The Evolving Landscape of Host Biomarkers for Diagnosis and Monitoring of Tuberculosis
Abstract
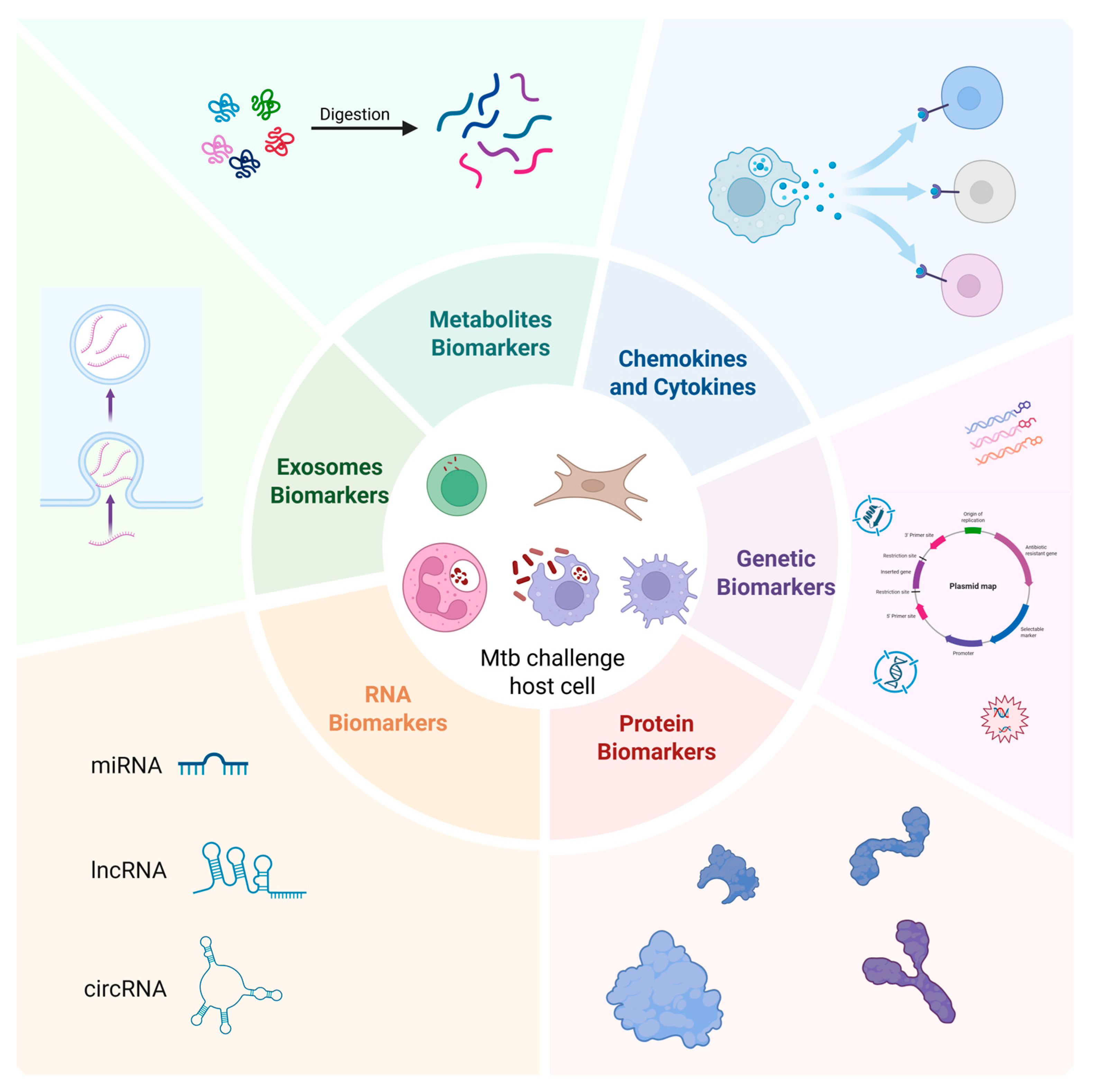
1. Introduction
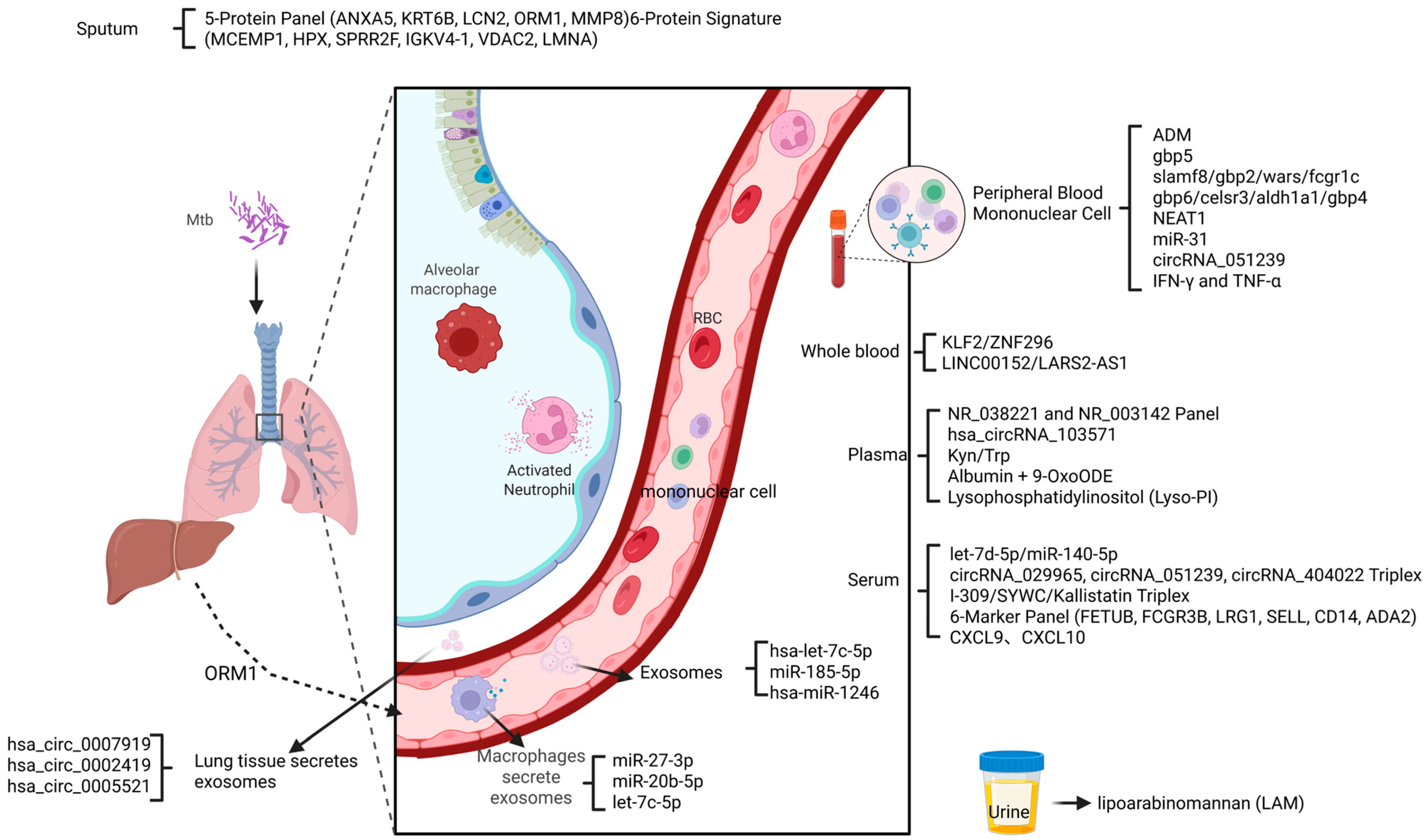
2. Genetic Biomarkers
3. RNA Biomarkers
4. Protein Biomarkers
5. Chemokine and Cytokine Biomarkers
6. Metabolites Biomarkers
7. Exosome Biomarkers
8. Expectations
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Global Tuberculosis Report 2024; World Health Organ: Geneva, Switzerland, 2024. [Google Scholar]
- Boom, W.H.; Schaible, U.E.; Achkar, J.M. The knowns and unknowns of latent Mycobacterium tuberculosis infection. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Zhang, H.; Guan, W.; Zhou, J. Advances in the Diagnosis of Latent Tuberculosis Infection. Infect. Drug Resist. 2025, 18, 483–493. [Google Scholar] [CrossRef]
- Asgharzadeh, V.; Seyyed Rezaei, S.A.; Asgharzadeh, M.; Rashedi, J.; Kafil, H.S.; Nobari, H.J.; Khalili, A.A.; Raeisi, M.; Ozma, M.A.; Poor, B.M. Host Risk Factors for Tuberculosis. Infect. Disord. Drug Targets 2025, 25, e18715265304343. [Google Scholar] [CrossRef] [PubMed]
- Palanivel, J.; Sounderrajan, V.; Thangam, T.; Rao, S.S.; Harshavardhan, S.; Parthasarathy, K. Latent Tuberculosis: Challenges in Diagnosis and Treatment, Perspectives, and the Crucial Role of Biomarkers. Curr. Microbiol. 2023, 80, 392. [Google Scholar] [CrossRef] [PubMed]
- Mohammadnabi, N.; Shamseddin, J.; Emadi, M.; Bodaghi, A.B.; Varseh, M.; Shariati, A.; Rezaei, M.; Dastranj, M.; Farahani, A. Mycobacterium tuberculosis: The Mechanism of Pathogenicity, Immune Responses, and Diagnostic Challenges. J. Clin. Lab. Anal. 2024, 38, e25122. [Google Scholar] [CrossRef] [PubMed]
- Suárez, I.; Fünger, S.M.; Kröger, S.; Rademacher, J.; Fätkenheuer, G.; Rybniker, J. The Diagnosis and Treatment of Tuberculosis. Dtsch. Arztebl. Int. 2019, 116, 729–735. [Google Scholar]
- Goletti, D.; Delogu, G.; Matteelli, A.; Migliori, G.B. The role of IGRA in the diagnosis of tuberculosis infection, differentiating from active tuberculosis, and decision making for initiating treatment or preventive therapy of tuberculosis infection. Int. J. Infect. Dis. 2022, 124, S12–S19. [Google Scholar] [CrossRef]
- Ludi, Z.; Sule, A.A.; Samy, R.P.; Putera, I.; Schrijver, B.; Hutchinson, P.E.; Gunaratne, J.; Verma, I.; Singhal, A.; Nora, R.L.D.; et al. Diagnosis and biomarkers for ocular tuberculosis: From the present into the future. Theranostics 2023, 13, 2088–2113. [Google Scholar] [CrossRef]
- Rahlwes, K.C.; Dias, B.R.; Campos, P.C.; Alvarez-Arguedas, S.; Shiloh, M.U. Pathogenicity and virulence of Mycobacterium tuberculosis. Virulence 2023, 14, 2150449. [Google Scholar] [CrossRef]
- Collins, J.M.; Walker, D.I.; Jones, D.P.; Tukvadze, N.; Liu, K.H.; Tran, V.T.; Uppal, K.; Frediani, J.K.; Easley, K.A.; Shenvi, N.; et al. High-resolution plasma metabolomics analysis to detect Mycobacterium tuberculosis-associated metabolites that distinguish active pulmonary tuberculosis in humans. PLoS ONE 2018, 13, e0205398. [Google Scholar] [CrossRef]
- Carranza, C.; Herrera, M.T.; Guzmán-Beltrán, S.; Salgado-Cantú, M.G.; Salido-Guadarrama, I.; Santiago, E.; Chávez-Galán, L.; Gutiérrez-González, L.H.; González, Y. A Dual Marker for Monitoring MDR-TB Treatment: Host-Derived miRNAs and M. tuberculosis-Derived RNA Sequences in Serum. Front. Immunol. 2021, 12, 760468. [Google Scholar] [CrossRef]
- Gong, W.; Wu, X. Differential Diagnosis of Latent Tuberculosis Infection and Active Tuberculosis: A Key to a Successful Tuberculosis Control Strategy. Front. Microbiol. 2021, 12, 745592. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hu, Y.; Wang, W.; Zou, F.; Yang, J.; Gao, W.; Feng, S.; Chen, G.; Shi, C.; Cai, Y.; et al. Integrating pathogen- and host-derived blood biomarkers for enhanced tuberculosis diagnosis: A comprehensive review. Front. Immunol. 2024, 15, 1438989. [Google Scholar] [CrossRef] [PubMed]
- Sivakumaran, D.; Jenum, S.; Vaz, M.; Selvam, S.; Ottenhoff, T.H.M.; Haks, M.C.; Malherbe, S.T.; Doherty, T.M.; Ritz, C.; Grewal, H.M.S. Combining host-derived biomarkers with patient characteristics improves signature performance in predicting tuberculosis treatment outcomes. Commun. Biol. 2020, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zafari, P.; Golpour, M.; Hafezi, N.; Bashash, D.; Esmaeili, S.; Tavakolinia, N.; Rafiei, A. Tuberculosis comorbidity with rheumatoid arthritis: Gene signatures, associated biomarkers, and screening. IUBMB Life 2021, 73, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, B. Hepatocellular carcinoma: Molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. 2023, 42, 629–652. [Google Scholar] [CrossRef]
- Lee, S.W.; Wu, L.S.; Huang, G.M.; Huang, K.Y.; Lee, Z.Y.; Weng, J.T.Y. Gene expression profiling identifies candidate biomarkers for active and latent tuberculosis. BMC Bioinform. 2016, 17, 3. [Google Scholar] [CrossRef]
- John, S.H.; Kenneth, J.; Gandhe, A.S. Host biomarkers of clinical relevance in tuberculosis: Review of gene and protein expression studies. Biomarkers 2012, 17, 1–8. [Google Scholar] [CrossRef]
