Bullous Congenital Ichthyosiform Erythroderma with Tinea Capitis in Half-Siblings: Rare Phenomenon in Ichthyosis with Co-Existing Trichophyton rubrum Infection and Blocker Displacement Amplification for Mosaic Mutation Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and DNA Extraction
2.3. Whole Exome Sequencing
2.4. Conventional PCR
2.5. Blocker Displacement Amplification-Based PCR
2.6. Sanger Sequencing
2.7. Statistical Analysis
3. Results
3.1. Clinical Manifestations
3.2. Mutation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Correction Statement
References
- Peter Rout, D.; Nair, A.; Gupta, A.; Kumar, P. Epidermolytic hyperkeratosis: Clinical update. Clin. Cosmet. Investig. Dermatol. 2019, 12, 333–344. [Google Scholar] [CrossRef]
- Hernández-Martín, Á.; Paller, A.S.; Sprecher, E.; Akiyama, M.; Granier Tournier, C.; Aldwin-Easton, M.; Bodemer, C.; Choate, K.; Fischer, J.; Gostynski, A.; et al. A Proposal for a New Pathogenesis-guided Classification for Inherited Epidermal Differentiation Disorders. Br. J. Dermatol. 2025; [Epub ahead of print]. [Google Scholar] [CrossRef]
- Foster, M.L.; Jones, J.M.; Schadt, C.R. Epidermolytic Hyperkeratosis. JAMA Dermatol. 2021, 157, 1114. [Google Scholar] [CrossRef]
- Mondal, A.K.; Kumar, P.; Mondal, A. Bullous congenital ichthyosiform erythroderma. Indian Pediatr. 2011, 48, 968. [Google Scholar]
- Gånemo, A.; Lindholm, C.; Lindberg, M.; Sjödén, P.O.; Vahlquist, A. Quality of life in adults with congenital ichthyosis. J. Adv. Nurs. 2003, 44, 412–419. [Google Scholar] [CrossRef]
- Bygum, A.; Virtanen, M.; Brandrup, F.; Gånemo, A.; Sommerlund, M.; Strauss, G.; Vahlquist, A. Generalized and naevoid epidermolytic ichthyosis in Denmark: Clinical and mutational findings. Acta Derm.-Venereol. 2013, 93, 309–313. [Google Scholar] [CrossRef]
