Comparative Evaluation of Computational Methods for Validating Housekeeping Gene RT-qPCR Data in 3T3-L1 Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Propagation and Adipogenic Induction of Preadipocytes
2.2. L. paracasei Cell-Free Supernatants Preparation and Application on Mature Adipocytes
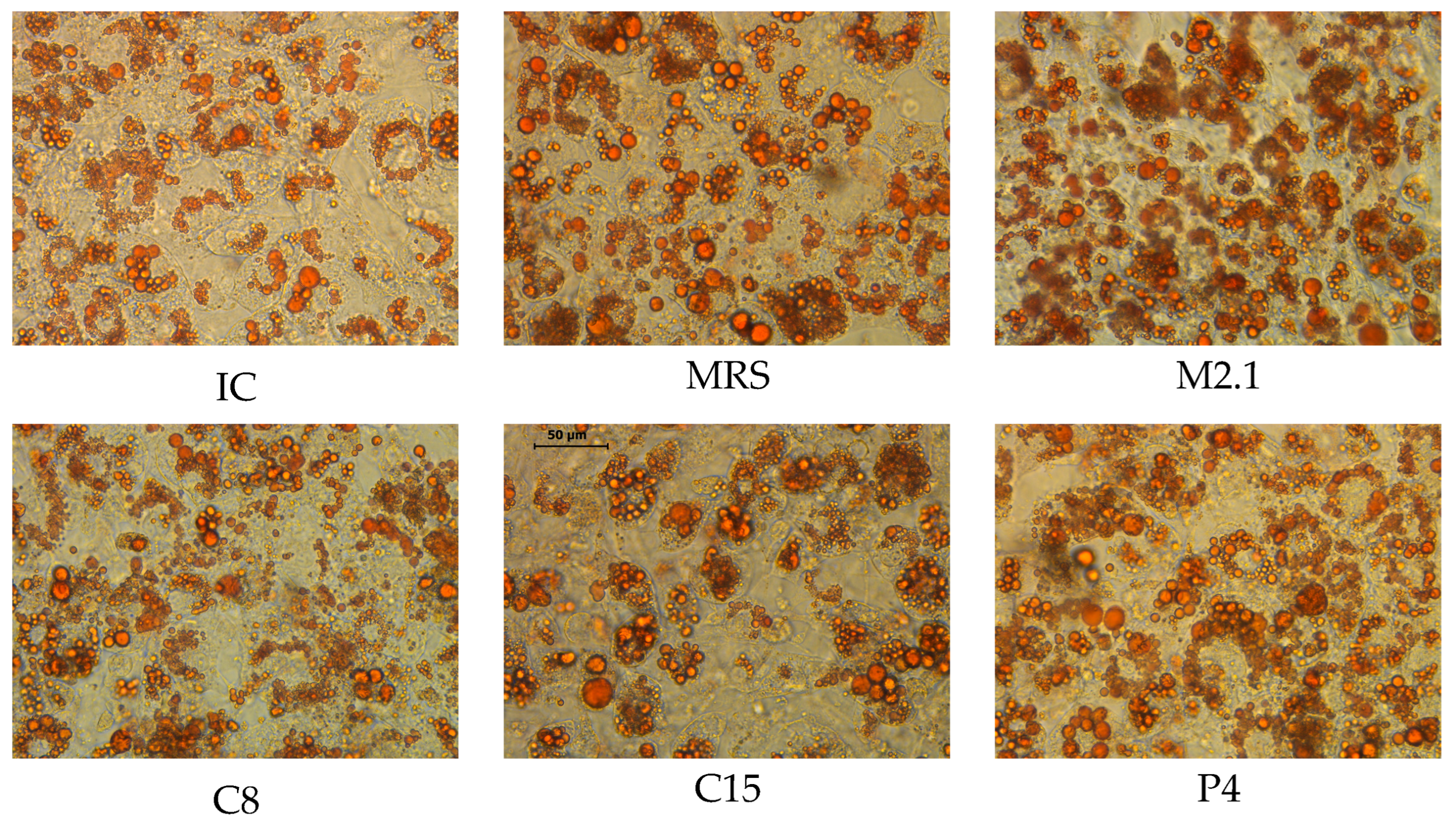
2.3. Intracellular Lipid Deposition Visualization
2.4. Gene Expression Assays
2.5. Primer Design
2.6. Analysis of Expression Stability Among Selected HKGs
2.7. Statistical Analysis
3. Results
3.1. Adipogenic Induction and LB Treatment
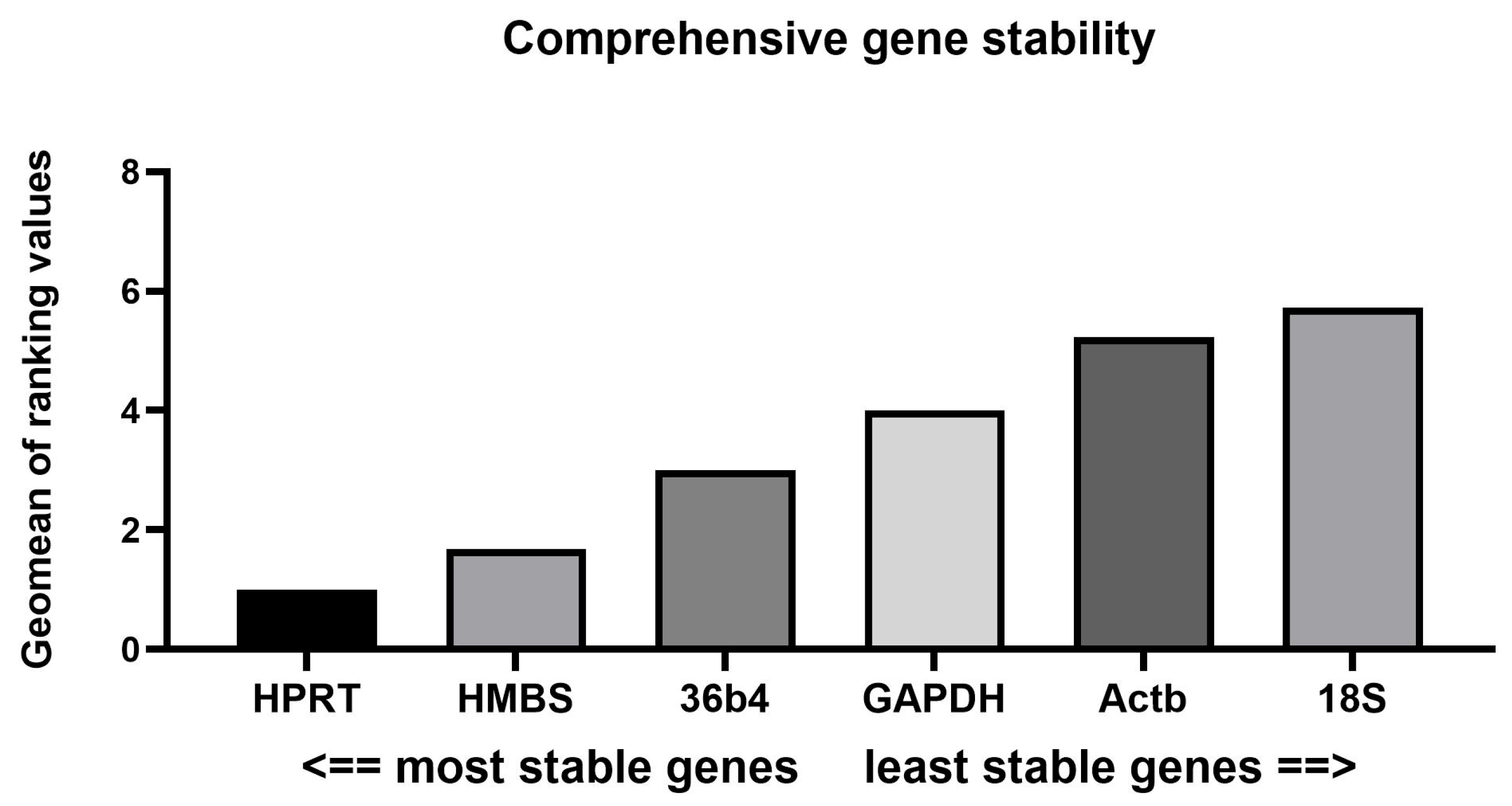
3.2. Expression Stability Assessment via Four Popular Algorithms
3.2.1. NormFinder and geNorm
3.2.2. BestKeeper and RefFinder
3.3. Pairwise ΔCt Analysis
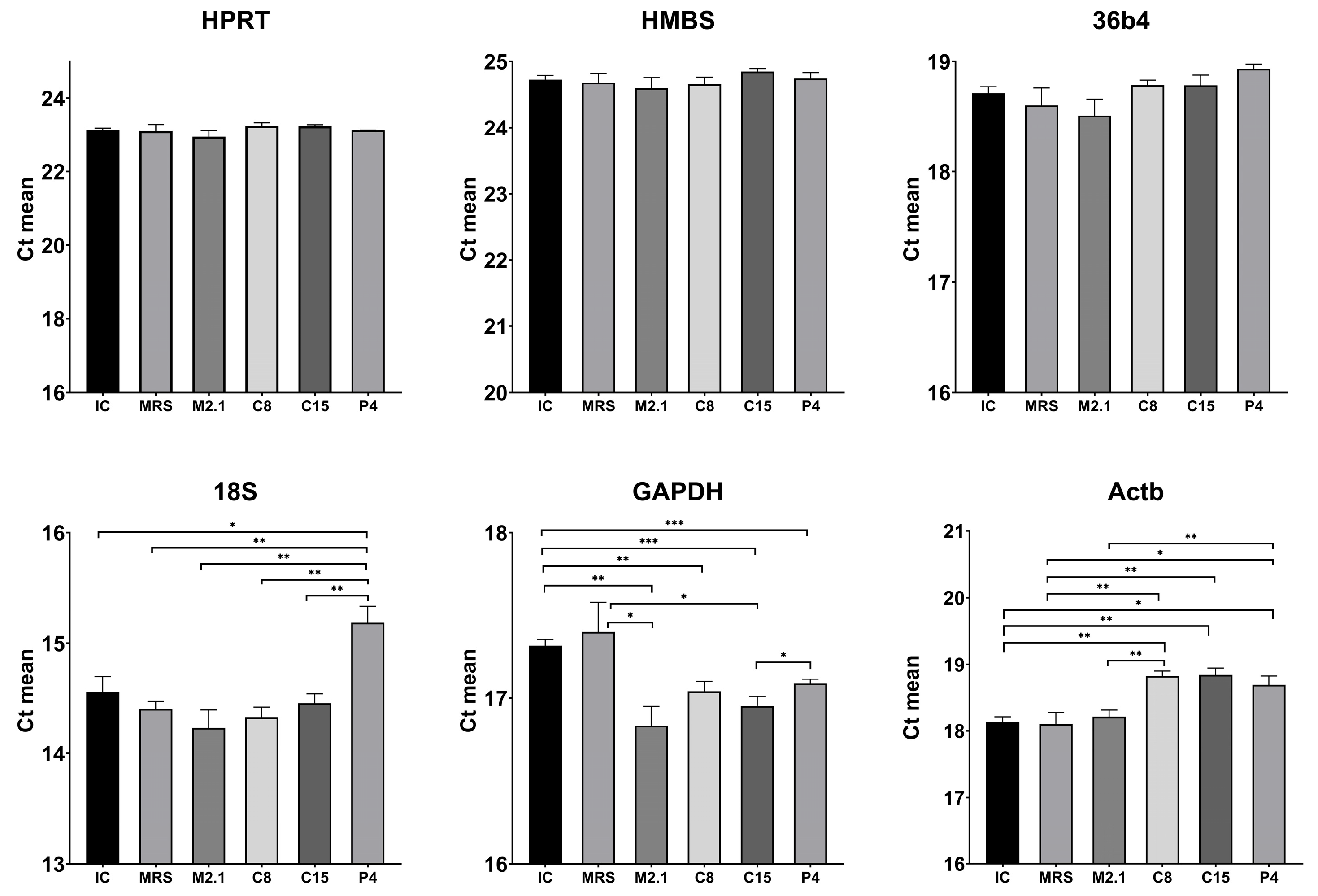
3.4. Inter-Group Statistical Analysis of Raw Ct Values for Reference Gene Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HKG(s) | Housekeeping gene(s) |
| RT-qPCR | Reverse transcription quantitative polymerase chain reaction |
| RNA | Ribonucleic acid |
| cDNA | Complementary deoxyribonucleic acid |
| 36B4 | Ribosomal protein, large, P0 |
| HPRT | Hypoxanthine-guanine phosphoribosyl transferase |
| Actb | Actin, beta |
| HMBS | Hydroxymethylbilane synthase |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| 18S | 18S ribosomal RNA |
| PPARγ | Peroxisome proliferator-activated receptor gamma, transcript variant 2 |
| L. paracasei | Lacticaseibacillus paracasei |
| MIQE | Minimum Information for Publication of Quantitative Real-Time PCR Experiments |
| DMEM | Dulbecco’s modified Eagle’s medium |
| FBS | Fetal bovine serum |
| PBS | Phosphate-buffered saline |
| BM | Basal medium |
| AIM | Adipogenic induction media |
| AMM | Maintenance media |
| MRS | de Man, Rogosa, and Sharpe broth-treated induced control group |
| IC | Untreated, induced control group |
| M2.1, C8, C15, P4 | Experimental group of induced adipocytes treated with 10% v/v supernatants from the respective L. paracasei strain |
| SD | Standard deviation |
| CV | Coefficient of variation |
| r | Pearson correlation coefficient |
| Ct | Cycle threshold |
| ∆Ct | delta Ct |
| ∆∆Ct | delta delta Ct |
References
- Vallianou, N.G.; Kounatidis, D.; Tsilingiris, D.; Panagopoulos, F.; Christodoulatos, G.S.; Evangelopoulos, A.; Karampela, I.; Dalamaga, M. The Role of Next-Generation Probiotics in Obesity and Obesity-Associated Disorders: Current Knowledge and Future Perspectives. Int. J. Mol. Sci. 2023, 24, 6755. [Google Scholar] [CrossRef] [PubMed]
- Sankararaman, S.; Noriega, K.; Velayuthan, S.; Sferra, T.; Martindale, R. Gut Microbiome and Its Impact on Obesity and Obesity-Related Disorders. Curr. Gastroenterol. Rep. 2023, 25, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Oudat, Q.; Okour, A. The Role of Probiotics in Modulating Gut Microbiota and Metabolic Health for Weight Management: A Mini Review. Acta Microbiol. Hell. 2025, 70, 5. [Google Scholar] [CrossRef]