- Krishnan, P.; Bobak, C.A.; Hill, J.E. Sex-specific blood-derived RNA biomarkers for childhood tuberculosis. Sci. Rep. 2024, 14, 16. [Google Scholar] [CrossRef]
- Kasule, G.W.; Hermans, S.; Semugenze, D.; Wekiya, E.; Nsubuga, J.; Mwachan, P.; Kabugo, J.; Joloba, M.; García-Basteiro, A.L.; Ssengooba, W. Non-sputum-based samples and biomarkers for detection of Mycobacterium tuberculosis: The hope to improve childhood and HIV-associated tuberculosis diagnosis. Eur. J. Med. Res. 2024, 29, 1–13. [Google Scholar] [CrossRef]
- Xu, Y.; Tan, Y.; Zhang, X.; Cheng, M.; Hu, J.; Liu, J.; Chen, X.; Zhu, J. Comprehensive identification of immuno-related transcriptional signature for active pulmonary tuberculosis by integrated analysis of array and single cell RNA-seq. J. Infect. 2022, 85, 534–544. [Google Scholar] [CrossRef]
- Huynh, J.; Nhat, L.H.T.; Bao, N.L.H.; Hai, H.T.; Thu, D.D.A.; Tram, T.T.B.; Dung, V.T.M.; Vinh, D.D.; Ngoc, N.M.; Donovan, J.; et al. The Ability of a 3-Gene Host Signature in Blood to Distinguish Tuberculous Meningitis From Other Brain Infections. J. Infect. Dis. 2024, 230, e268–e278. [Google Scholar] [CrossRef]
- Ansari, A.; Singh, G.P.; Singh, M.; Singh, H. Identification of host immune-related biomarkers in active tuberculosis: A comprehensive analysis of differentially expressed genes. Tuberculosis 2024, 148, 102538. [Google Scholar] [CrossRef] [PubMed]
- Komakech, K.; Semugenze, D.; Joloba, M.; Cobelens, F.; Ssengooba, W. Diagnostic accuracy of point-of-care triage tests for pulmonary tuberculosis using host blood protein biomarkers: A systematic review and meta-analysis. eClinicalMedicine 2025, 84, 103257. [Google Scholar] [CrossRef]
- Nakiboneka, R.; Walbaum, N.; Musisi, E.; Nevels, M.; Nyirenda, T.; Nliwasa, M.; Msefula, C.L.; Sloan, D.; Sabiiti, W. Specific human gene expression in response to infection is an effective marker for diagnosis of latent and active tuberculosis. Sci. Rep. 2024, 14, 26884. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.; Loy, C.J.; Eweis-LaBolle, D.; Lenz, J.S.; Steadman, A.; Andgrama, A.; Nhung, N.V.; Yu, C.; Worodria, W.; Denkinger, C.M.; et al. Circulating cell-free RNA in blood as a host response biomarker for detection of tuberculosis. Nat. Commun. 2024, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Nakiboneka, R.; Margaritella, N.; Nyirenda, T.; Chaima, D.; Walbaum, N.; Musisi, E.; Tionge, S.; Msosa, T.; Nliwasa, M.; Msefula, C.L.; et al. Suppression of host gene expression is associated with latent TB infection: A possible diagnostic biomarker. Sci. Rep. 2024, 14, 15621. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, Q.; Ma, Q.; Chen, J.; Kong, X.; Zeng, Y.; Liu, L.; Lu, S.; Wang, X. Identification of core biomarkers for tuberculosis progression through bioinformatics analysis and in vitro research. Sci. Rep. 2025, 15, 3137. [Google Scholar] [CrossRef]
- Ding, S.; Huang, C.; Gao, J.; Bi, C.; Zhou, Y.; Cai, Z. Exploring T-cell metabolism in tuberculosis: Development of a diagnostic model using metabolic genes. Eur. J. Med. Res. 2025, 30, 1–24. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, X.; Xu, J.; He, J.; Lu, X. Identification and functional characterization of glycosyltransferase-related biomarkers for tuberculosis diagnosis. AMB Express 2025, 15, 56. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, X.; Wang, B.; Tang, L.; Liu, X.; Ding, X.; Dong, Y.; Lei, H.; Wang, D.; Feng, H. Single-cell transcriptomic analysis reveals a decrease in the frequency of macrophage-RGS1high subsets in patients with osteoarticular tuberculosis. Mol. Med. 2024, 30, 118. [Google Scholar] [CrossRef]
- Kanabalan, R.D.; Lee, L.J.; Lee, T.Y.; Chong, P.P.; Hassan, L.; Ismail, R.; Chin, V.K. Human tuberculosis and Mycobacterium tuberculosis complex: A review on genetic diversity, pathogenesis and omics approaches in host biomarkers discovery. Microbiol. Res. 2021, 246, 126674. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, Y.; Li, H. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018, 257, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wu, K.J.; Jia, Q.J.; Ding, X.F. Roles of miRNA and lncRNA in triple-negative breast Cancer. J. Zhejiang Univ. Sci. B 2020, 21, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.F.; Yuan, W.; Du, Y.L.; Wang, B.L. Research progress of lncRNA and miRNA in hepatic ischemia-reperfusion injury. Hepatobiliary Pancreat. Dis. Int. 2023, 22, 45–53. [Google Scholar] [CrossRef]
- Kundu, M.; Basu, J. The Role of microRNAs and Long Non-Coding RNAs in the Regulation of the Immune Response to Mycobacterium tuberculosis Infection. Front. Immunol. 2021, 12, 687962. [Google Scholar] [CrossRef]
- Fathizadeh, H.; Hayat, S.M.G.; Dao, S.; Ganbarov, K.; Tanomand, A.; Asgharzadeh, M.; Kafil, H.S. Long non-coding RNA molecules in tuberculosis. Int. J. Biol. Macromol. 2020, 156, 340–346. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Ma, J.; Gong, H.; Shao, J.; Li, J.; Zhan, Y.; Wang, Z.; Wang, C.; Li, W. Immune regulation and emerging roles of noncoding RNAs in Mycobacterium tuberculosis infection. Front. Immunol. 2022, 13, 987018. [Google Scholar] [CrossRef]