- Arin, M.J.; Oji, V.; Emmert, S.; Hausser, I.; Traupe, H.; Krieg, T.; Grimberg, G. Expanding the keratin mutation database: Novel and recurrent mutations and genotype-phenotype correlations in 28 patients with epidermolytic ichthyosis. Br. J. Dermatol. 2011, 164, 442–447. [Google Scholar] [CrossRef]
- Severino-Freire, M.; Jonca, N.; Pichery, M.; Tournier, E.; Chassaing, N.; Mazereeuw-Hautier, J. Extensive Post-zygotic Mosaicism of KRT1 or KRT10 Mutation Mimicking Classical Epider-molytic Ichthyosis. Acta Derm.-Venereol. 2017, 97, 387–388. [Google Scholar] [CrossRef]
- Samuelov, L.; Sarig, O.; Gat, A.; Halachmi, S.; Shalev, S.; Sprecher, E. Extensive lentigo simplex, linear epidermolytic naevus and epidermolytic naevus comedonicus caused by a somatic mutation in KRT10. Br. J. Dermatol. 2015, 173, 293–296. [Google Scholar] [CrossRef]
- Wu, L.R.; Chen, S.X.; Wu, Y.; Patel, A.A.; Zhang, D.Y. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 2017, 1, 714–723. [Google Scholar] [CrossRef]
- Gambin, T.; Liu, Q.; Karolak, J.A.; Grochowski, C.M.; Xie, N.G.; Wu, L.R.; Yan, Y.H.; Cao, Y.; Coban Akdemir, Z.H.; Wilson, T.A.; et al. Low-level parental somatic mosaic SNVs in exomes from a large cohort of trios with diverse suspected Mendelian conditions. Genet. Med. 2020, 22, 1768–1776. [Google Scholar] [CrossRef]
- Barac, A.; Stjepanovic, M.; Krajisnik, S.; Stevanovic, G.; Paglietti, B.; Milosevic, B. Dermatophytes: Update on Clinical Epidemiology and Treatment. Mycopathologia 2024, 189, 101. [Google Scholar] [CrossRef] [PubMed]
- Freitas, C.F.; Mulinari-Brenner, F.; Fontana, H.R.; Gentili, A.C.; Hammerschmidt, M. Ichthyosis associated with widespread tinea corporis: Report of three cases. An. Bras. Dermatol. 2013, 88, 627–630. [Google Scholar] [CrossRef]
- Sheetz, K.; Lynch, P.J. Ichthyosis and dermatophyte fungal infection. J. Am. Acad. Dermatol. 1991, 24, 321. [Google Scholar] [CrossRef]