- Kamble, N.S.; Thomas, S.; Madaan, T.; Ehsani, N.; Sange, S.; Tucker, K.; Muhumure, A.; Kunkler, S.; Kotagiri, N. Engineered Bacteria as an Orally Administered Anti-Viral Treatment and Immunization System. Gut Microbes 2025, 17, 2500056. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jeong, Y.; Kim, J.-E.; Kim, Y.; Paek, N.-S.; Kang, C.-H. Anti-Obesity Potential of Lactobacillus Spp. Isolated from Infant Feces. Biotechnol. Bioproc. E 2021, 26, 575–585. [Google Scholar] [CrossRef]
- Li, S.; Liu, Z.; Zhang, Q.; Su, D.; Wang, P.; Li, Y.; Shi, W.; Zhang, Q. The Antidiabetic Potential of Probiotics: A Review. Nutrients 2024, 16, 2494. [Google Scholar] [CrossRef]
- Kadeer, G.; Fu, W.; He, Y.; Feng, Y.; Liu, W.-H.; Hung, W.-L.; Feng, H.; Zhao, W. Effect of Different Doses of Lacticaseibacillus Paracasei K56 on Body Fat and Metabolic Parameters in Adult Individuals with Obesity: A Pilot Study. Nutr. Metab. 2023, 20, 16. [Google Scholar] [CrossRef]
- Sitdhipol, J.; Niwasabutra, K.; Chaiyawan, N.; Nuankham, K.; Thanagornyothin, T.; Tanasupawat, S.; Chanput, W.P.; Phapugrangkul, P.; Chaipanya, C.; Phuengjayaem, S.; et al. Evaluating the Safety and Efficacy of Lacticaseibacillus Paracasei TISTR 2593 as a Therapeutic Probiotic for Obesity Prevention. Front. Microbiol. 2025, 16, 1501395. [Google Scholar] [CrossRef]
- Scott, E.; De Paepe, K.; Van de Wiele, T. Postbiotics and Their Health Modulatory Biomolecules. Biomolecules 2022, 12, 1640. [Google Scholar] [CrossRef]
- Lee, H.B.; Kang, S.-S. Inhibitory Effect of Bacterial Lysates Extracted from Pediococcus Acidilactici on the Differentiation of 3T3-L1 Pre-Adipocytes. Int. J. Mol. Sci. 2022, 23, 11614. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Barros, C.P.; Guimarães, J.T.; Esmerino, E.A.; Duarte, M.C.K.; Silva, M.C.; Silva, R.; Ferreira, B.M.; Sant’Ana, A.S.; Freitas, M.Q.; Cruz, A.G. Paraprobiotics and Postbiotics: Concepts and Potential Applications in Dairy Products. Curr. Opin. Food Sci. 2020, 32, 1–8. [Google Scholar] [CrossRef]
- Tomičić, Z.; Šarić, L.; Tomičić, R. Potential Future Applications of Postbiotics in the Context of Ensuring Food Safety and Human Health Improvement. Antibiotics 2025, 14, 674. [Google Scholar] [CrossRef] [PubMed]
- Thorakkattu, P.; Khanashyam, A.C.; Shah, K.; Babu, K.S.; Mundanat, A.S.; Deliephan, A.; Deokar, G.S.; Santivarangkna, C.; Nirmal, N.P. Postbiotics: Current Trends in Food and Pharmaceutical Industry. Foods 2022, 11, 3094. [Google Scholar] [CrossRef]
- Molina, D.; Marinas, I.C.; Angamarca, E.; Hanganu, A.; Stan, M.; Chifiriuc, M.C.; Tenea, G.N. Postbiotic-Based Extracts from Native Probiotic Strains: A Promising Strategy for Food Preservation and Antimicrobial Defense. Antibiotics 2025, 14, 318. [Google Scholar] [CrossRef]