- Xu, W.; Yang, J.; Yu, H.; Li, S. Diagnostic value of lncRNAs LINC00152 and LARS2-AS1 and their regulatory roles in macrophage immune response in tuberculosis. Tuberculosis 2024, 148, 102530. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cui, J.; Niu, W.; Huang, J.; Feng, T.; Sun, B.; Yao, H. Long non-coding PCED1B-AS1 regulates macrophage apoptosis and autophagy by sponging miR-155 in active tuberculosis. Biochem. Biophys. Res. Commun. 2019, 509, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Huang, Z.; Luo, Q.; Qing, C. The Expression of lncRNA NEAT1 in Human Tuberculosis and Its Antituberculosis Effect. BioMed Res. Int. 2018, 2018, 9529072. [Google Scholar] [CrossRef]
- Xu, S.; Yuan, H.; Li, L.; Yang, K.; Zhao, L. Identification of N6-methylandenosine-related lncRNA for tuberculosis diagnosis in person living with human immunodeficiency virus. Tuberculosis 2023, 140, 102337. [Google Scholar] [CrossRef]
- Chen, Z.L.; Wei, L.L.; Shi, L.Y.; Li, M.; Jiang, T.T.; Chen, J.; Liu, C.M.; Yang, S.; Tu, H.H.; Hu, Y.T.; et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci. Rep. 2017, 7, 16751. [Google Scholar] [CrossRef]
- Hu, X.; Liao, S.; Bai, H.; Gupta, S.; Zhou, Y.; Zhou, J.; Jiao, L.; Wu, L.; Wang, M.; Chen, X.; et al. Long Noncoding RNA and Predictive Model To Improve Diagnosis of Clinically Diagnosed Pulmonary Tuberculosis. J. Clin. Microbiol. 2020, 58, 7. [Google Scholar] [CrossRef]
- Kesheh, M.M.; Bayat, M.; Kobravi, S.; Lotfalizadeh, M.H.; Heydari, A.; Memar, M.Y.; Baghi, H.B.; Kermanshahi, A.Z.; Ravaei, F.; Taghavi, S.P.; et al. MicroRNAs and human viral diseases: A focus on the role of microRNA-29. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2025, 1871, 167500. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, Y.; Liu, T.; Shi, Y.; Wang, Y.; Wu, J.; Qi, Y. Novel Biomarker Panel of Let-7d-5p and MiR-140-5p Can Distinguish Latent Tuberculosis Infection from Active Tuberculosis Patients. Infect. Drug Resist. 2023, 16, 3847–3859. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.; Nair, V.; Gcanga, L.; Lakshmanan, V.; Kalamuddin, M.; Anang, V.; Rathore, S.; Dhawan, S.; Alam, T.; Khanna, V.; et al. Identifying quantitative sncRNAs signature using global sequencing as a potential biomarker for tuberculosis diagnosis and their role in regulating host response. Int. J. Biol. Macromol. 2024, 271, 132714. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.K.; Yao, F.Y.; Xu, J.Q.; Deng, Z.; Su, R.G.; Peng, Y.P.; Luo, Q.; Li, J.M. Microarray Expression Profile of Circular RNAs in Peripheral Blood Mononuclear Cells from Active Tuberculosis Patients. Cell Physiol. Biochem. 2018, 45, 1230–1240. [Google Scholar] [CrossRef]
- Yi, Z.; Gao, K.; Li, R.; Fu, Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J. Cell. Mol. Med. 2018, 22, 4076–4084. [Google Scholar] [CrossRef]
- Kazemi, S.; Mirzaei, R.; Karampoor, S.; Hosseini-Fard, S.R.; Ahmadyousefi, Y.; Soltanian, A.R.; Keramat, F.; Saidijam, M.; Alikhani, M.Y. Circular RNAs in tuberculosis: From mechanism of action to potential diagnostic biomarker. Microb. Pathog. 2023, 185, 106459. [Google Scholar] [CrossRef]
- Huang, Z.; Su, R.; Yao, F.; Peng, Y.; Luo, Q.; Li, J. Circulating circular RNAs hsa_circ_0001204 and hsa_circ_0001747 act as diagnostic biomarkers for active tuberculosis detection. Int. J. Clin. Exp. Pathol. 2018, 11, 586–594. [Google Scholar]
- Sampath, P.; Periyasamy, K.M.; Ranganathan, U.D.; Bethunaickan, R. Monocyte and Macrophage miRNA: Potent Biomarker and Target for Host-Directed Therapy for Tuberculosis. Front. Immunol. 2021, 12, 667206. [Google Scholar] [CrossRef]
- Daniel, E.A.; Sathiyamani, B.; Thiruvengadam, K.; Vivekanandan, S.; Vembuli, H.; Hanna, L.E. MicroRNAs as diagnostic biomarkers for Tuberculosis: A systematic review and meta- analysis. Front. Immunol. 2022, 13, 954396. [Google Scholar] [CrossRef]
- Hemati, Z.; Neamati, F.; Khaledi, M.; Gheibihayat, S.M.; Jafarzadeh, L.; Momen-Heravi, M.; Haddadi, M.H.; Sameni, F.; Fathizadeh, H. Circular RNAs and tuberculosis infection. Int. J. Biol. Macromol. 2022, 226, 1218–1225. [Google Scholar] [CrossRef]
- Arroyo, E.; Oliveira-Alves, M.G.; Chamorro-Petronacci, C.M.; Marichalar-Mendia, X.; Bravo-López, S.B.; Blanco-Carrión, J.; Pérez-Sayáns, M. Protein-based salivary biomarkers for the diagnosis of periodontal diseases: Systematic review and meta-analysis. J. Taibah Univ. Med. Sci. 2023, 18, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Cereda, E.; Pisati, R.; Rondanelli, M.; Caccialanza, R. Whey Protein, Leucine- and Vitamin-D-Enriched Oral Nutritional Supplementation for the Treatment of Sarcopenia. Nutrients 2022, 14, 1524. [Google Scholar] [CrossRef]
- Xu, F.; Ni, M.; Qu, S.; Duan, Y.; Zhang, H.; Qin, Z. Molecular markers of tuberculosis and their clinical relevance: A systematic review and meta-analysis. Ann. Palliat. Med. 2022, 11, 532–543. [Google Scholar] [CrossRef]