- Szlávicz, E.; Németh, C.; Szepes, É.; Gyömörei, C.; Gyulai, R.; Lengyel, Z. Congenital ichthyosis associated with Trichophyton rubrum tinea, imitating drug hypersensitivity reaction. Med. Mycol. Case Rep. 2020, 29, 15–17. [Google Scholar] [CrossRef]
- Karolak, J.A.; Liu, Q.; Xie, N.G.; Wu, L.R.; Rocha, G.; Fernandes, S.; Ho-Ming, L.; Lo, I.F.; Mowat, D.; Fiorino, E.K.; et al. Highly Sensitive Blocker Displacement Amplification and Droplet Digital PCR Reveal Low-Level Parental FOXF1 Somatic Mosaicism in Families with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins. J. Mol. Diagn. 2020, 22, 447–456. [Google Scholar] [CrossRef]
- Michaels, B.D.; Del Rosso, J.Q. Tinea capitis in infants: Recognition, evaluation, and management suggestions. J. Clin. Aesthet. Dermatol. 2012, 5, 49–59. [Google Scholar]
- Mayser, P.; Nenoff, P.; Reinel, D.; Abeck, D.; Brasch, J.; Daeschlein, G.; Effendy, I.; Ginter-Hanselmayer, G.; Gräser, Y.; Hipler, U.C.; et al. S1 guidelines: Tinea capitis. J. Dtsch. Dermatol. Ges. J. Ger. Soc. Dermatol. JDDG 2020, 18, 161–179. [Google Scholar] [CrossRef]
- Schøsler, L.; Andersen, L.K.; Arendrup, M.C.; Sommerlund, M. Recurrent terbinafine resistant Trichophyton rubrum infection in a child with congenital ichthyosis. Pediatr. Dermatol. 2018, 35, 259–260. [Google Scholar] [CrossRef]
- Gupta, A.K.; Mays, R.R.; Versteeg, S.G.; Piraccini, B.M.; Shear, N.H.; Piguet, V.; Tosti, A.; Friedlander, S.F. Tinea capitis in children: A systematic review of management. J. Eur. Acad. Dermatol. Venereol. JEADV 2018, 32, 2264–2274. [Google Scholar] [CrossRef] [PubMed]
- Waśkiel-Burnat, A.; Rakowska, A.; Sikora, M.; Ciechanowicz, P.; Olszewska, M.; Rudnicka, L. Trichoscopy of Tinea Capitis: A Systematic Review. Dermatol. Ther. 2020, 10, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, C.; Zhou, X.; Li, C.; Zhang, H.; Lian, B.Q.; Lee, J.J.; Shen, J.; Liu, Y.; Lian, C.G. Homozygous