral 0.357 | Neutral 0.167 | Neutral 0.190 | Neutral 0.180 | Neutral 0.212 | 6 |
| rs558477960 | G480E | Neutral 0.319 | Neutral 0.258 | Disease 0.020 | Disease 0.550 | Neutral 0.260 | 5 |
| rs104893688 | T532M | Disease 0.686 | Disease 0.602 | Disease 0.000 | Disease 0.740 | Disease 0.720 | 4 |
| rs771537277 | L535V | Disease 0.537 | Neutral 0.327 | Disease 0.000 | Disease 0.700 | Neutral 0.361 | 3 |
| rs1050035768 | D543G | Neutral 0.330 | Neutral 0.295 | Disease 0.0000 | Disease 0.670 | Neutral 0.356 | 3 |
| BTD Enzyme | I-Mutant 2.0 | MuPro | DDGun | |||
|---|---|---|---|---|---|---|
| Stability Effect | ΔΔG (kcal/mol) | Stability Effect | ΔΔG (kcal/mol) | Stability Effect | ΔΔG (kcal/mol) | |
| BTD_C186Y | Decrease | −0.20 | Decrease | −1.33 | Decrease | −0.3 |
| BTD_R209C | Decrease | −1.63 | Decrease | −1.07 | Decrease | −1.6 |
| BTD_D444H | Decrease | −0.12 | Decrease | −0.92 | Decrease | 0 |
| BTD_Q456H | Decrease | −0.76 | Decrease | −1.14 | Decrease | −0.1 |
| BTD_P497S | Decrease | −1.65 | Decrease | −0.83 | Decrease | −0.5 |
| BTD_L535V | Decrease | −2.91 | Decrease | −0.93 | Decrease | −0.1 |
| BTD_D543G | Decrease | −1.31 | Decrease | −1.94 | Decrease | −0.2 |
| BTD_I108V | Decrease | −1.43 | Decrease | −0.39 | Decrease | −0.4 |
| BTD_H323R | Increase | 0.42 | Decrease | −0.83 | Decrease | −0.2 |
| BTD_G480E | Increase | 0.43 | Decrease | 0.60 | Neutral | 0 |
| BTD_Q511E | Increase | 0.26 | Decrease | −0.40 | Increase | −0.1 |
| BTD_A478T | Decrease | −2.05 | Decrease | −1.03 | Neutral | 0 |
| BTD_T479M | Decrease | −0.54 | Decrease | −0.37 | Neutral | 0 |
| BTD_T532M | Decrease | −1.08 | Increase | 0.63 | Neutral | 0 |
| BTD Enzyme | Aliphatic Index | Instability Index | * pI | Extension Coefficient | * GRAVY |
|---|---|---|---|---|---|
| BTD_WT | 86.37 | 31.95 | 5.81 | 75,260 | −0.032 |
| BTD_C186Y | 31.68 | 5.81 | 75,750 | −0.039 | |
| BTD_R209C | 32.23 | 5.75 | 75,260 | −0.019 | |
| BTD_H323R | 31.41 | 5.83 | 74,260 | −0.034 | |
| BTD_D444H | 31.53 | 5.90 | 74,260 | −0.031 | |
| BTD_Q456H | 32.32 | 5.84 | 74,260 | −0.031 | |
| BTD_P497S | 31.60 | 5.81 | 74,260 | −0.030 | |
| BTD_Q511E | 31.95 | 5.76 | 74,260 | −0.032 | |
| BTD_A478T | 31.95 | 5.81 | 75,260 | −0.036 | |
| BTD_T479M | 32.11 | 5.81 | 74,260 | −0.027 | |
| BTD_T532M | 32.33 | 5.81 | 74,260 | −0.027 | |
| BTD_D543G | 31.79 | 5.87 | 74,260 | −0.026 | |
| BTD_I108V | 31.95 | 5.81 | 74,260 | −0.032 | |
| BTD_G480E | 32.97 | 5.76 | 74,260 | −0.037 | |
| BTD_L535V | 31.81 | 5.81 | 74,260 | −0.031 |
| BTD Enzyme | ERRAT Overall Quality Factor | ProSA-Web (Z-Score) * | Ramachandran Plot | ||
|---|---|---|---|---|---|
| Favored Region † | Allowed Region † | Disallowed Region † | |||
| BTD_WT | 92.02 | −8.75 | 84% | 15% | 1.1% |
| BTD_C186Y | 88.79 | −8.69 | 84% | 15% | 1.1% |
| BTD_R209C | 92.24 | −8.77 | 84% | 15% | 1.1% |
| BTD_H323R | 91.16 | −8.72 | 84% | 15% | 1.1% |
| BTD_D444H | 92.04 | −8.76 | 84% | 15% | 1.1% |
| BTD_Q456H | 92.02 | −8.79 | 84% | 15% | 1.1% |
| BTD_P497S | 92.02 | −8.76 | 83.8% | 15.2% | 1.1% |
| BTD_Q511E | 92.02 | −8.74 | 84% | 15% | 1.1% |