- Schiff, H.F.; Walker, N.F.; Ugarte-Gil, C.; Tebruegge, M.; Manousopoulou, A.; Garbis, S.D.; Mansour, S.; Wong, P.H.; Rockett, G.; Piazza, P.; et al. Integrated plasma proteomics identifies tuberculosis-specific diagnostic biomarkers. JCI Insight 2024, 9, e173273. [Google Scholar] [CrossRef] [PubMed]
- Mutavhatsindi, H.; Calder, B.; McAnda, S.; Malherbe, S.T.; Stanley, K.; Kidd, M.; Walzl, G.; Chegou, N.N. Identification of novel salivary candidate protein biomarkers for tuberculosis diagnosis: A preliminary biomarker discovery study. Tuberculosis 2021, 130, 102118. [Google Scholar] [CrossRef] [PubMed]
- HaileMariam, M.; Eguez, R.V.; Singh, H.; Bekele, S.; Ameni, G.; Pieper, R.; Yu, Y. S-Trap, an Ultrafast Sample-Preparation Approach for Shotgun Proteomics. J. Proteome Res. 2018, 17, 2917–2924. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wei, L.L.; Shi, L.Y.; Pan, Z.F.; Yu, X.M.; Li, T.Y.; Liu, C.M.; Ping, Z.P.; Jiang, T.T.; Chen, Z.L.; et al. Screening and identification of five serum proteins as novel potential biomarkers for cured pulmonary tuberculosis. Sci. Rep. 2015, 5, 15615. [Google Scholar] [CrossRef]
- Yu, X.W.; Zhang, J.A.; Xie, J.P. Progress in PD-1/PD-L1, PD-L2 signaling pathway and its role in host anti-tuberculosis immunity. Chin. J. Tuberc. Respir. Dis. 2024, 47, 485–489. [Google Scholar]
- Wong, E.A.; Joslyn, L.; Grant, N.L.; Klein, E.; Lin, P.L.; Kirschner, D.E.; Flynn, J.L.; Bäumler, A.J. Low Levels of T Cell Exhaustion in Tuberculous Lung Granulomas. Infect. Immun. 2018, 86, 9. [Google Scholar] [CrossRef]
- Koeppel, L.; Denkinger, C.M.; Wyss, R.; Broger, T.; Chegou, N.N.; Dunty, J.M.; Scott, K.; Cáceres, T.; Dutoit, E.; Ugarte-Gil, C.; et al. Diagnostic performance of host protein signatures as a triage test for active pulmonary TB. J. Clin. Microbiol. 2023, 61, e0026423. [Google Scholar] [CrossRef]
- Singh, H.; Gonzalez-Juarbe, N.; Pieper, R.; Yu, Y.; Vashee, S. Predictive biomarkers for latent Mycobacterium tuberculosis infection. Tuberculosis 2024, 147, 102399. [Google Scholar] [CrossRef]
- Núñez-Jurado, D.; Rodríguez-Martín, I.; Guerrero, J.M.; Santotoribio, J.D. LDH/ADA ratio in pleural fluid for the diagnosis of infectious pleurisy. Clin. Exp. Med. 2023, 23, 5201–5213. [Google Scholar] [CrossRef]
- Sun, H.; Pan, L.; Jia, H.; Zhang, Z.; Gao, M.; Huang, M.; Wang, J.; Sun, Q.; Wei, R.; Du, B.; et al. Label-Free Quantitative Proteomics Identifies Novel Plasma Biomarkers for Distinguishing Pulmonary Tuberculosis and Latent Infection. Front. Microbiol. 2018, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Pan, L.; Han, F.; Luo, B.; Jia, H.; Xing, A.; Li, Q.; Zhang, Z. Proteomic profiling for plasma biomarkers of tuberculosis progression. Mol. Med. Rep. 2018, 18, 1551–1559. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.; Ghosh, D. Proteomics-based host-specific biomarkers for tuberculosis: The future of TB diagnosis. J. Proteom. 2024, 305, 105245. [Google Scholar] [CrossRef] [PubMed]
- Franco Fontes, C.; Silva Bidu, N.; Rodrigues Freitas, F.; Maranhão, R.C.; Santos Monteiro, A.D.S.; Couto, R.D.; Netto, E.M. Changes in serum amyloid A, plasma high-density lipoprotein cholesterol and apolipoprotein A-I as useful biomarkers for Mycobacterium tuberculosis infection. J. Med. Microbiol. 2023, 72, 6. [Google Scholar] [CrossRef]
- Liu, Y.; Ndumnego, O.C.; Chen, T.; Kim, R.S.; Jenny-Avital, E.R.; Ndung’u, T.; Wilson, D.; Achkar, J.M. Soluble CD14 as a Diagnostic Biomarker for Smear-Negative HIV-Associated Tuberculosis. Pathogens 2018, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Vito, O.; Psarras, S.; Syggelou, A.; Wright, V.J.; Amanatidou, V.; Newton, S.M.; Shailes, H.; Trochoutsou, K.; Tsagaraki, M.; Levin, M.; et al. Novel RNA biomarkers improve discrimination of children with tuberculosis disease from those with non-TB pneumonia after in vitro stimulation. Front. Immunol. 2024, 15, 1401647. [Google Scholar] [CrossRef] [PubMed]
- Fajgenbaum, D.C.; June, C.H. Cytokine Storm. N. Engl. J. Med. 2020, 383, 2255–2273. [Google Scholar] [CrossRef] [PubMed]
- Selimov, P.; Karalilova, R.; Damjanovska, L.; Delcheva, G.; Stankova, T.; Stefanova, K.; Maneva, A.; Selimov, T.; Batalov, A. Rheumatoid arthritis and the proinflammatory cytokine IL-17. Folia Medica 2023, 65, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Harsanyi, S.; Kupcova, I.; Danisovic, L.; Klein, M. Selected Biomarkers of Depression: What Are the Effects of Cytokines and Inflammation? Int. J. Mol. Sci. 2022, 24, 578. [Google Scholar] [CrossRef]
- Sampath, P.; Rajamanickam, A.; Thiruvengadam, K.; Natarajan, A.P.; Hissar, S.; Dhanapal, M.; Thangavelu, B.; Jayabal, L.; Ramesh, P.M.; Ranganathan, U.D.; et al. Plasma chemokines CXCL10 and CXCL9 as potential diagnostic markers of drug-sensitive and drug-resistant tuberculosis. Sci. Rep. 2023, 13, 7404. [Google Scholar] [CrossRef]
- Shiratori, B.; Leano, S.; Nakajima, C.; Chagan-Yasutan, H.; Niki, T.; Ashino, Y.; Suzuki, Y.; Telan, E.; Hattori, T. Elevated OPN, IP-10, and Neutrophilia in Loop-Mediated Isothermal Amplification Confirmed Tuberculosis Patients. Mediat. Inflamm. 2014, 2014, 513263. [Google Scholar] [CrossRef]