ALOXE3 Nonsense Variant Identified in a Patient with Non-Bullous Congenital Ichthyosiform Erythroderma Complicated by Superimposed Bullous Majocchi’s Granuloma: The Consequences of Skin Barrier Dysfunction. Int. J. Mol. Sci. 2015, 16, 21791–21801. [Google Scholar] [CrossRef]
- Agostini, G.; Geti, V.; Difonzo, E.M.; Giannotti, B. Dermatophyte infection in ichthyosis vulgaris. Mycoses 1992, 35, 197–199. [Google Scholar] [CrossRef]
- Youssefian, L.; Khodavaisy, S.; Khosravi-Bachehmir, F.; Park, J.S.; Saeidian, A.H.; Mahmoudi, H.; Saffarian, Z.; Naraghi, Z.S.; Kamyab-Hesari, K.; Zeinali, S.; et al. Ichthyosis, psoriasiform dermatitis, and recurrent fungal infections in patients with biallelic mutations in PERP. J. Eur. Acad. Dermatol. Venereol. JEADV 2022, 36, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, M.; Yadav, P.; Yadav, J.; Chander, R. Clear zone phenomenon: A rare phenomenon in ichthyosis with co-existing superficial fungal infection. Indian J. Dermatol. Venereol. Leprol. 2021, 87, 103–105. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Liang, P.; Chen, J.; Feng, P.; Lai, W. Keratitis-ichthyosis-deafness syndrome accompanied by disseminated cutaneous fungal infection. J. Dermatol. 2017, 44, 1255–1261. [Google Scholar] [CrossRef] [PubMed]
- Scheers, C.; Andre, J.; Thompson, C.; Rebuffat, E.; Harag, S.; Kolivras, A. Refractory Trichophyton rubrum infection in lamellar ichthyosis. Pediatr. Dermatol. 2013, 30, e200–e203. [Google Scholar] [CrossRef]
- Shirato, K.; Marshman, G. Dermatophytosis and Sjögren-Larsson syndrome: Foe or friend? Australas. J. Dermatol. 2011, 52, 231–232. [Google Scholar] [CrossRef]
- Hoetzenecker, W.; Schanz, S.; Schaller, M.; Fierlbeck, G. Generalized tinea corporis due to Trichophyton rubrum in ichthyosis vulgaris. J. Eur. Acad. Dermatol. Venereol. JEADV 2007, 21, 1129–1131. [Google Scholar] [CrossRef]