- Harat, S.G.; Pourjafar, H. Health Benefits and Safety of Postbiotics Derived from Different Probiotic Species. Curr. Pharm. Des. 2025, 31, 116–127. [Google Scholar] [CrossRef]
- Żółkiewicz, J.; Marzec, A.; Ruszczyński, M.; Feleszko, W. Postbiotics—A Step Beyond Pre- and Probiotics. Nutrients 2020, 12, 2189. [Google Scholar] [CrossRef]
- Hamdi, A.; Lloyd, C.; Eri, R.; Van, T.T.H. Postbiotics: A Promising Approach to Combat Age-Related Diseases. Life 2025, 15, 1190. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, S.; Li, S.; Jiang, W.; Wang, J.; Xiao, J.; Chen, T.; Ma, J.; Khan, M.Z.; Wang, W.; et al. Unlocking the Power of Postbiotics: A Revolutionary Approach to Nutrition for Humans and Animals. Cell Metab. 2024, 36, 725–744. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Tu, H.; Chen, T. Postbiotics in Human Health: A Narrative Review. Nutrients 2023, 15, 291. [Google Scholar] [CrossRef]
- Colautti, A.; Ginaldi, F.; Camprini, L.; Comi, G.; Reale, A.; Iacumin, L. Investigating Safety and Technological Traits of a Leading Probiotic Species: Lacticaseibacillus Paracasei. Nutrients 2024, 16, 2212. [Google Scholar] [CrossRef]
- Shah, A.B.; Baiseitova, A.; Zahoor, M.; Ahmad, I.; Ikram, M.; Bakhsh, A.; Shah, M.A.; Ali, I.; Idress, M.; Ullah, R.; et al. Probiotic significance of Lactobacillus strains: A comprehensive review on health impacts, research gaps, and future prospects. Gut Microbes 2024, 16, 2431643. [Google Scholar] [CrossRef]
- Bengoa, A.A.; Dardis, C.; Garrote, G.L.; Abraham, A.G. Health-Promoting Properties of Lacticaseibacillus Paracasei: A Focus on Kefir Isolates and Exopolysaccharide-Producing Strains. Foods 2021, 10, 2239. [Google Scholar] [CrossRef]
- Li, C.-H.; Chen, T.-Y.; Wu, C.-C.; Cheng, S.-H.; Chang, M.-Y.; Cheng, W.-H.; Chiu, S.-H.; Chen, C.-C.; Tsai, Y.-C.; Yang, D.-J.; et al. Safety Evaluation and Anti-Inflammatory Efficacy of Lacticaseibacillus Paracasei PS23. Int. J. Mol. Sci. 2023, 24, 724. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-T.; Chao, W.-Y.; Lin, C.-H.; Shih, T.-W.; Pan, T.-M. Comprehensive Safety Assessment of Lacticaseibacillus Paracasei Subsp. Paracasei NTU 101 Through Integrated Genotypic and Phenotypic Analysis. Curr. Issues Mol. Biol. 2024, 46, 12354–12374. [Google Scholar] [CrossRef]
- Lee, H.B.; Bang, W.Y.; Shin, G.R.; Jeon, H.J.; Jung, Y.H.; Yang, J. Isolation, Characterization, and Safety Evaluation of the Novel Probiotic Strain Lacticaseibacillus Paracasei IDCC 3401 via Genomic and Phenotypic Approaches. Microorganisms 2024, 12, 85. [Google Scholar] [CrossRef]
- Dufau, J.; Shen, J.X.; Couchet, M.; De Castro Barbosa, T.; Mejhert, N.; Massier, L.; Griseti, E.; Mouisel, E.; Amri, E.-Z.; Lauschke, V.M.; et al. In Vitro and Ex Vivo Models of Adipocytes. Am. J. Physiol.-Cell Physiol. 2021, 320, C822–C841. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-J.; Sharma, A.; Lee, H.-J. Postbiotics against Obesity: Perception and Overview Based on Pre-Clinical and Clinical Studies. Int. J. Mol. Sci. 2023, 24, 6414. [Google Scholar] [CrossRef] [PubMed]