- Yoon, C.; Semitala, F.C.; Atuhumuza, E.; Katende, J.; Mwebe, S.; Asege, L.; Armstrong, D.T.; Andama, A.O.; Dowdy, D.W.; Davis, J.L.; et al. Point-of-care C-reactive protein-based tuberculosis screening for people living with HIV: A diagnostic accuracy study. Lancet Infect. Dis. 2017, 17, 1285–1292. [Google Scholar] [CrossRef]
- Dreesman, A.; Corbière, V.; Libin, M.; Racapé, J.; Collart, P.; Singh, M.; Locht, C.; Mascart, F.; Dirix, V. Specific Host Signatures for the Detection of Tuberculosis Infection in Children in a Low TB Incidence Country. Front. Immunol. 2021, 12, 575519. [Google Scholar] [CrossRef]
- Chetty, S.; Govender, P.; Zupkosky, J.; Pillay, M.; Ghebremichael, M.; Moosa, M.-Y.S.; Ndung’u, T.; Porichis, F.; Kasprowicz, V.O.; Paxton, W.A. Co-Infection with Mycobacterium tuberculosis Impairs HIV-Specific CD8+ and CD4+ T Cell Functionality. PLoS ONE 2015, 10, e0118654. [Google Scholar] [CrossRef]
- Koyuncu, D.; Niazi, M.K.K.; Tavolara, T.; Abeijon, C.; Ginese, M.L.; Liao, Y.; Mark, C.; Specht, A.; Gower, A.C.; Restrepo, B.I.; et al. CXCL1: A new diagnostic biomarker for human tuberculosis discovered using Diversity Outbred mice. PLoS Pathog. 2021, 17, e1009773. [Google Scholar] [CrossRef]
- Li, H.; Ren, W.; Liang, Q.; Zhang, X.; Li, Q.; Shang, Y.; Ma, L.; Li, S.; Pang, Y. A novel chemokine biomarker to distinguish active tuberculosis from latent tuberculosis: A cohort study. Qjm Int. J. Med. 2023, 116, 1002–1009. [Google Scholar] [CrossRef]
- Maulina, N.; Hayati, Z.; Hasballah, K.; Zulkarnain, Z. Tryptophan and Its Derived Metabolites as Biomarkers for Tuberculosis Disease: A Systematic Review. Iran. Biomed. J. 2024, 28, 140–147. [Google Scholar] [CrossRef]
- Druszczynska, M.; Seweryn, M.; Wawrocki, S.; Kowalewska-Pietrzak, M.; Pankowska, A.; Rudnicka, W. Cytokine Biosignature of Active and Latent Mycobacterium Tuberculosis Infection in Children. Pathogens 2021, 10, 517. [Google Scholar] [CrossRef]
- Wang, L.; Yang, G.; Guo, L.; Yao, L.; Liu, Y.; Sha, W. Olink proteomics and lipidomics analysis of serum from patients infected with non-tuberculous mycobacteria and Mycobacterium tuberculosis. Inflamm. Res. 2024, 73, 1945–1960. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kang, Y.A.; Park, J.H.; Cha, H.H.; Jeon, N.Y.; Lee, S.W.; Lee, S.O.; Choi, S.H.; Kim, Y.S.; Woo, J.H.; et al. An IFN-γ and TNF-α dual release fluorospot assay for diagnosing active tuberculosis. Clin. Microbiol. Infect. 2020, 26, 928–934. [Google Scholar] [CrossRef]
- Shaik, J.; Pillay, M.; Jeena, P. A Review of Host-Specific Diagnostic and Surrogate Biomarkers in Children with Pulmonary Tuberculosis. Paediatr. Respir. Rev. 2024, 52, 44–50. [Google Scholar] [CrossRef]
- Qu, R.; Zhang, Y.; Ma, Y.; Zhou, X.; Sun, L.; Jiang, C.; Zhang, Z.; Fu, W. Role of the Gut Microbiota and Its Metabolites in Tumorigenesis or Development of Colorectal Cancer. Adv. Sci. 2023, 10, e2205563. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, K.; Wang, X.; Pang, Y.; Jiang, C. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 2021, 12, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Magdalena, D.; Michal, S.; Marta, S.; Magdalena, K.-P.; Anna, P.; Magdalena, G.; Rafał, S. Targeted metabolomics analysis of serum and Mycobacterium tuberculosis antigen-stimulated blood cultures of pediatric patients with active and latent tuberculosis. Sci. Rep. 2022, 12, 4131. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, J.; Ju, Y.; Zhang, X.; Qimanguli, W.; Li, C.; Yue, L.; Tuohetaerbaike, B.; Li, Y.; Wen, H.; et al. Combining metabolome and clinical indicators with machine learning provides some promising diagnostic markers to precisely detect smear-positive/negative pulmonary tuberculosis. BMC Infect. Dis. 2022, 22, 707. [Google Scholar] [CrossRef]
- Duffy, F.J.; Weiner, J., 3rd; Hansen, S.; Tabb, D.L.; Suliman, S.; Thompson, E.; Maertzdorf, J.; Shankar, S.; Tromp, G.; Parida, S.; et al. Immunometabolic Signatures Predict Risk of Progression to Active Tuberculosis and Disease Outcome. Front. Immunol. 2019, 10, 527. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-De La Cruz, M.L.; Salinas-Carmona, M.C. The immune exhaustion paradox: Activated functionality during chronic bacterial infections. J. Infect. Dev. Ctries 2024, 18, 1824–1836. [Google Scholar] [CrossRef]
- Che, N.; Cheng, J.; Li, H.; Zhang, Z.; Zhang, X.; Ding, Z.; Dong, F.; Li, C. Decreased serum 5-oxoproline in TB patients is associated with pathological damage of the lung. Clin. Chim. Acta 2013, 423, 5–9. [Google Scholar] [CrossRef]
- Liu, Y.; Mei, B.; Chen, D.; Cai, L. GC-MS metabolomics identifies novel biomarkers to distinguish tuberculosis pleural effusion from malignant pleural effusion. J. Clin. Lab. Anal. 2021, 35, e23706. [Google Scholar] [CrossRef] [PubMed]