- Ludwig, R.J.; Woodfolk, J.A.; Grundmann-Kollmann, M.; Enzensberger, R.; Runne, U.; Platts-Mills, T.A.; Kaufmann, R.; Zollner, T.M. Chronic dermatophytosis in lamellar ichthyosis: Relevance of a T-helper 2-type immune response to Trichophyton rubrum. Br. J. Dermatol. 2001, 145, 518–521. [Google Scholar] [CrossRef] [PubMed]
- Oztürkcan, S.; Parlak, A.H.; Marufi, M.; Bakici, M.Z.; Egilmez, R.; Gültekin, A. Congenital ichthyosiform erythroderma with Trichophyton rubrum infection. Indian Pediatr. 1994, 31, 317–320. [Google Scholar]
- Shelley, E.D.; Shelley, W.B.; Schafer, R.L. Generalized Trichophyton rubrum infection in congenital ichthyosiform erythroderma. J. Am. Acad. Dermatol. 1989, 20, 1133–1134. [Google Scholar] [CrossRef]
- Kamalam, A.; Thambiah, A.S. Genetic ichthyosis and Trichophyton rubrum infection in infants. Mykosen 1982, 25, 281–283. [Google Scholar] [CrossRef]
- Haruna, K.; Suga, Y.; Mizuno, Y.; Hasegawa, T.; Kourou, K.; Matsuba, S.; Muramatsu, S.; Ikeda, S. R156C mutation of keratin 10 causes mild form of epidermolytic hyperkeratosis. J. Dermatol. 2007, 34, 545–548. [Google Scholar] [CrossRef]
- Cheraghlou, S.; Atzmony, L.; Roy, S.F.; McNiff, J.M.; Choate, K.A. Mutations in KRT10 in epidermolytic acanthoma. J. Cutan. Pathol. 2020, 47, 524–529. [Google Scholar] [CrossRef]
- Kono, M.; Suga, Y.; Akashi, T.; Ito, Y.; Takeichi, T.; Muro, Y.; Akiyama, M. A Child with Epidermolytic Ichthyosis from a Parent with Epidermolytic Nevus: Risk Evaluation of Transmission from Mosaic to Germline. J. Investig. Dermatol. 2017, 137, 2024–2026. [Google Scholar] [CrossRef]
- Campbell, I.M.; Yuan, B.; Robberecht, C.; Pfundt, R.; Szafranski, P.; McEntagart, M.E.; Nagamani, S.C.; Erez, A.; Bartnik, M.; Wiśniowiecka-Kowalnik, B.; et al. Parental somatic mosaicism is underrecognized and influences recurrence risk of genomic disorders. Am. J. Hum. Genet. 2014, 95, 173–182. [Google Scholar] [CrossRef]