- Kober, A.K.M.H.; Saha, S.; Ayyash, M.; Namai, F.; Nishiyama, K.; Yoda, K.; Villena, J.; Kitazawa, H. Insights into the Anti-Adipogenic and Anti-Inflammatory Potentialities of Probiotics against Obesity. Nutrients 2024, 16, 1373. [Google Scholar] [CrossRef]
- Song, S.Y.; Kim, J.-W.; Lee, N.-K.; Paik, H.-D. Lactiplantibacillus Plantarum WB4201, WB4202, and WB4203 Modulated Adipogenesis, Lipogenesis, and Fatty Acid β-Oxidation in 3T3-L1 Cells. Probiotics Antimicro. Prot. 2025, 1–13. [Google Scholar] [CrossRef]
- Wang, S.; Li, J.; Liu, W.-H.; Li, N.; Liang, H.; Hung, W.; Jiang, Q.; Cheng, R.; Shen, X.; He, F. Lacticaseibacillus paracasei K56 Inhibits Lipid Accumulation in Adipocytes by Promoting Lipolysis. Food Sci. Hum. Wellness 2024, 13, 3511–3521. [Google Scholar] [CrossRef]
- Jeong, Y.; Kim, H.; Lee, J.Y.; Won, G.; Choi, S.-I.; Kim, G.-H.; Kang, C.-H. The Antioxidant, Anti-Diabetic, and Anti-Adipogenesis Potential and Probiotic Properties of Lactic Acid Bacteria Isolated from Human and Fermented Foods. Fermentation 2021, 7, 123. [Google Scholar] [CrossRef]
- Hyun, I.K.; Lee, J.S.; Yoon, J.-W.; Kang, S.-S. Skimmed Milk Fermented by Lactic Acid Bacteria Inhibits Adipogenesis in 3T3-L1 Pre-Adipocytes by Downregulating PPARγ via TNF-α Induction in Vitro. Food Funct. 2021, 12, 8605–8614. [Google Scholar] [CrossRef]
- Han, K.J.; Lee, N.-K.; Yu, H.-S.; Park, H.; Paik, H.-D. Anti-Adipogenic Effects of the Probiotic Lactiplantibacillus Plantarum KU15117 on 3T3-L1 Adipocytes. Probiotics Antimicro. Prot. 2022, 14, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA Using Real-Time RT-PCR. Nat. Protoc. 2006, 1, 1559–1582. [Google Scholar] [CrossRef]
- Grätz, C.; Bui, M.L.U.; Thaqi, G.; Kirchner, B.; Loewe, R.P.; Pfaffl, M.W. Obtaining Reliable RT-qPCR Results in Molecular Diagnostics—MIQE Goals and Pitfalls for Transcriptional Biomarker Discovery. Life 2022, 12, 386. [Google Scholar] [CrossRef]
- Tang, Q.; Zhou, G.-C.; Liu, S.-J.; Li, W.; Wang, Y.-L.; Xu, G.-Y.; Li, T.-F.; Meng, G.-Q.; Xue, J.-Y. Selection and Validation of Reference Genes for qRT-PCR Analysis of Gene Expression in Tropaeolum Majus (Nasturtium). Horticulturae 2023, 9, 1176. [Google Scholar] [CrossRef]
- Li, S.; Ge, X.; Bai, G.; Chen, C. Selection of Reference Genes for Expression Normalization by RT-qPCR in Dracocephalum moldavica L. Curr. Issues Mol. Biol. 2024, 46, 6284–6299. [Google Scholar] [CrossRef] [PubMed]
- Cahyadi, D.D.; Warita, T.; Irie, N.; Mizoguchi, K.; Tashiro, J.; Hosaka, Y.Z.; Warita, K. Housekeeping Gene Expression Variability in Differentiating and Non-Differentiating 3T3-L1 Cells. Adipocyte 2023, 12, 2235081. [Google Scholar] [CrossRef]