- Isa, F.; Collins, S.; Lee, M.H.; Decome, D.; Dorvil, N.; Joseph, P.; Smith, L.; Salerno, S.; Wells, M.T.; Fischer, S.; et al. Mass Spectrometric Identification of Urinary Biomarkers of Pulmonary Tuberculosis. EBioMedicine 2018, 31, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Jiang, J.; Qian, H.; Yan, Y.; Xu, W. Exosomal circRNA: Emerging insights into cancer progression and clinical application potential. J. Hematol. Oncol. 2023, 16, 67. [Google Scholar] [CrossRef]
- Mukhtar, F.; Guarnieri, A.; Brancazio, N.; Falcone, M.; Di Naro, M.; Azeem, M.; Zubair, M.; Nicolosi, D.; Di Marco, R.; Petronio, G.P.; et al. The role of Mycobacterium tuberculosis exosomal miRNAs in host pathogen cross-talk as diagnostic and therapeutic biomarkers. Front. Microbiol. 2024, 15, 1441781. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, Z.; Fu, Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. J. Cell Biochem. 2019, 120, 5889–5896. [Google Scholar] [CrossRef]
- Zhan, X.; Yuan, W.; Zhou, Y.; Ma, R.; Ge, Z. Small RNA sequencing and bioinformatics analysis of RAW264.7-derived exosomes after Mycobacterium Bovis Bacillus Calmette-Guérin infection. BMC Genom. 2022, 23, 355. [Google Scholar] [CrossRef]
- Kaushik, A.C.; Wu, Q.; Lin, L.; Li, H.; Zhao, L.; Wen, Z.; Song, Y.; Wu, Q.; Wang, J.; Guo, X.; et al. Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis. Brief Bioinform. 2021, 22, 6. [Google Scholar] [CrossRef]
- Lyu, L.; Zhang, X.; Li, C.; Yang, T.; Wang, J.; Pan, L.; Jia, H.; Li, Z.; Sun, Q.; Yue, L.; et al. Small RNA Profiles of Serum Exosomes Derived from Individuals with Latent and Active Tuberculosis. Front. Microbiol. 2019, 10, 1174. [Google Scholar] [CrossRef]
- Liu, H.; Lu, G.; Wang, W.; Jiang, X.; Gu, S.; Wang, J.; Yan, X.; He, F.; Wang, J. A Panel of CircRNAs in the Serum Serves as Biomarkers for Mycobacterium tuberculosis Infection. Front. Microbiol. 2020, 11, 1215. [Google Scholar] [CrossRef]
- Yuan, Q.; Wen, Z.; Yang, K.; Zhang, S.; Zhang, N.; Song, Y.; Chen, F.; Zeng, Z.-Y. Identification of Key CircRNAs Related to Pulmonary Tuberculosis Based on Bioinformatics Analysis. BioMed Res. Int. 2022, 2022, 1717784. [Google Scholar] [CrossRef]
- Luo, H.L.; Peng, Y.; Luo, H.; Zhang, J.; Liu, G.B.; Xu, H.; Huang, G.X.; Sun, Y.F.; Huang, J.; Zheng, B.Y.; et al. Circular RNA hsa_circ_0001380 in peripheral blood as a potential diagnostic biomarker for active pulmonary tuberculosis. Mol. Med. Rep. 2020, 21, 1890–1896. [Google Scholar] [CrossRef]
- Zhang, M.; Xie, Y.; Li, S.; Ye, X.; Jiang, Y.; Tang, L.; Wang, J. Proteomics Analysis of Exosomes From Patients With Active Tuberculosis Reveals Infection Profiles and Potential Biomarkers. Front. Microbiol. 2021, 12, 800807. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Xin, H.; Cao, X.; Liu, Z.; He, Y.; Zhang, B.; Yan, J.; Wang, D.; Guan, L.; Shen, F.; et al. Association Between Plasma Exosomes S100A9/C4BPA and Latent Tuberculosis Infection Treatment: Proteomic Analysis Based on a Randomized Controlled Study. Front. Microbiol. 2022, 13, 934716. [Google Scholar] [CrossRef] [PubMed]
- Dahiya, B.; Khan, A.; Mor, P.; Kamra, E.; Singh, N.; Gupta, K.B.; Sheoran, A.; Sreenivas, V.; Mehta, P.K. Detection of Mycobacterium tuberculosis lipoarabinomannan and CFP-10 (Rv3874) from urinary extracellular vesicles of tuberculosis patients by immuno-PCR. Pathog. Dis. 2019, 77, ftz049. [Google Scholar] [CrossRef] [PubMed]
| Biomarker | AUC | Sensitivity (%) | Specificity (%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| slamf8 (↑), gbp2 (↑), wars (↑), and fcgr1c (↑) Panel | 0.86 | 85 | 73 | Children with active TB (Male): Multicenter (Kenya, South Africa, Malawi), total n = 85 (TB:39; Other diseases:46) Includes HIV+/− subgroups. | Whole-blood transcriptome (PAX gene RNA tube) | [20] |
| gbp6 (↑), celsr3 (↓), aldh1a1 (↑), and gbp4 (↑) Panel | 0.83 | 85 | 69 | Children with active TB (Female): Multicenter (as above) Total n = 61 (TB:27; Other diseases:34) Includes HIV+/− subgroups. | ||
| ADM (↑) | 0.786–0.899 | 74–95 | 52–78 | TB: 46 (Adult), 9 (Child) LTBI: 25, 9 HC: 37, 9 (Multicenter, China) | PBMC sc RNA-seq & Whole Blood | [22] |
| gbp5 (↑) (single) | 0.88 | 83 | 83 | Multicenter (Uganda, Vietnam, Philippines); Adult PTB (n = 251, TB+ = 142) | Plasma cfRNA | [23] |
| batf2(↑) | - | - | - | ATB: 61; ORD/LTBI/HC: 143 (Single-center) | Whole blood | [24] |
| CD64 (↑) | - | - | - | ATB: 61; ORD/LTBI/HC: 143 (Single-center) | Whole blood | [24] |
| gbp5/dusp3/klf2 Panel (↓) | 0.9 | 83.3–90.7 | 59.8–75.6 | LTBI: 24; HC: 37 (Single-center) | Whole blood | [25] |
| Biomarker | AUC | Sensitivity (%) | Specificity (%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| LINC00152 (↑) | 0.919 | 76.36 | 72.73 | LTB vs. ATB (55 LTB, 55 ATB); Single-center | Plasma | [40] |
| LARS2-AS1 (↑) | 0.782 | 63.64 | 94.55 | LTB vs. HC (55 LTB, 55 HC); Single-center | Plasma | [40] |
| LINC00152 (↑) + LARS2-AS1 | 0.829 | 68.18 | 83.64 | LTB vs. HC (Combined model); Single-center | Plasma | [40] |
| m6A-modification-related lncRNAs (e.g., LINC00460, LINC01116) | 0.935 | 92.9 | 90.9 | HIV/TB vs. HIV (Training set: 14 HIV/TB, 11 HIV); Single-center | Whole blood | [43] |