| Origin | Phenotype | Age of Onset | N | Clinical Presentation | Method Of Diagnosis | Nucleotide Change | Amino Acid Changes | Ref |
|---|---|---|---|---|---|---|---|---|
| UAE | BTD | <1 years | 7 | Not reported | Biochemical tests (NBS) and molecular genetics tests | c.1330G>C † c.1207T>G c.968A>G c.1489C>T c.557G>A | p.D444H † p.F403V p.H323R p.P497S p.C186Y | [17] |
| UAE | BTD | <1.5 years | 13 | Not reported | Biochemical tests and molecular genetics tests | c.1330G>C c.557G>A | p.D444H p.C186Y | [72] |
| UAE | BTD | 12 years | 1 | Global developmental delay, seizure, blind sleep disturbance | Biochemical tests and molecular genetics tests | c.560del | p.P187Qfs*77 | [75] |
| 11 years | 1 | Speech delay, learning difficulty | c.1207T>G c.1330G>C | p.F403V p.D444H | ||||
| 8 years | 1 | Strabismus, delayed speech, hyperactivity, hyperpigmented and hypopigmented macules, hearing loss | c.557G>A c.1330G>C | p.C186Y p.D444H | ||||
| 6 years | 1 | Speech delay, learning difficulty | c.1330G>C | p.D444H | ||||
| UAE | BTD | ≤3 years | 2 | Speech delays and eczema | Biochemical tests and molecular genetics tests | c.557G>A c.1330G>C | p.F403V p.D444H | [79] |
| UAE | BTD | <1 year | 2 | Not reported | Biochemical tests (NBS) and molecular genetics tests | c.322A>G c.1595C>T | p.I108V p.T532M | [73] |
| UAE | BTD | <1 year | 14 ‡ | Global developmental delay, seizure, blindness, sleep disturbance, hearing impairment | Biochemical tests and molecular genetics tests | c.476G> c.1330G>C c.1595C>T c.968A>G c.1207T>G c.557G>A c.1489C>T c.257T>C | p.S159N p.D444H p.T532M p.H323R p.F403V p.C186Y p.P497S p.M86T | [74] |
| 34 § | c.626G>A c.1368A>C c.380C>T c.1420G>T c.470G>A c.557G>A c.1330G>C c.968A>G c.476G>A c.1595C>T c.424C>A c.476G>A c.922A>C c.1489C>T | p.R209H p.Q456H p.P127L p.E474X p.R157H p.C186Y p.D444H p.H323R p.S159N p.T532M p.P142T p.S159D p.M308L p.P497S | ||||||
| UAE | BTD | 13 years | 1 | Juvenile optic atrophy, functional visual loss, bilateral optic nerve head pallor, mild thinning of the orbital segments of the optic nerves, and hearing difficulty | Biochemical tests and molecular genetics tests | c.1213T.G c.1336G.C | p.F405V p.D446H | [76] |
| Saudi Arabia | BTD | <1 year | 1 | Degenerative brain disease, convulsive disorder, and seborrheic dermatitis | Biochemical tests (NBS) | Not reported | Not reported | [80] |
| 20 | Seizures, dermatitis, hypotonia, poor feeding, sensorineural deafness, poor speech and optic atrophy | [16] | ||||||
| 3 | Not reported | [78] | ||||||
| ≤17 years | 10 | Severe acidosis, seizures, coma, global developmental delay, generalized weakness, dysphasia, loss of hearing and vision, severe spastic quadriparesis, ophthalmoplegia, dysarthria, convulsions (generalized tonic/clonic), ataxia, lethargy, ataxia, slurred speech, severe hypotonia, hyporeflexia and choreoathetosis movements | [77] | |||||
| Qatar | BTD | <1 year | 2 | Not reported | Biochemical tests (NBS) | Not reported | Not reported | [71] |
| 12 | [25] | |||||||
| Lebanon | BTD | <1 year | 1 | Seizure; brain atrophy, global developmental delay, hyperreflexia, optic disc pallor, large for gestational age | Biochemical tests | Not reported | Not reported | [114] |
