Comprehensive Analysis of N6-Methyladenosine Methylation in Transverse Aortic Constriction-Induced Cardiac Fibrosis Based on MeRIP-Seq Analysis
Abstract
1. Introduction
2. Material and Methods
2.1. Animals and Transverse Aortic Constriction (TAC) Model
2.2. Echocardiographic Assessment
2.3. Histology Analysis
2.4. RNA Dot Blot
2.5. Real-Time Polymerase Chain Reaction (RT-PCR)
2.6. Western Blot Analysis (WB)
2.7. Methylated RNA Immunoprecipitation Sequencing (MeRIP-Seq)
2.8. Differential Expression Analysis and Functional Enrichment Analysis
2.9. Construction of Protein–Protein Interaction and Identification of Hub Genes
2.10. Establishment of Transcription Factors (TFs) and miRNA Coregulatory Network
2.11. Construction of Drug–Hub Genes Interaction Network and Molecular Docking Simulation
3. Results
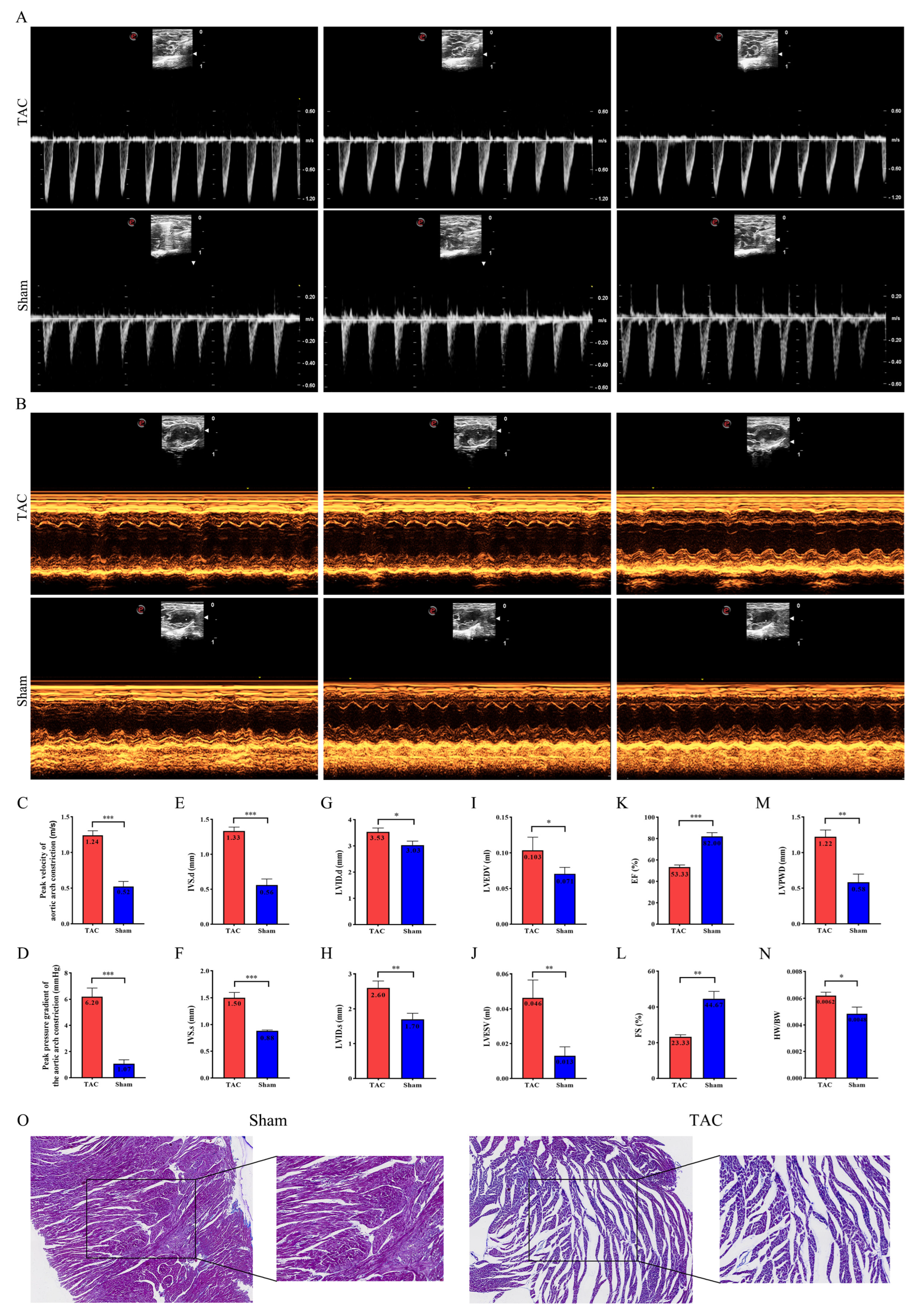
3.1. TAC Surgery Induced Ventricular Remodeling and Cardiac Fibrosis in Mice
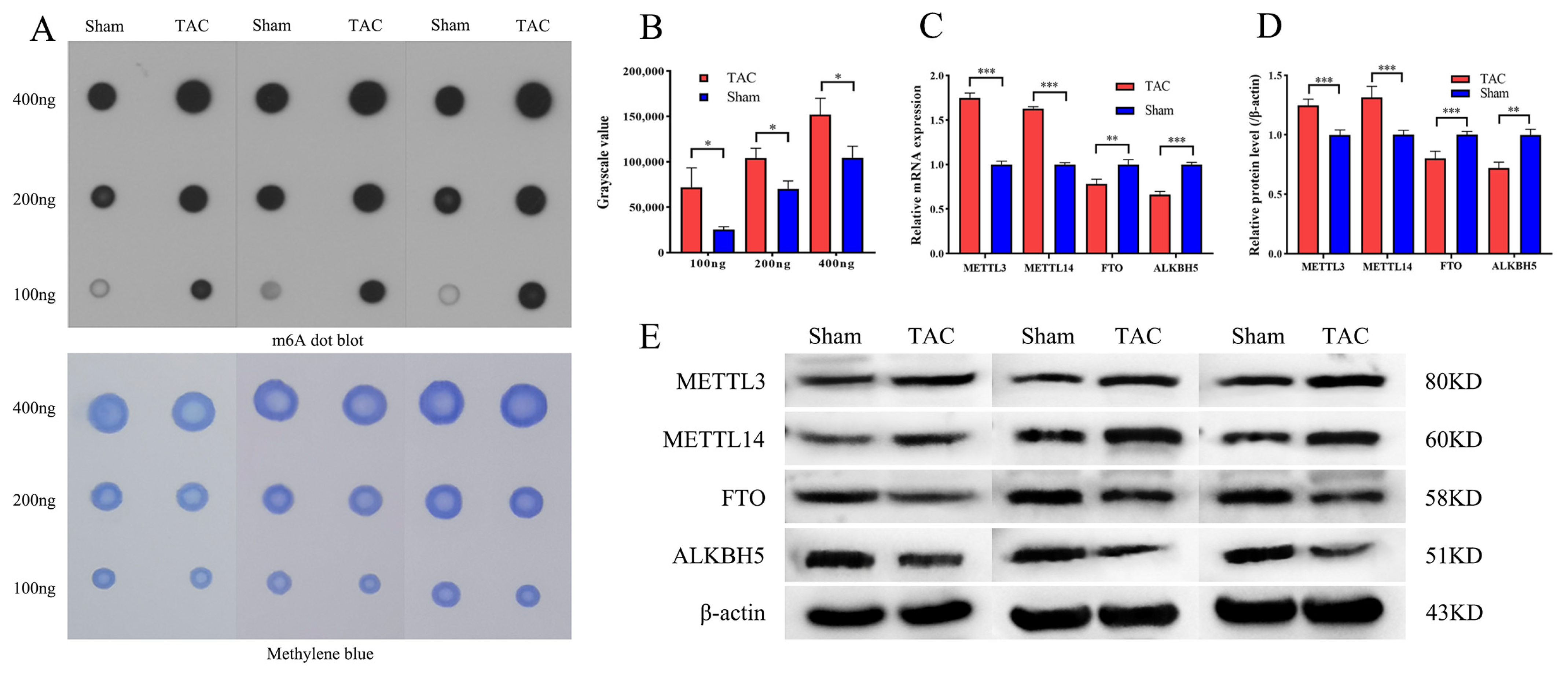
3.2. Excessive m6A Methylation and Alteration of m6A-Associated Modifying Enzymes
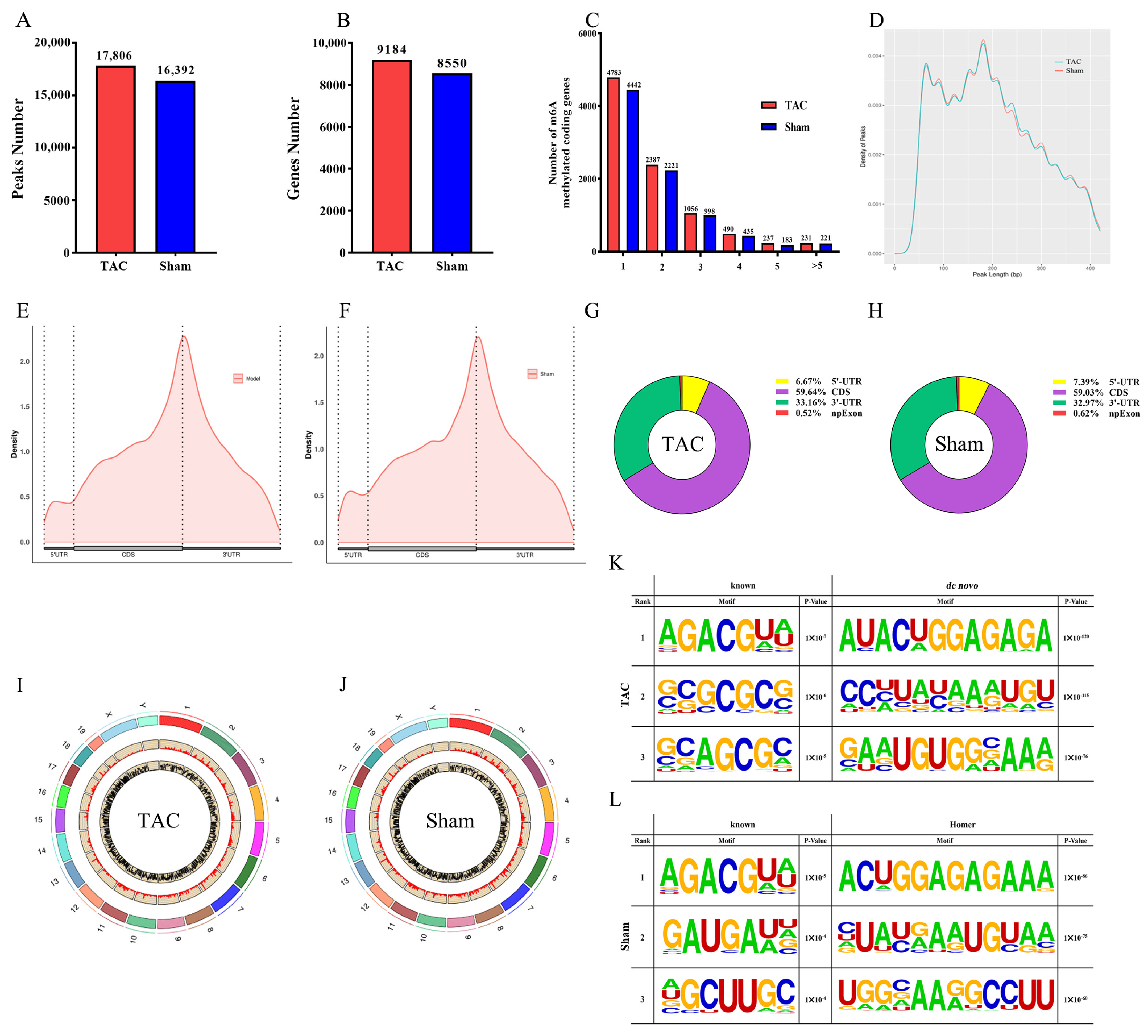
3.3. General and Topological Characteristics of m6A Methylation Modification in Cardiac Tissue-Based MeRIP-Seq Analysis
3.4. Conjoint Analysis and Functional Enrichment Analysis of MeRIP-Seq and RNA-Seq
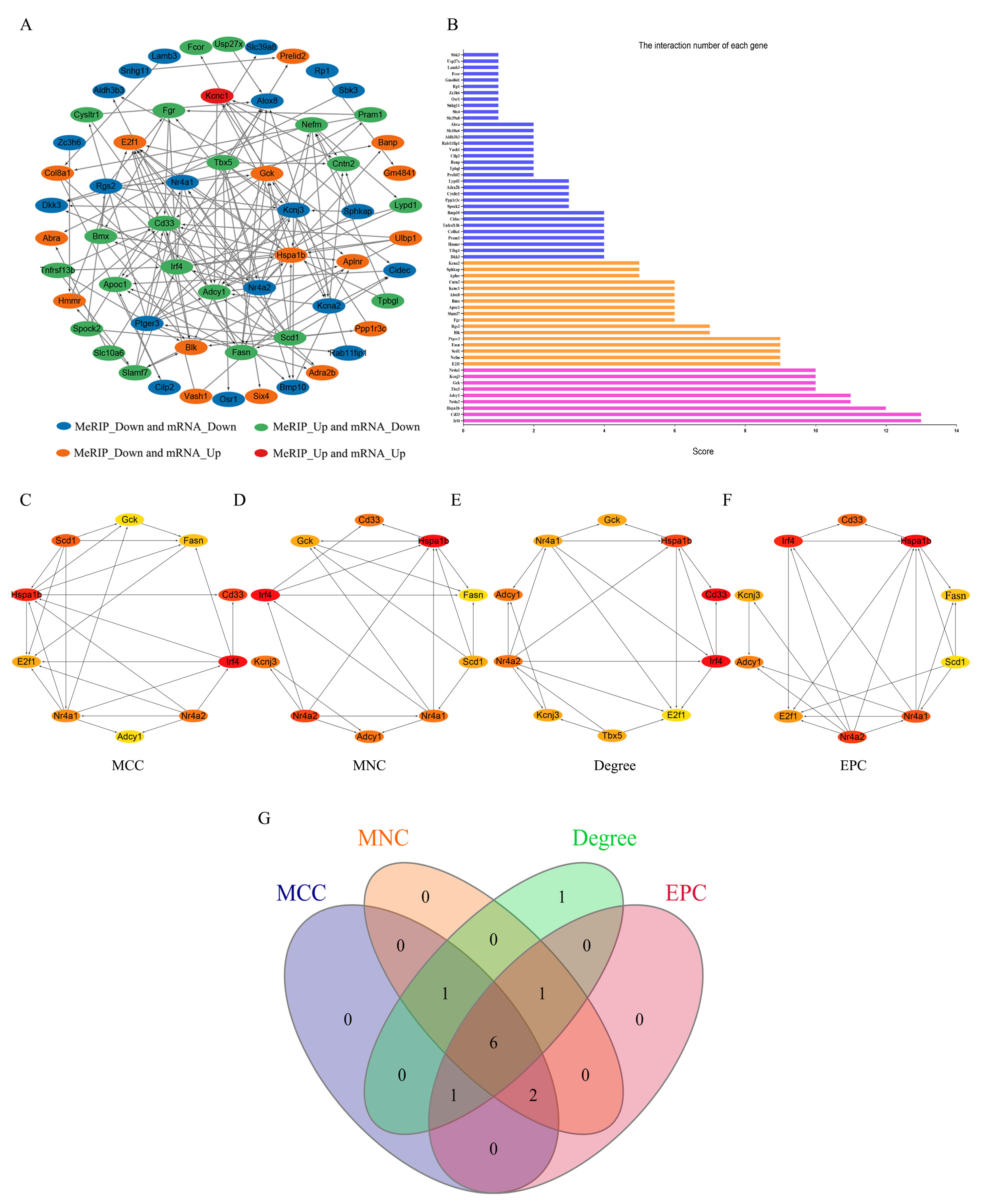
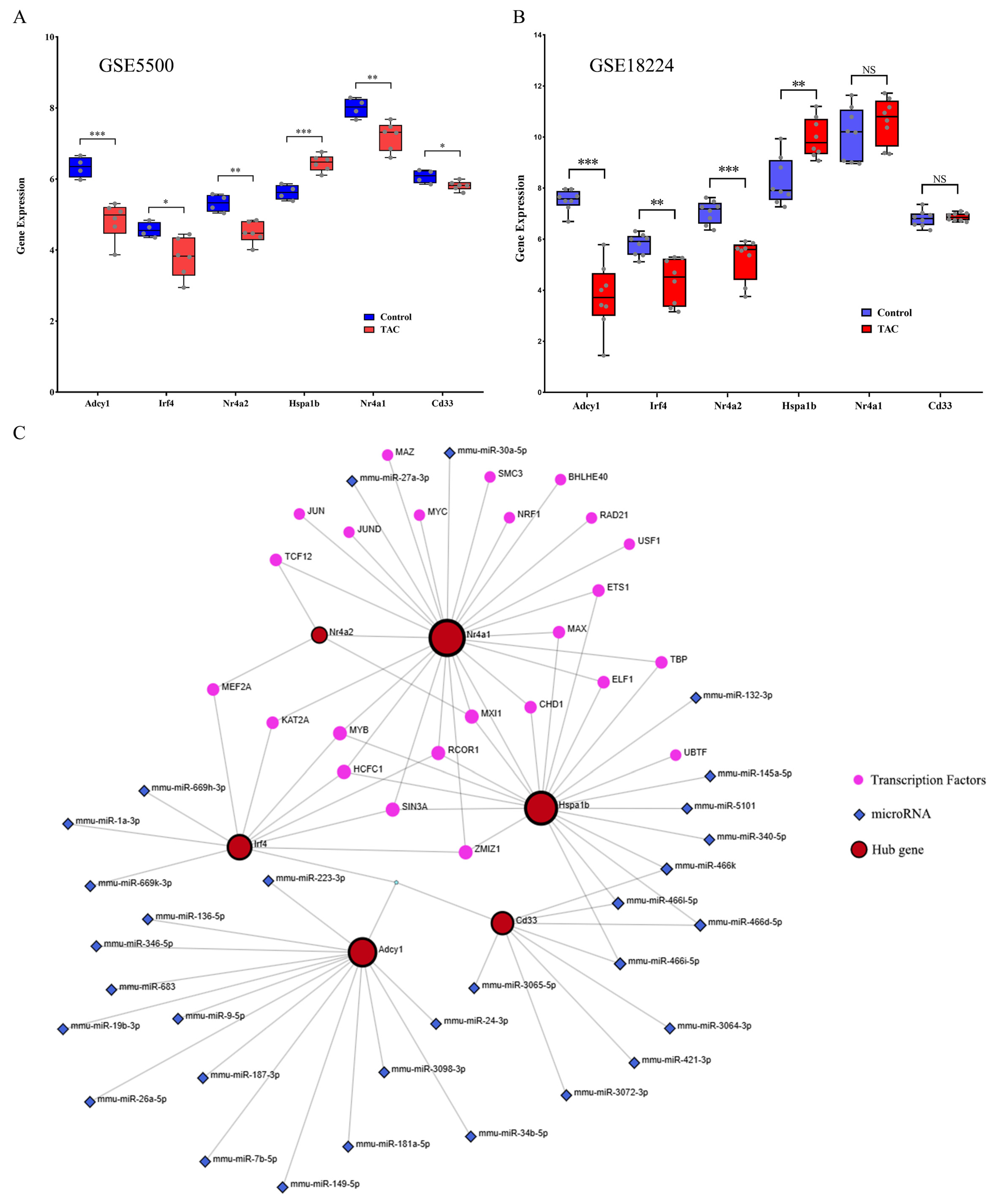
3.5. Construction of PPI Network and Identification of Hub Genes
3.6. Verification of the Six Hub Genes and Establishment of TF-miRNA Coregulatory Network
3.7. Drug–Hub Genes Interaction Network and Molecular Docking Simulation
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Frangogiannis, N.G. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol. Asp. Med. 2019, 65, 70–99. [Google Scholar] [CrossRef]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef] [PubMed]