- Wolter, M.; Felsberg, J.; Malzkorn, B.; Kaulich, K.; Reifenberger, G. Droplet digital PCR-based analyses for robust, rapid, and sensitive molecular diagnostics of gliomas. Acta Neuropathol. Commun. 2022, 10, 42. [Google Scholar] [CrossRef]
- Hu, X.; He, W.B.; Zhang, S.P.; Luo, K.L.; Gong, F.; Dai, J.; Zhang, Y.; Wan, Z.X.; Li, W.; Yuan, S.M.; et al. Next-generation sequence-based preimplantation genetic testing for monogenic disease resulting from maternal mosaicism. Mol. Genet. Genom. Med. 2021, 9, e1662. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Luo, J.; Yu, H.; Dong, B.; Zhang, Q.; Zhang, W.; Chen, K.; Xiang, Y.; Liu, D.; Huang, G. Blocker displacement amplification-based genetic diagnosis for autosomal dominant polycystic kidney disease and the clinical outcomes of preimplantation genetic testing. J. Assist. Reprod. Genet. 2023, 40, 783–792. [Google Scholar] [CrossRef] [PubMed]
- Akkhasutthikun, P.; Kaewsapsak, P.; Nimsamer, P.; Klomkliew, P.; Visedthorn, S.; Chanchaem, P.; Teerapakpinyo, C.; Payungporn, S.; Luangdilok, S. Tissue and Plasma-Based Highly Sensitive Blocker Displacement Amplicon Nanopore Sequencing for EGFR Mutations in Lung Cancer. Cancer Res. Treat. 2024, 56, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Su, X.; Leong, S.; Xiu, X.; Song, P.; Peng, J.; Si, Y. Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection. Biosensors 2025, 15, 308. [Google Scholar] [CrossRef] [PubMed]
- Schmuth, M.; Yosipovitch, G.; Williams, M.L.; Weber, F.; Hintner, H.; Ortiz-Urda, S.; Rappersberger, K.; Crumrine, D.; Feingold, K.R.; Elias, P.M. Pathogenesis of the permeability barrier abnormality in epidermolytic hyperkeratosis. J. Investig. Dermatol. 2001, 117, 837–847. [Google Scholar] [CrossRef] [PubMed]
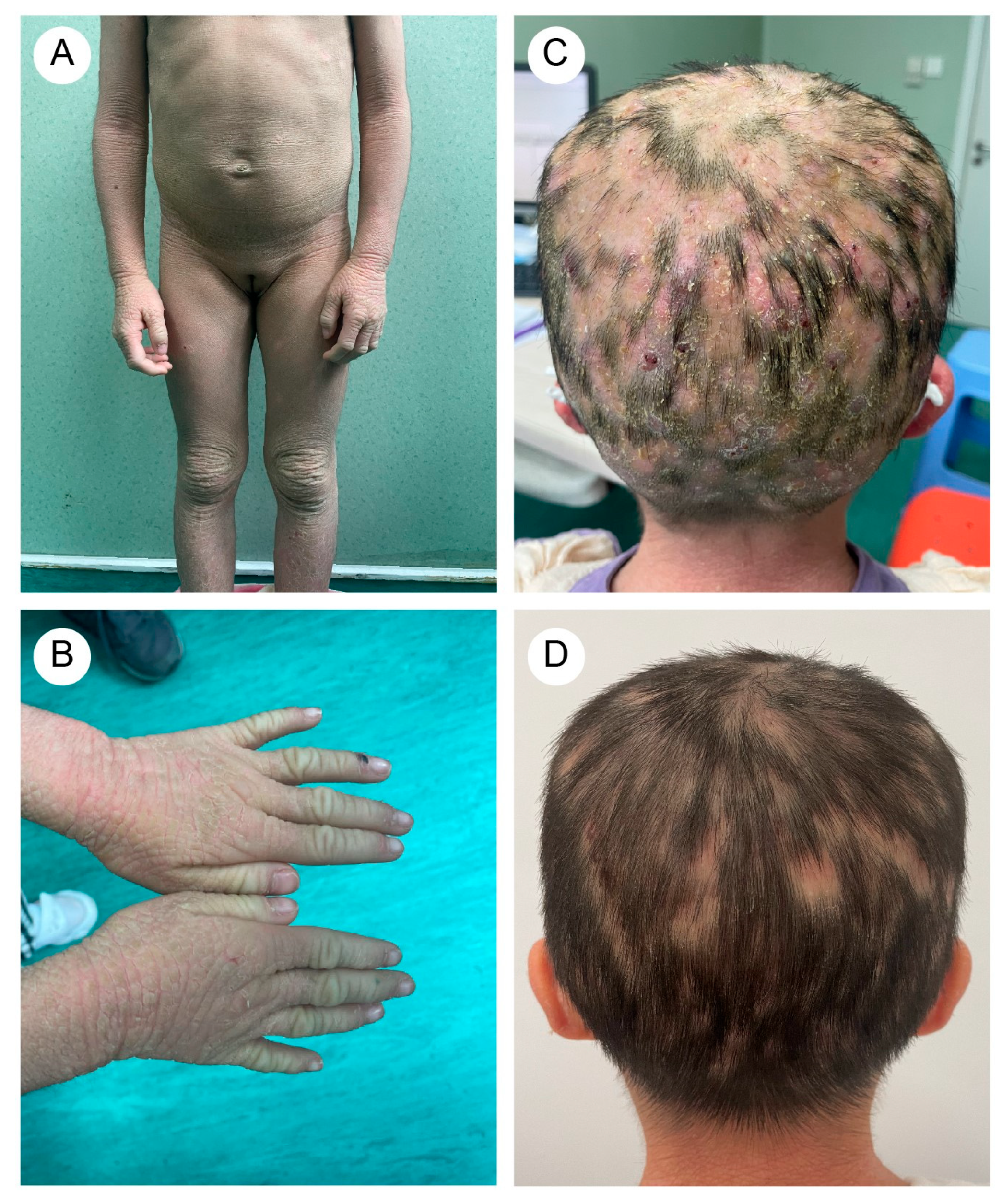
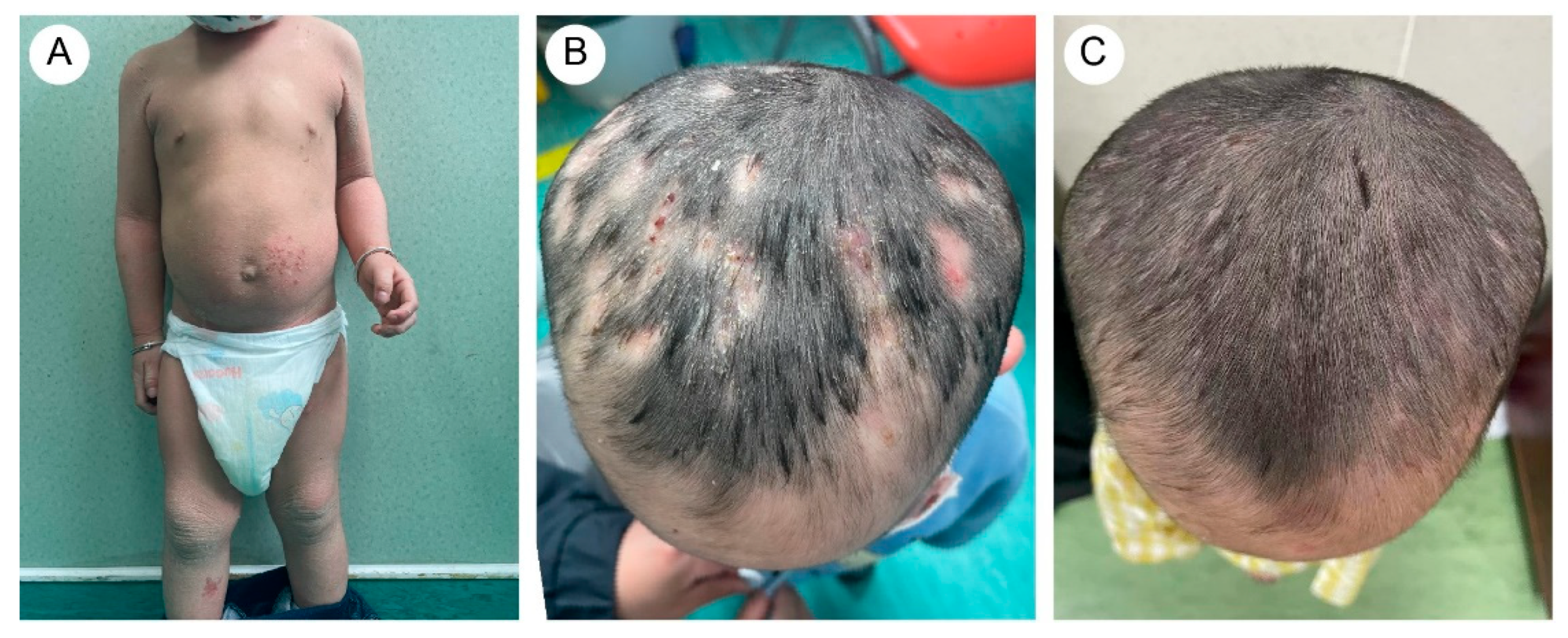