- Gong, H.; Sun, L.; Chen, B.; Han, Y.; Pang, J.; Wu, W.; Qi, R.; Zhang, T. Evaluation of Candidate Reference Genes for RT-qPCR Studies in Three Metabolism Related Tissues of Mice after Caloric Restriction. Sci. Rep. 2016, 6, 38513. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase Relative Quantification Framework and Software for Management and Automated Analysis of Real-Time Quantitative PCR Data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of Stable Housekeeping Genes, Differentially Regulated Target Genes and Sample Integrity: BestKeeper--Excel-Based Tool Using Pair-Wise Correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Lian, C.; Zhang, B.; Yang, J.; Lan, J.; Yang, H.; Guo, K.; Li, J.; Chen, S. Validation of suitable reference genes by various algorithms for gene expression analysis in Isodon rubescens under different abiotic stresses. Sci. Rep. 2022, 12, 19599. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Choi, S.-I.; Jang, M.; Jeong, Y.; Kang, C.-H.; Kim, G.-H. Anti-Adipogenic Effect of Lactobacillus Fermentum MG4231 and MG4244 through AMPK Pathway in 3T3-L1 Preadipocytes. Food Sci. Biotechnol. 2020, 29, 1541–1551. [Google Scholar] [CrossRef]
- Oh, N.; Lee, J.; Kim, H.; Kwon, M.; Seo, J.; Roh, S. Comparison of Cell-Free Extracts from Three Newly Identified Lactobacillus Plantarum Strains on the Inhibitory Effect of Adipogenic Differentiation and Insulin Resistance in 3T3-L1 Adipocytes. BioMed Res. Int. 2021, 2021, 6676502. [Google Scholar] [CrossRef]
- Lee, C.S.; Park, M.H.; Kim, S.H. Selection and Characterization of Probiotic Bacteria Exhibiting Antiadipogenic Potential in 3T3-L1 Preadipocytes. Probiotics Antimicrob. Proteins 2022, 14, 72–86. [Google Scholar] [CrossRef] [PubMed]
- Grigorova, N.; Ivanova, Z.; Vachkova, E.; Petrova, V.; Beev, G. Antidiabetic and Hypolipidemic Properties of Newly Isolated Wild Lacticaseibacillus Paracasei Strains in Mature Adipocytes. Appl. Sci. 2023, 13, 6489. [Google Scholar] [CrossRef]
- Arnhold, S.; Elashry, M.I.; Klymiuk, M.C.; Geburek, F. Investigation of Stemness and Multipotency of Equine Adipose-Derived Mesenchymal Stem Cells (ASCs) from Different Fat Sources in Comparison with Lipoma. Stem Cell Res. Ther. 2019, 10, 309. [Google Scholar] [CrossRef]
- Chechi, K.; Gelinas, Y.; Mathieu, P.; Deshaies, Y.; Richard, D. Validation of Reference Genes for the Relative Quantification of Gene Expression in Human Epicardial Adipose Tissue. PLoS ONE 2012, 7, e32265. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- Arora, R.; Malla, W.A.; Tyagi, A.; Saxena, S.; Mahajan, S.; Sajjanar, B.; Tiwari, A.K. Identification of Suitable Reference Genes for qPCR Analysis of 4T1 Mouse Mammary Tumor Cell Line. Indian J. Anim. Res. 2021, 58, 1872–1877. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. miRDeepFinder: A miRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Hubrecht, R.C.; Carter, E. The 3Rs and Humane Experimental Technique: Implementing Change. Animals 2019, 9, 754. [Google Scholar] [CrossRef] [PubMed]