- Berk, B.C.; Fujiwara, K.; Lehoux, S. ECM remodeling in hypertensive heart disease. J. Clin. Investig. 2007, 117, 568–575. [Google Scholar] [CrossRef]
- Bacmeister, L.; Schwarzl, M.; Warnke, S.; Stoffers, B.; Blankenberg, S.; Westermann, D.; Lindner, D. Inflammation and fibrosis in murine models of heart failure. Basic Res. Cardiol. 2019, 114, 19. [Google Scholar] [CrossRef]
- Moore-Morris, T.; Cattaneo, P.; Puceat, M.; Evans, S.M. Origins of cardiac fibroblasts. J. Mol. Cell. Cardiol. 2016, 91, 1–5. [Google Scholar] [CrossRef]
- Wu, Q.; Song, J.; Liu, W.; Li, L.; Li, S. Recent advances in positron emission tomography for detecting early fibrosis after myocardial infarction. Front. Cardiovasc. Med. 2024, 11, 1479777. [Google Scholar] [CrossRef]
- Wei, X.; Mao, Y.; Chen, Z.; Kang, L.; Xu, B.; Wang, K. Exercise-induced myocardial hypertrophy preconditioning promotes fibroblast senescence and improves myocardial fibrosis through Nrf2 signaling pathway. Cell Cycle. 2023, 22, 1529–1543. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, Y.; Jia, Q.; Wang, X.; Lu, Y.; Zhang, A.; Lv, S.; Zhang, J. Morphological and Functional Characteristics of Animal Models of Myocardial Fibrosis Induced by Pressure Overload. Int. J. Hypertens. 2020, 2020, 3014693. [Google Scholar] [CrossRef]
- Moore-Morris, T.; Guimarães-Camboa, N.; Yutzey, K.E.; Pucéat, M.; Evans, S.M. Cardiac fibroblasts: From development to heart failure. J. Mol. Med. 2015, 93, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Zhang, H.; Huang, S.; Yin, L.; Wang, F.; Luo, P.; Huang, H. Epigenetic regulation in cardiovascular disease: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2022, 7, 200. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5’ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Zhao, K.; Yang, C.X.; Li, P.; Sun, W.; Kong, X.Q. Epigenetic role of N6-methyladenosine (m6A) RNA methylation in the cardiovascular system. J. Zhejiang Univ. Sci. B. 2020, 21, 509–523. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Fu, Y.; Jia, G.; Pang, X.; Wang, R.N.; Wang, X.; Li, C.J.; Smemo, S.; Dai, Q.; Bailey, K.A.; Nobrega, M.A.; et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun. 2013, 4, 1798. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Flamand, M.N.; Meyer, K.D. m6A and YTHDF proteins contribute to the localization of select neuronal mRNAs. Nucleic Acids Res. 2022, 50, 4464–4483. [Google Scholar] [CrossRef] [PubMed]
- Ramesh-Kumar, D.; Guil, S. The IGF2BP family of RNA binding proteins links epitranscriptomics to cancer. Semin. Cancer Biol. 2022, 86 Pt 3, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Berulava, T.; Buchholz, E.; Elerdashvili, V.; Pena, T.; Islam, M.R.; Lbik, D.; Mohamed, B.A.; Renner, A.; von Lewinski, D.; Sacherer, M.; et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur. J. Heart Fail. 2020, 22, 54–66. [Google Scholar] [CrossRef]
- Li, T.; Zhuang, Y.; Yang, W.; Xie, Y.; Shang, W.; Su, S.; Dong, X.; Wu, J.; Jiang, W.; Zhou, Y.; et al. Silencing of METTL3 attenuates cardiac fibrosis induced by myocardial infarction via inhibiting the activation of cardiac fibroblasts. FASEB J. 2021, 35, e21162. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Y.; Deng, L.; Zha, Y.; Zhang, S. FTO suppresses cardiac fibrosis after myocardial infarction via m6A-mediated epigenetic modification of EPRS. Mol. Med. 2024, 30, 213. [Google Scholar] [CrossRef]