| 0.904 | 93.8 | 80 | HIV/TB vs. HIV (Validation set: 15 HIV/TB, 16 HIV) | |||
| NR_038221 and NR_003142 Panel (↑) | 0.845 | 79.2 | 75 | Active TB (TB): 52; Healthy Controls (HC): 52; Cohort: TB vs. HC; Single-center (Shaoxing Sixth Hospital) | Plasma | [44] |
| NR_038221 (↑) | 0.677 | - | - | Active TB (TB): 52; Healthy Controls (HC): 52; Cohort: TB vs. HC; Single-center (Shaoxing Sixth Hospital) | Plasma | [44] |
| Lung tissue-specific lncRNAs (e.g., ENST00000497872, n333737) (↓) | 0.89 | 86 | 82 | Clinically diagnosed PTB (no micro evidence), microbiologically confirmed PTB, non-TB disease controls, healthy controls; n = 1764 total (Validation cohort: 97 Clin Dx PTB + 140 Non-TB) | PBMC | [45] |
| miR-29a (↓) | - | - | - | HIV/HCV co-infected patients (n = 121) | Serum | [46] |
| miR-29a (↑) | - | - | - | COVID-19 patients (n = 20) | PBMC | [46] |
| hsa-let-7d-5p and hsa-miR-140-5p Panel (↓) | 0.930 (Train) | 100 | 88.5 (Train) | Train/Val Set: ATB:29, LTBI:25, HC:30; Cohort: ATB vs. LTBI vs. HC | Serum | [47] |
| 0.923 (Val) | - | 92.3(Val) | [47] | |||
| miR-223-5p and miR-10b-5p Panel | 0.79 | - | - | ATB (drug-sensitive) vs. HC (55 ATB, 24 HC); Single-center | Serum | [48] |
| hsa_circ_001937(↑) | 0.873 | 85 | 77.5 | 40 TB vs. 40 HC (adult active TB); Single-center | PBMC | [49] |
| 0.85 | 72.2 | 90 | 115 TB vs. 90 HC (independent validation cohort) | PBMC | ||
| hsa_circRNA_103571 (↓) | 0.838 | ATB vs. HC (32 ATB, 29 HC); Single-center | Plasma | [50] | ||
| circRNA_051239 (↑) | 0.85 | 71.43 | 66.67 | DR-TB vs. DS-TB; Single-center | Serum | [51] |
| hsa_circ_0001204 and hsa_circ_0001747 Panel | 0.92 | NP | NP | APTB patients vs. HC; Single-center | Plasma | [52] |
| Biomarker | AUC | Sensitivity (%) | Specificity(%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| Serum ADA2 + CD14 Panel | - | - | - | HIV+/HIV− TB patients (n = 209) | Serum | [59] |
| I-309/SYWC (↑)/Kallistatin Triplex (↓) | 0.9 | 90 | 70 | 479 Adults (177 TB, 302 Non-TB); Multicenter (South Africa, Peru, Vietnam; Performance ↓ in Vietnam) | Serum | [65] |
| I-309/SYWC Panel (↑) | 0.88 | 89 | 74 | 479 Adults (177 TB, 302 Non-TB); Multicenter (South Africa, Peru, Vietnam; Performance ↓ in Vietnam) | Serum | [65] |
| 5-Protein Panel (ANXA5, KRT6B, LCN2, ORM1, MMP8) | MCC = 0.767 | 84 | 84 | Mixed cohort: PTB (n = 31), LTBI (n = 25), Healthy (n = 19) | Sputum | [66] |
| 6-Protein Signature (MCEMP1, HPX, SPRR2F, IGKV4-1, VDAC2, LMNA) | MCC = 0.954 | 97.7 | 97.7 | LTBI (n = 25) vs. Healthy (n = 19) | Sputum | [66] |
| 6-Marker Panel (FETUB, FCGR3B, LRG1, SELL, CD14, ADA2) | 0.972 | 90.6 | 90 | PTB vs. HC (UK MIMIC cohort, n = 62) | Serum | [59] |
| Adenosine Deaminase (ADA) (↑) | - | 40–100 | 68–100 | 259 Adults with pleural effusion (incl. 41 TPE) | Pleural Fluid | [67] |
| AGP1 (↑) | 0.816 | 63.5 | 91.8 | PTB vs. LTBI (Training set, n = 169) | Plasma | [68] |
| ACT (↑) | 0.835 | 68.2 | 92.9 | PTB vs. LTBI (Training set, n = 169) | Plasma | [68] |
| ACT, AGP1 and CDH1 Panel | 0.946 | 82.3 | 92.8 | PTB vs. LTBI (Training set PTB =85, LTBI = 84) | Plasma | [68] |
| 0.989 | 96.5 | 95.8 | PTB vs. HC (Training set PTB = 85, HC = 71) | |||
| S100A9 (↑) | 0.891 | 86 | 90 | STB group vs. MTB/HC (81 STB, 80 MTB, 50 HC) | Plasma | [69] |
| SOD1 (↓) | 0.525 | 79 | 32 | STB group vs. MTB/HC (81 STB, 80 MTB, 50 HC) | Plasma | [69] |
| TIMP-2 + TSP4 | 0.878 | 75 | 87.5 | TB treatment 8 weeks vs. Baseline (n = 39) | Serum | [70] |
| SAA (↑) | 0.98 | 96.88 | 78.43 | 129 Symptomatic Adults (97 TB, 32 Non-TB); Brazil, Single-center | Plasma | [71] |
| HDL-C (↓) | 0.84 | 75 | 72.16 | 129 Symptomatic Adults (97 TB, 32 Non-TB); Brazil, Single-center | Plasma | [71] |
| Biomarker | AUC | Sensitivity (%) | Specificity (%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| CXCL10 (IP-10) (↑) | 0.84 (DR-TB vs. DS-TB) | - | - | Indian Adults: DR-TB (n = 40), DS-TB (n = 40), LTB (n = 40), HC (n = 40); Single-center | Plasma | [77] |
| - | 77 | 94 | Belgian Children: Active TB (n = 12), LTBI (n = 18), Uninfected (n = 17); Single-center (Discovery cohort) | PBMC supernatant | ||
| CXCL9 (MIG) (↑) | 0.82 (DR-TB vs. DS-TB) | - | - | Indian Adults: DR-TB (n = 40), DS-TB (n = 40), LTB (n = 40), HC (n = 40); LTBI vs. ATB (Beijing Chest Hospital cohort, n = 208) | Plasma | [77] |
| > 0.9 | 97 | 100 | Belgian children: Active TB (n = 12), LTBI (n = 18), Uninfected (n = 17); Single-center (Discovery cohort) | PBMC supernatant | ||
| CCL8 | 0.89 | 90.79 | 100 | ATB vs. LTBI (IGRA-positive) (Beijing Chest Hospital cohort, n = 208) Single-center | Plasma | [83] |
| CCL8 + CXCL9 | 0.958 | 96 | 84.37 | ATB vs. LTBI (IGRA-positive) (Beijing Chest Hospital cohort, n = 208) Single-center | Plasma | [83] |
| CXCL1 (↑) | 0.80 (DR-TB vs. LTB) | - | - | Indian Adults: DR-TB (n = 40), DS-TB (n = 40), LTBI(n = 40), HC (n = 40); Single-center | Plasma | [82] |
| IFN-γ and TNF-α (↑) | - | 100 | 100 | Belgian Children Discovery cohort (n = 47) | PBMC supernatant | [84] |