- Assessment, U.E.N.C. for E. The Principles of Humane Experimental Technique. Available online: https://hero.epa.gov/hero/index.cfm/reference/details/reference_id/4219117 (accessed on 29 July 2025).
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering Probiotic Microorganisms: In Vitro, in Vivo, Genetic and Omics Approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef]
- Vinderola, G.; Gueimonde, M.; Gomez-Gallego, C.; Delfederico, L.; Salminen, S. Correlation between in Vitro and in Vivo Assays in Selection of Probiotics from Traditional Species of Bacteria. Trends Food Sci. Technol. 2017, 68, 83–90. [Google Scholar] [CrossRef]
- Institute of Medicine (US) and National Research Council (US) Committee on the Framework for Evaluating the Safety of Dietary Supplements. Dietary Supplements: A Framework for Evaluating Safety; National Academies Press (US): Washington, DC, USA, 2005. [Google Scholar] [CrossRef]
- Hays, H.; Gu, Z.; Mai, K.; Zhang, W. Transcriptome-Based Nutrigenomics Analysis Reveals the Roles of Dietary Taurine in the Muscle Growth of Juvenile Turbot (Scophthalmus maximus). Comp. Biochem. Physiol. Part D Genom. Proteom. 2023, 47, 101120. [Google Scholar] [CrossRef] [PubMed]
- Martín-Alonso, S.; Frutos-Beltrán, E.; Menéndez-Arias, L. Reverse Transcriptase: From Transcriptomics to Genome Editing. Trends Biotechnol. 2021, 39, 194–210. [Google Scholar] [CrossRef]
- Mehta, N. RT-qPCR Made Simple: A Comprehensive Guide on the Methods, Advantages, Disadvantages, and Everything in Between. Undergrad. Res. Nat. Clin. Sci. Technol. J. 2022, 6, 1–6. [Google Scholar] [CrossRef]
- Bong, D.; Sohn, J.; Lee, S.-J.V. Brief Guide to RT-qPCR. Mol. Cells 2024, 47, 100141. [Google Scholar] [CrossRef]
- Zhao, F.; Maren, N.A.; Kosentka, P.Z.; Liao, Y.Y.; Lu, H.; Duduit, J.R.; Huang, D.; Ashrafi, H.; Zhao, T.; Huerta, A.I.; et al. An optimized protocol for stepwise optimization of real-time RT-PCR analysis. Hortic. Res. 2021, 8, 179. [Google Scholar] [CrossRef]
- Harshitha, R.; Arunraj, D.R. Real-Time Quantitative PCR: A Tool for Absolute and Relative Quantification. Biochem. Mol. Biol. Educ. 2021, 49, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative Expression Software Tool (REST) for Group-Wise Comparison and Statistical Analysis of Relative Expression Results in Real-Time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Mao, Y.; Huang, S.; Ni, J.; Lu, W.; Hou, J.; Wang, Y.; Zhao, W.; Li, M.; Wang, Q.; et al. Selection of Suitable Reference Genes for Quantitative Real-Time PCR in Sapium Sebiferum. Front. Plant Sci. 2017, 8, 637. [Google Scholar] [CrossRef]
- Tontonoz, P.; Spiegelman, B.M. Fat and Beyond: The Diverse Biology of PPARγ. Annu. Rev. Biochem. 2008, 77, 289–312. [Google Scholar] [CrossRef] [PubMed]
- Lehrke, M.; Lazar, M.A. The Many Faces of PPARγ. Cell 2005, 123, 993–999. [Google Scholar] [CrossRef]