- Chen, L.; Zhao, Y.; Wang, W.; Zhang, S.; Zhou, X. Effect of FTO on cardiac hypertrophy through the regulation of OBSCN expression. Genes Dis. 2024, 11, 101165. [Google Scholar] [CrossRef]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8 (Suppl. 4), S11. [Google Scholar] [CrossRef]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef]
- Cotto, K.C.; Wagner, A.H.; Feng, Y.Y.; Kiwala, S.; Coffman, A.C.; Spies, G.; Wollam, A.; Spies, N.C.; Griffith, O.L.; Griffith, M. DGIdb 3.0: A redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018, 46, D1068–D1073. [Google Scholar] [CrossRef] [PubMed]
- Vietor, J.; Gege, C.; Stiller, T.; Busch, R.; Schallmayer, E.; Kohlhof, H.; Höfner, G.; Pabel, J.; Marschner, J.A.; Merk, D. Development of a potent nurr1 agonist tool for in vivo applications. J. Med. Chem. 2023, 66, 6391–6402. [Google Scholar] [CrossRef]
- Stiller, T.; Merk, D. Exploring fatty acid mimetics as NR4A ligands. J. Med. Chem. 2023, 66, 15362–15369. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Song, K.; Tu, B.; Sun, H.; Ding, J.F.; Luo, Y.; Sha, J.-M.; Li, R.; Zhang, Y.; Zhao, J.-Y.; et al. METTL3 boosts glycolysis and cardiac fibroblast proliferation by increasing AR methylation. Int. J. Biol. Macromol. 2022, 223 Pt A, 899–915. [Google Scholar] [CrossRef]
- Ding, J.F.; Sun, H.; Song, K.; Zhou, Y.; Tu, B.; Shi, K.-H.; Lu, D.; Xu, S.-S.; Tao, H. IGFBP3 epigenetic promotion induced by METTL3 boosts cardiac fibroblast activation and fibrosis. Eur. J. Pharmacol. 2023, 942, 175494. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, P.; Xiao, Y.; Zha, L.; Ma, J.; Xiao, H. METTL14 Promotes Lipopolysaccharide-Induced Myocardial Damage via m6A-Dependent Stabilization of TRPM7 mRNA. Int. Heart J. 2024, 65, 1118–1127. [Google Scholar] [CrossRef]