| 0.918 | 84 | 94 | 153 Adults (45 Active TB, 108 Non-active TB incl. 38 LTBI); Single-center prospective cohort | PBMC (stimulated) | ||
| BAFF/TNFSF13B (↑) | 0.809 (HC vs. TB) | - | - | 216 Polish children (aged 1–17 years) who received BCG vaccination (TB, n = 15; LTBI, n = 50; HC, n = 151) | Serum | [85] |
| MMP-2 (↓) | 0.848 (HC vs. TB) | - | - | 216 Polish children (aged 1–17 years) who received BCG vaccination (TB, n = 15; LTBI, n = 50; HC, n = 151) | Serum | [85] |
| FAHFAs (e.g., FAHFA 18:2) (↓) + IL-8(↑) Model | 0.9754 | 92.3 | 96 | Adult PTB (MTB, n = 26) vs. HC (n = 26); Single-center (Shanghai Pulmonary Hospital) | Serum | [86] |
| CXCL9, CXCL10, CXCL1 Triplex (↑) | Overall AUC 0.80 | - | - | Indian Adults: DR-TB (n = 40), DS-TB (n = 40), LTB (n = 40), HC (n = 40); Single-center | Plasma | [77] |
| CXCL9 (↑) | 0.8876 (RGM) | - | - | Indian Adults: DR-TB (n = 40), DS-TB (n = 40), LTB (n = 40), HC (n = 40); Single-center | Serum | [77] |
| 0.9042 (SGM) | ||||||
| CXCL10 (↑) | 0.8649 (MTB) | - | - | Adult PTB (MTB, n = 26) vs. HC (n = 26); Single-center (Shanghai Pulmonary Hospital) | Serum | [77] |
| IFN-γ (↑) | 0.8387 (MTB) | - | - | Adult PTB (MTB, n = 26) vs. HC (n = 26); Single-center (Shanghai Pulmonary Hospital) | Serum | [86] |
| IL-8 (↑) | 0.9186 (MTB) | - | - | Adult PTB (MTB, n = 26) vs. HC (n = 26); Single-center (Shanghai Pulmonary Hospital) | Serum | [86] |
| FAHFA 18:2 (↓) | 0.8708 (MTB) | - | - | Adult PTB (MTB, n = 26) vs. HC (n = 26); Single-center (Shanghai Pulmonary Hospital) | Serum | [86] |
| 0.9440 (RGM) | - | - |
| Biomarker | AUC | Sensitivity (%) | Specificity (%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| Glutamine (Gln) (↓) | 0.581 | - | - | Polish children (TB:15, LTBI:52, NMP:20, HC:149) | QFT TB1 Supernatant | [91] |
| Citrulline (Cit) (↓) | 0.848 | 82 | 88 | 17 TBPE vs. 17 MPE (Adults); Single-center | Pleural Fluid | [91] |
| Lysophosphatidylinositol (Lyso-PI) (18:0) (↑) | 0.94 | - | - | 17 Active TB vs. 16 household contacts (Adults); Single-center | Plasma | [11] |
| Albumin + 9-OxoODE | 0.83 | 80 | 86 | 27 SPPT vs. 36 Controls (Adults); Single-center | Plasma | [92] |
| l-Pyroglutamic acid (PGA) + Secretin | 0.93 | 86 | 100 | 37 SNPT vs. 36 Controls (Adults); Single-center | Plasma | [92] |
| MLP Model (20 Metabolites) | 0.95 | 100 | 100 | 27 SPPT, 37 SNPT, 36 Controls (3-class); Single-center | Plasma | [93] |
| PD-L1 + IDO-1(↑) | - | - | - | TB patient granuloma tissue | Granuloma Tissue | [94] |
| 5-Oxoproline (↑) | 0.7 | - | - | Discovery cohort: Haitian Active TB (n = 102) vs. HC (n = 102) | Serum | [95] |
| 5-Oxoproline (l-5-Oxoproline) (↓) | 0.709 | 47 | 94 | 17 TBPE vs. 17 MPE (Adults); Single-center | Pleural Fluid | [96] |
| Biomarker | AUC | Sensitivity (%) | Specificity (%) | Cohort Characteristics | Sample | Reference |
|---|---|---|---|---|---|---|
| miR-20b-5p (↓) | - | - | - | RAW 264.7 macrophages (in vitro infection model) | Macrophages | [99] |
| hsa-let-7c-5p (↑) | - | - | - | Adult TB (n = 60), LTBI (n = 60), HC (n = 60) | Serum exosomes | [100] |
| mmu-miR-27-3p (↑) | - | - | - | Murine RAW264.7 macrophages (BCG infection model) | Macrophage exosomes | [100] |
| mmu-miR-25-3p (↑) | - | - | - | Murine RAW264.7 macrophages (BCG infection model) | Macrophage exosomes | [100] |
| miR-185-5p (↑) | 0.75 | 50 | 93.75 | 20 Active TB Adults vs. 17 Healthy; Single-center | Plasma exosomes | [101] |
| hsa-miR-1246 | - | - | - | Adult TB (n = 60), LTBI (n = 60), HC (n = 60) | Serum exosomes | [102] |
| mmu-let-7c-5p (↑) | - | - | - | Murine RAW264.7 macrophages (BCG infection model) | Macrophage exosomes | [100] |
| circRNA_051239 (↑) | 0.9738 | - | - | Active TB (n = 128) vs. CAP (n = 50) vs. HC (n = 50) | Serum | [103] |
| hsa_circ_0007919 (↑) | - | - | - | PTB lung tissue samples (n = 9 patients) | Lung tissue | [104] |
| hsa_circ_0002419 (↓) | - | - | - | PTB lung tissue samples (n = 9 patients) | Lung tissue | [104] |
| hsa_circ_0005521 (↓) | - | - | - | PTB lung tissue samples (n = 9 patients) | Lung tissue | [104] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, Y.; Li, H.; Liu, T.; Zhong, R.; Guo, J.; Du, J.; Pang, Y. The Evolving Landscape of Host Biomarkers for Diagnosis and Monitoring of Tuberculosis. Biomedicines 2025, 13, 2076. https://doi.org/10.3390/biomedicines13092076
Cui Y, Li H, Liu T, Zhong R, Guo J, Du J, Pang Y. The Evolving Landscape of Host Biomarkers for Diagnosis and Monitoring of Tuberculosis. Biomedicines. 2025; 13(9):2076. https://doi.org/10.3390/biomedicines13092076
Chicago/Turabian StyleCui, Yang, Haoran Li, Tianhui Liu, Rujie Zhong, Jiaying Guo, Jian Du, and Yu Pang. 2025. "The Evolving Landscape of Host Biomarkers for Diagnosis and Monitoring of Tuberculosis" Biomedicines 13, no. 9: 2076. https://doi.org/10.3390/biomedicines13092076
APA StyleCui, Y., Li, H., Liu, T., Zhong, R., Guo, J., Du, J., & Pang, Y. (2025). The Evolving Landscape of Host Biomarkers for Diagnosis and Monitoring of Tuberculosis. Biomedicines, 13(9), 2076. https://doi.org/10.3390/biomedicines13092076