- Nobusue, H.; Onishi, N.; Shimizu, T.; Sugihara, E.; Oki, Y.; Sumikawa, Y.; Chiyoda, T.; Akashi, K.; Saya, H.; Kano, K. Regulation of MKL1 via Actin Cytoskeleton Dynamics Drives Adipocyte Differentiation. Nat. Commun. 2014, 5, 3368. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human Housekeeping Genes, Revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Waldenström, J. With Reference to Reference Genes: A Systematic Review of Endogenous Controls in Gene Expression Studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef]
- You, S.; Cao, K.; Chen, C.; Li, Y.; Wu, J.; Zhu, G.; Fang, W.; Wang, X.; Wang, L. Selection and Validation Reference Genes for qRT-PCR Normalization in Different Cultivars during Fruit Ripening and Softening of Peach (Prunus Persica). Sci. Rep. 2021, 11, 7302. [Google Scholar] [CrossRef] [PubMed]
- Almeida-Oliveira, F.; Leandro, J.G.B.; Ausina, P.; Sola-Penna, M.; Majerowicz, D. Reference Genes for Quantitative PCR in the Adipose Tissue of Mice with Metabolic Disease. Biomed. Pharmacother. 2017, 88, 948–955. [Google Scholar] [CrossRef] [PubMed]
| Abbreviation | Full Name | Forward | Reverse | Product Length |
|---|---|---|---|---|
| 36B4 | Ribosomal protein, large, P0 | TTATAACCCTGAAGTGCTCGAC | CGCTTGTACCCATTGATGATG | 147 |
| HPRT | Hypoxanthine-guanine phosphoribosyl transferase | ACAGGCCAGACTTTGTTGGA | ACTTGCGCTCATCTTAGGCT | 150 |
| Actb | Actin, beta | CCTCTATGCCAACACAGTGC | GTACTCCTGCTTGCTGATCC | 211 |
| HMBS | Hydroxymethylbilane synthase | CCTGAAGGATGTGCCTACCA | CCACTCGAATCACCCTCATCT | 175 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | AAATGGTGAAGGTCGGTGTG | TGAATTTGCCGTGAGTGGAG | 583 |
| 18S | 18S ribosomal RNA | ATGCGGCGGCGTTATTCC | GCTATCAATCTGTCAATCCTGTC | 204 |
| PPARγ | Peroxisome proliferator-activated receptor gamma, transcript var. 2 | AGGGCGATCTTGACAGGAAA | CGAAACTGGCACCCTTGAAA | 164 |
| Gene Abbr. | Stability M-Value (geNorm) | Ranking | Stability Value (NormFinder) | Ranking |
|---|---|---|---|---|
| 36B4 | 0.325 | 3 | 0.082 | 2 |
| HPRT | 0.299 | 1 | 0.071 | 1 |
| Actb | 0.452 | 5 | 0.202 | 5 |
| HMBS | 0.324 | 2 | 0.109 | 3 |
| GAPDH | 0.402 | 4 | 0.181 | 4 |
| 18S | 0.510 | 6 | 0.215 | 6 |
| 18S | 36B4 | GAPDH | HMBS | HPRT | Actb | |
|---|---|---|---|---|---|---|
| n | 36 | 36 | 36 | 36 | 36 | 36 |
| geo Mean [CP] | 14.52 | 18.72 | 17.10 | 24.71 | 23.14 | 18.46 |
| ar Mean [CP] | 14.52 | 18.72 | 17.11 | 24.71 | 23.14 | 18.47 |
| min [CP] | 13.58 | 18.03 | 16.34 | 23.91 | 22.11 | 17.30 |
| max [CP] | 15.56 | 19.21 | 18.01 | 25.11 | 23.745 | 19.34 |
| std dev [±CP] | 0.31 | 0.20 | 0.21 | 0.19 | 0.16 | 0.33 |
| CV [% CP] | 2.13 | 1.08 | 1.25 | 0.77 | 0.70 | 1.80 |
| min [x-fold] | −73.63 | −24.12 | −33.87 | −38.69 | −113.1 | −211.3 |
| max [x-fold] | 119.8 | 9.39 | 62.82 | 6.23 | 16.44 | 54.91 |
| std dev [±x-fold] | 4.13 | 2.53 | 2.67 | 2.40 | 2.11 | 4.56 |
| Regression Analysis: HKG vs. BestKeeper | ||||||