- Mathiyalagan, P.; Adamiak, M.; Mayourian, J.; Sassi, Y.; Liang, Y.; Agarwal, N.; Jha, D.; Zhang, S.; Kohlbrenner, E.; Chepurko, E.; et al. FTO-Dependent N6-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019, 139, 518–532. [Google Scholar] [CrossRef]
- Ju, W.; Liu, K.; Ouyang, S.; Liu, Z.; He, F.; Wu, J. Changes in N6-Methyladenosine Modification Modulate Diabetic Cardiomyopathy by Reducing Myocardial Fibrosis and Myocyte Hypertrophy. Front. Cell Dev. Biol. 2021, 9, 702579. [Google Scholar] [CrossRef]
- Zhuang, T.; Chen, M.H.; Wu, R.X.; Wang, J.; Hu, X.D.; Meng, T.; Wu, A.-H.; Yang, Y.-F.; Lei, Y.; Hu, D.-H.; et al. ALKBH5-mediated m6A modification of IL-11 drives macrophage-to-myofibroblast transition and pathological cardiac fibrosis in mice. Nat. Commun. 2024, 15, 1995. [Google Scholar] [CrossRef]
- Li, W.; Xing, C.; Bao, L.; Han, S.; Luo, T.; Wang, Z.; Fan, H. Comprehensive analysis of RNA m6A methylation in pressure overload-induced cardiac hypertrophy. BMC Genom. 2022, 23, 576. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Möller, A.; Jauch-Speer, S.L.; Gandhi, S.; Vogl, T.; Roth, J.; Fehler, O. The roles of toll-like receptor 4, CD33, CD68, CD69, or CD147/EMMPRIN for monocyte activation by the DAMP S100A8/S100A9. Front. Immunol. 2023, 14, 1110185. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Wang, R.; Song, C.; Liu, J.; Chen, R.; Zheng, M.; Liu, W.; Jiang, G.; Mao, W. Exploring the causal relationship between immune cells and idiopathic pulmonary fibrosis: A bi-directional Mendelian randomization study. BMC Pulm. Med. 2024, 24, 145. [Google Scholar] [CrossRef]