| Somalia | BTD | <1 year | 26/33 | Not reported | Biochemical tests and molecular genetics tests | c.424C>A c.1489C>T c.1284C>T c.1432G>C | p.P142T p.P497S p.Y428Y p.A478P | [115] |
| 7/33 | p.H65R, p.D444H ‡‡ 98:d7i3, p.D444H ‡‡ p.A171T, p.D444H ‡‡ p.V417F, p.D444H ‡‡ p.Y210C, p.D444H ‡‡ p.R538H | |||||||
| Türkiye | BTD | <1.5 years (symptomatic) | 13 | Seizures, hypotonia; lethargy; mental retardation, ataxia, vision impairment, hearing impairment alopecia, rash, conjunctivitis Respiratory problems, GI, gastrointestinal, metabolic acidosis, hyperammonaemia, organic aciduria (elevated concentrations of 3 hydroxyisovalerate, with and without methylcitrate, 3-hydroxypropionate, 3-methylcrotonylglycine, and/or lactate) | Biochemical tests and molecular genetics tests Biochemical tests and molecular genetics tests | c.171T>G c.98G:del7ins3 c.587C>G c.235C>T c.612C>T c.1368A>C c.100G>A c.1369G>A | p.Y57Ter Frameshift p.T196R p.R79C p.R538C p.Q456H 3′Splice site p.V457M | [18] |
| <1.5 years ascertained by newborn screening (NBS) | 18 | Not Reported | c.235C>T c.470G>A c.1595C>T c.557G>A c.98G:del7ins3 c.100G>A c.1330G>C, c.511G>A c.929G>A c.1368A>C | p.R79C p.R157H p.T532M p.C186Y Frameshift 3′Splice site p.D444H, p.A171T G310E Q456H |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ibrahim, F.E.; Gattu Linga, B.; Samara, M.; Roshanuddin, J.; Younes, S.; Nasrallah, G.K.; Zayed, H.; Qoronfleh, M.W.; Mohammed, S.G.A.A.; El Khoury, D.; et al. Genomic and Structural Investigation of Mutations in Biotinidase (BTD) Gene Deficiency in Greater Middle Eastern Cohort: Insights from Molecular Dynamics Study. Biomedicines 2025, 13, 2210. https://doi.org/10.3390/biomedicines13092210
Ibrahim FE, Gattu Linga B, Samara M, Roshanuddin J, Younes S, Nasrallah GK, Zayed H, Qoronfleh MW, Mohammed SGAA, El Khoury D, et al. Genomic and Structural Investigation of Mutations in Biotinidase (BTD) Gene Deficiency in Greater Middle Eastern Cohort: Insights from Molecular Dynamics Study. Biomedicines. 2025; 13(9):2210. https://doi.org/10.3390/biomedicines13092210
Chicago/Turabian StyleIbrahim, Faisal E., BalaSubramani Gattu Linga, Muthanna Samara, Jameela Roshanuddin, Salma Younes, Gheyath K. Nasrallah, Hatem Zayed, M. Walid Qoronfleh, Sawsan G. A. A. Mohammed, Dalia El Khoury, and et al. 2025. "Genomic and Structural Investigation of Mutations in Biotinidase (BTD) Gene Deficiency in Greater Middle Eastern Cohort: Insights from Molecular Dynamics Study" Biomedicines 13, no. 9: 2210. https://doi.org/10.3390/biomedicines13092210
APA StyleIbrahim, F. E., Gattu Linga, B., Samara, M., Roshanuddin, J., Younes, S., Nasrallah, G. K., Zayed, H., Qoronfleh, M. W., Mohammed, S. G. A. A., El Khoury, D., Velayutham, D., Abdoh, G., Al Rifai, H., & Al-Dewik, N. (2025). Genomic and Structural Investigation of Mutations in Biotinidase (BTD) Gene Deficiency in Greater Middle Eastern Cohort: Insights from Molecular Dynamics Study. Biomedicines, 13(9), 2210. https://doi.org/10.3390/biomedicines13092210