|---|---|---|---|---|---|---|
| 18S | 36B4 | GAPDH | HMBS | HPRT | Actb | |
| coeff. of corr. [r] | 0.48 | 0.80 | 0.57 | 0.74 | 0.79 | 0.67 |
| coeff. of det. [r2] | 0.23 | 0.63 | 0.33 | 0.54 | 0.63 | 0.45 |
| intercept [CP] | −3.64 | −0.41 | 2.06 | 7.98 | 4.16 | −6.64 |
| slope [CP] | 0.95 | 1.00 | 0.79 | 0.87 | 0.99 | 1.31 |
| SE [CP] | ±0.374 | ±0.165 | ±0.246 | ±0.175 | ±0.167 | ±0.316 |
| p-value | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| Power [x-fold] | 78.00 | 99.01 | 36.89 | 55.28 | 94.92 | 411.45 |
| 18S | HPRT | HMBS | 36B4 | GAPDH | Actb | |
|---|---|---|---|---|---|---|
| PPARγ coeff. of correlation. [r] | −0.130 | 0.463 | 0.342 | 0.512 | −0.002 | 0.759 |
| PPARγ p-value | 0.452 | 0.004 | 0.041 | 0.001 | 0.992 | 0.001 |
| Gene Names | Mean Δ Ct | Std.Dev. | Mean STD. Dev. * |
|---|---|---|---|
| HPRT/18S | 8.61 | 0.50 | |
| HPRT/36B4 | 4.42 | 0.21 | |
| HPRT/GAPDH | 6.03 | 0.24 | |
| HPRT/HMBS | 1.57 | 0.17 | |
| HPRT/Actb | 4.67 | 0.34 | 0.29 a |
| 18S/36B4 | 4.20 | 0.48 | |
| 18S/GAPDH | 2.58 | 0.51 | |
| 18S/HMBS | 10.18 | 0.47 | |
| 18S/Actb | 3.94 | 0.58 | |
| 18S/HPRT | 8.61 | 0.50 | 0.51 b |
| 36B4/GAPDH | 1.61 | 0.29 | |
| 36B4/HMBS | 5.99 | 0.24 | |
| 36B4/Actb | 0.32 | 0.24 | |
| 36B4/HPRT | 4.42 | 0.21 | |
| 36B4/18S | 4.20 | 0.48 | 0.29 a |
| GAPDH/HMBS | 7.60 | 0.28 | |
| GAPDH/Actb | 1.36 | 0.51 | |
| GAPDH/HPRT | 6.03 | 0.24 | |
| GAPDH/18S | 2.58 | 0.51 | |
| GAPDH/36B4 | 1.61 | 0.29 | 0.37 ab |
| HMBS/Actb | 6.24 | 0.38 | |
| HMBS/HPRT | 1.57 | 0.17 | |
| HMBS/18S | 10.18 | 0.47 | |
| HMBS/36B4 | 5.99 | 0.24 | |
| HMBS/GAPDH | 7.60 | 0.28 | 0.31 a |
| Actb/HPRT | 4.67 | 0.34 | |
| Actb/18S/ | 3.94 | 0.58 | |
| Actb/36B4 | 0.32 | 0.24 | |
| Actb/GAPDH | 1.36 | 0.51 | |
| Actb/HMBS | 6.24 | 0.38 | 0.41 ab |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivanova, Z.; Grigorova, N.; Petrova, V.; Vachkova, E.; Beev, G. Comparative Evaluation of Computational Methods for Validating Housekeeping Gene RT-qPCR Data in 3T3-L1 Cells. Biomedicines 2025, 13, 2036. https://doi.org/10.3390/biomedicines13082036
Ivanova Z, Grigorova N, Petrova V, Vachkova E, Beev G. Comparative Evaluation of Computational Methods for Validating Housekeeping Gene RT-qPCR Data in 3T3-L1 Cells. Biomedicines. 2025; 13(8):2036. https://doi.org/10.3390/biomedicines13082036
Chicago/Turabian StyleIvanova, Zhenya, Natalia Grigorova, Valeria Petrova, Ekaterina Vachkova, and Georgi Beev. 2025. "Comparative Evaluation of Computational Methods for Validating Housekeeping Gene RT-qPCR Data in 3T3-L1 Cells" Biomedicines 13, no. 8: 2036. https://doi.org/10.3390/biomedicines13082036
APA StyleIvanova, Z., Grigorova, N., Petrova, V., Vachkova, E., & Beev, G. (2025). Comparative Evaluation of Computational Methods for Validating Housekeeping Gene RT-qPCR Data in 3T3-L1 Cells. Biomedicines, 13(8), 2036. https://doi.org/10.3390/biomedicines13082036