- Tamura, T.; Yanai, H.; Savitsky, D.; Taniguchi, T. The IRF family transcription factors in immunity and oncogenesis. Annu. Rev. Immunol. 2008, 26, 535–584. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, G.; Moschovaki-Filippidou, F.; Würf, V.; Metzger, P.; Steiger, S.; Batz, F.; Carbajo-Lozoya, J.; Koziel, J.; Schnurr, M.; Cohen, C.D.; et al. IFN Regulatory Factor 4 Controls Post-ischemic Inflammation and Prevents Chronic Kidney Disease. Front. Immunol. 2019, 10, 2162. [Google Scholar] [CrossRef]
- Liu, H.; Liu, P.; Shi, X.; Yin, D.; Zhao, J. NR4A2 protects cardiomyocytes against myocardial infarction injury by promoting autophagy. Cell Death Discov. 2018, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, S.S.; Rezq, S.; Alsemeh, A.E.; Mahmoud, M.F. Moxonidine ameliorates cardiac injury in rats with metabolic syndrome by regulating autophagy. Life Sci. 2023, 312, 121210. [Google Scholar] [CrossRef]
- Tao, H.; Xu, W.; Qu, W.; Gao, H.; Zhang, J.; Cheng, X.; Liu, N.; Chen, J.; Xu, G.-L.; Li, X.; et al. Loss of ten-eleven translocation 2 induces cardiac hypertrophy and fibrosis through modulating ERK signaling pathway. Hum. Mol. Genet. 2021, 30, 865–879. [Google Scholar] [CrossRef]
- Wilson, A.J.; Arango, D.; Mariadason, J.M.; Heerdt, B.G.; Augenlicht, L.H. TR3/Nur77 in colon cancer cell apoptosis. Cancer Res. 2003, 63, 5401–5407. [Google Scholar]
- Ma, M.F.; Chen, Z.Y.; Wang, L.J.; Li, N.; Guo, B.Y. Orphan nuclear receptor 4 A1 involvement in transforming growth factor beta1-induced myocardial fibrosis in diabetic mice. J. Physiol. Pharmacol. 2023, 74, 633–646. [Google Scholar]
- Yang, H.X.; Sun, J.H.; Yao, T.T.; Li, Y.; Xu, G.R.; Zhang, C.; Liu, X.-C.; Zhou, W.-W.; Song, Q.-H.; Zhang, Y.; et al. Bellidifolin Ameliorates Isoprenaline-Induced Myocardial Fibrosis by Regulating TGF-β1/Smads and p38 Signaling and Preventing NR4A1 Cytoplasmic Localization. Front. Pharmacol. 2021, 12, 644886. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zeng, F.; Chen, Z.; Qin, Z.; Liu, Z. Regulation of cardiac fibrosis in mice with TAC/DOCA-induced HFpEF by resistin-like molecule gamma and adenylate cyclase 1. FEBS Open Bio 2024, 14, 1101–1115. [Google Scholar] [CrossRef] [PubMed]



| Genes | Primer Sequence 5′-3′ | Amplified Fragments/bp |
|---|---|---|
| β-actin | CATCCGTAAAGACCTCTATGCC AACATGGAGCCACCGATCCACA | 171 |
| METTL3 | GGACTCTGGGCACTTGGATTTA | 248 |
| CAGGTGCATCTGGCGTAGAG | ||
| METTL14 | GACTGGCATCACTGCGAATG | 126 |
| AGGTCCAATCCTTCCCCAGA | ||
| ALKBH5 | CACGTTGACCCCATCCACAT | 240 |
| CCTGAGAATGATGACCGCCC | ||
| FTO | TCTGTGTGTTGGGTGTCCTTT | 143 |
| AAAACGACAGCGGTGCTTAC |
| Antibody | Dilution | Source | Article No. |
|---|---|---|---|
| METTL14 | 1/1000 | Fine Test, Wuhan Fine Biotech Co., Wuhan, China | FNab10824 |
| METTL3 | 1/1000 | Fine Test, Wuhan Fine Biotech Co., Wuhan, China | FNab05139 |
| ALKBH5 | 1/1000 | Fine Test, Wuhan Fine Biotech Co., Wuhan, China | FNab00314 |
| FTO | 1/1000 | Fine Test, Wuhan Fine Biotech Co., Wuhan, China | FNab09787 |
| β-actin | 1/5000 | ImmunoWay, Plano, TX, USA | YT0099 |
| Gene | Drug Name | Molecular Formula | Structure | Docking Score (kcal/mol) |
|---|---|---|---|---|
| Nr4a2 | Compound 29 Vietor et al., 2023 [31] | C22H18FNO4 |  | −8.3 |
| Compound 108 PMID 37918435 [32] | C15H11F3O3 |  | −7.3 | |
| Compound 111 PMID 37918435 [32] | C14H10Cl2O3 |  | −6.8 | |
| Vidofludimus | C20H18FNO4 |  | −7.8 | |
| Adcy1 | Adenosine Monophosphate | C10H14N5O7P |  | −9.1 |
| Nr4a1 | Acetylcysteine | C5H9NO3S | 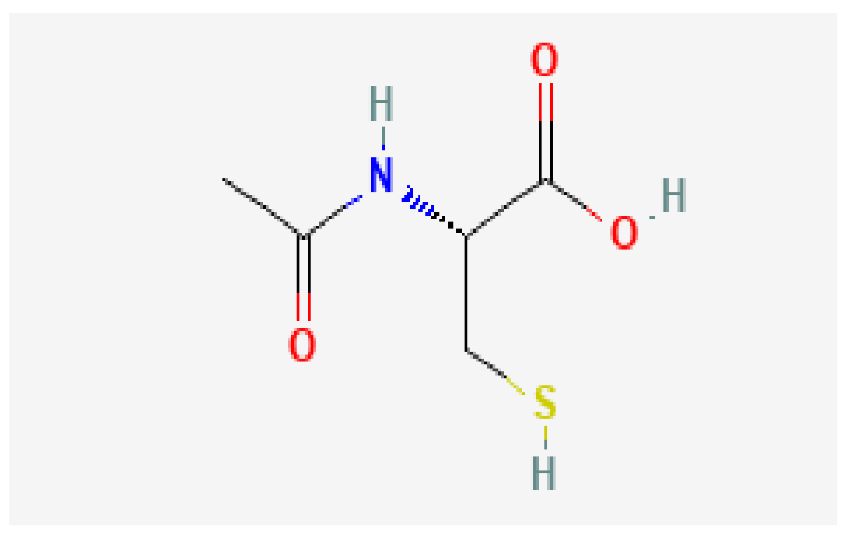 | −4.1 |
| CHEMBL547833 | C18H19ClN4O |  | −6.9 | |
| Cytosporone B | C18H26O5 |  | −5.9 | |
| Etoposide | C29H32O13 | 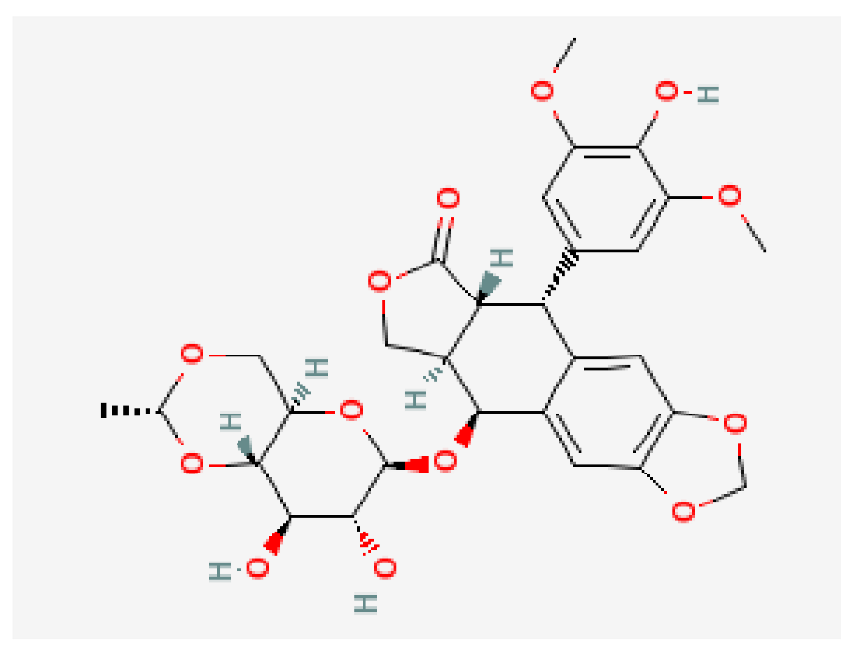 | −8.4 | |
| Histone deacetylase inhibitor | C24H28ClN3O4 | 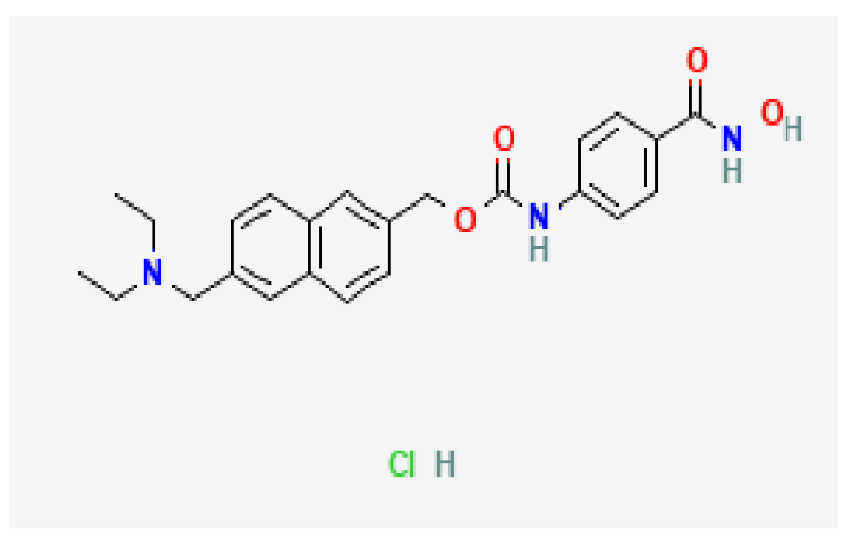 | −7.5 | |
| Levodopa | C9H11NO4 | 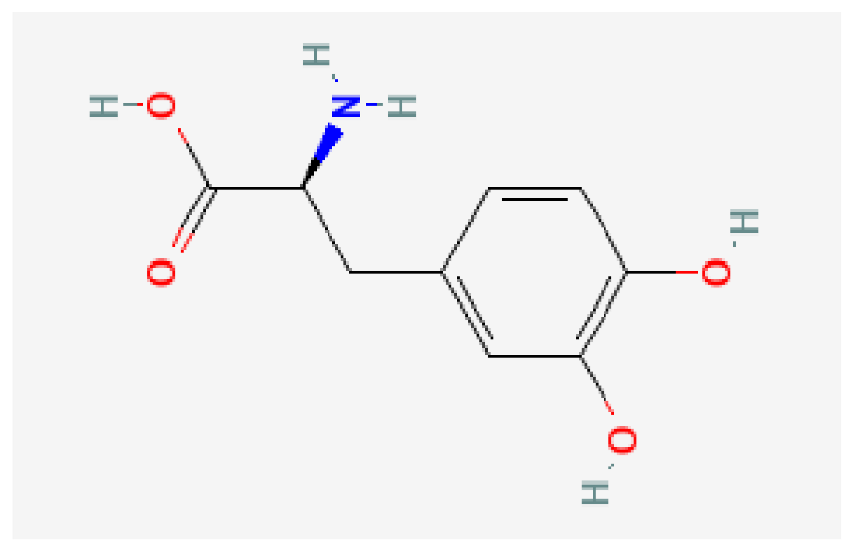 | −6.2 | |
| PD-98059 | C16H13NO3 | 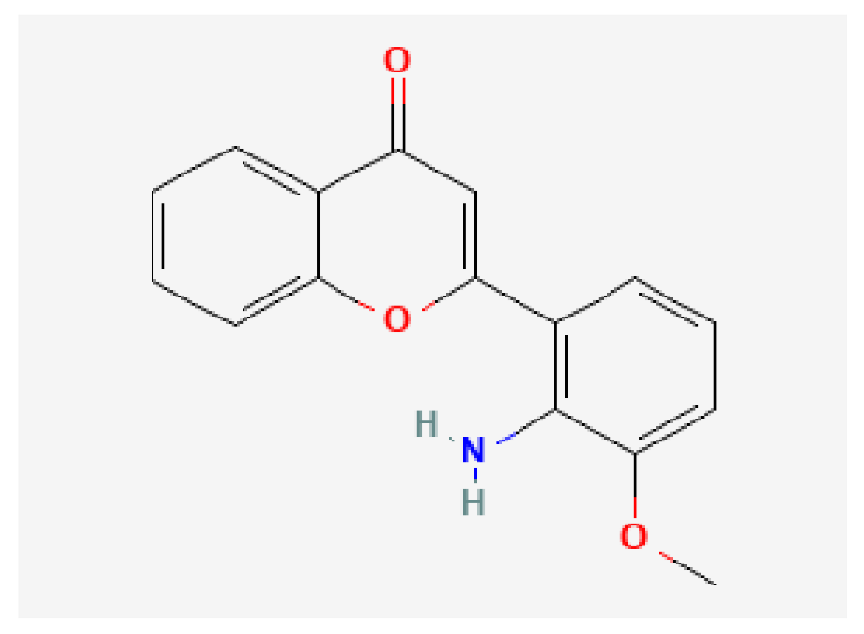 | −7.3 | |
| Tretinoin | C20H28O2 |  | −6.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Zhao, P.; He, Y.; Wang, J.; Song, B.; Yu, C. Comprehensive Analysis of N6-Methyladenosine Methylation in Transverse Aortic Constriction-Induced Cardiac Fibrosis Based on MeRIP-Seq Analysis. Biomedicines 2025, 13, 2092. https://doi.org/10.3390/biomedicines13092092
Liu S, Zhao P, He Y, Wang J, Song B, Yu C. Comprehensive Analysis of N6-Methyladenosine Methylation in Transverse Aortic Constriction-Induced Cardiac Fibrosis Based on MeRIP-Seq Analysis. Biomedicines. 2025; 13(9):2092. https://doi.org/10.3390/biomedicines13092092
Chicago/Turabian StyleLiu, Shidong, Pengying Zhao, Yuyuan He, Jieneng Wang, Bing Song, and Cuntao Yu. 2025. "Comprehensive Analysis of N6-Methyladenosine Methylation in Transverse Aortic Constriction-Induced Cardiac Fibrosis Based on MeRIP-Seq Analysis" Biomedicines 13, no. 9: 2092. https://doi.org/10.3390/biomedicines13092092
APA StyleLiu, S., Zhao, P., He, Y., Wang, J., Song, B., & Yu, C. (2025). Comprehensive Analysis of N6-Methyladenosine Methylation in Transverse Aortic Constriction-Induced Cardiac Fibrosis Based on MeRIP-Seq Analysis. Biomedicines, 13(9), 2092. https://doi.org/10.3390/biomedicines13092092