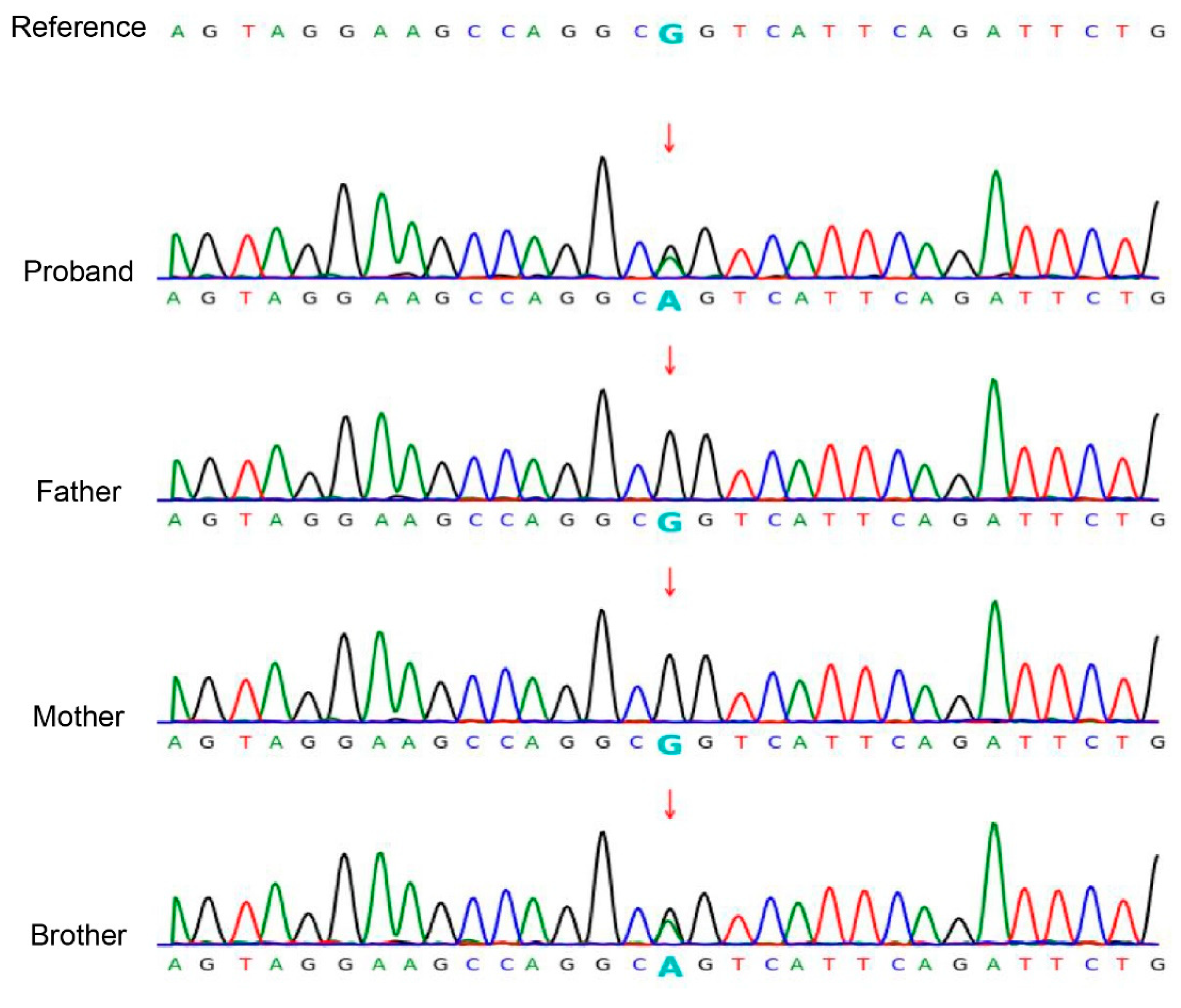

| Reference | Country | Age * | Type | Variant * | Dermatophyte |
|---|---|---|---|---|---|
| This study | China | 5Y8M, 2Y4M | BCIE | KRT10 | T. rubrum |
| Youssefian et al. (2022) [24] | Iran | 26Y, 35Y | Ichthyosis, psoriasiform dermatitis | PERP | T. rubrum, E. floccosum |
| Agrawal et al. (2021) [25] | Indian | 16Y, 19Y | X-linked recessive ichthyosis | STS | T. rubrum |
| Szlávicz et al. (2020) [15] | Hungary | 54Y | Ichthyosis | N.A. | T. rubrum |
| Schøsler et al. (2018) [19] | Denmark | 9Y | Congenital ichthyosiform | N.A. | T. rubrum |
| Ma et al. (2017) [26] | China | 35Y | Keratitis–ichthyosis–deafness syndrome | GJB2 | T. rubrum |
| Wang et al. (2015) [22] | China | 11Y | Non-bullous congenital ichthyosiform erythroderma | ALOXE3 | T. rubrum |
| Freitas et al. (2013) [13] | Brazil | 87Y, 73Y, 27Y | Congenital ichthyosiform erythroderma; ichthyosis linearis circumflexa; Sjögren–Larsson Syndrome | N.A. | T. rubrum |
| Scheers et al. (2013) [27] | Belgium | 10M | Congenital lamellar ichthyosis | N.A. | T. rubrum |
| Shirato et al. (2011) [28] | Dutch | 35Y, 40Y | Sjögren–Larsson syndrome, followed by a return of the lamellar ichthyosis | FALDH | T. rubrum |
| Hoetzenecker et al. (2007) [29] | Germany | 38Y | Ichthyosis vulgaris | N.A. | T. rubrum |
| Ludwig et al. (2001) [30] | USA | 2Y | Lamellar ichthyosis | N.A. | T. rubrum |
| Oztürkcan et al. (1994) [31] | Tiirkiye | 10Y | Non-bullous congenital ichthyosiform erythroderma | N.A. | T. rubrum |
| Agostini et al. (1992) [23] | Italy | 31Y | Ichthyosis vulgaris | N.A. | T. rubrum |
| Shelley et al. (1989) [32] | USA | 41Y | Congenital ichthyosiform erythroderma | N.A. | Staphylococcus aureus, Pseudomonasaeruginos, T. rubrum |
| Kamalam et al. (1982) [33] | Madras/India | 6M, 10M | X-linked recessive ichthyosis; ichthyosis vulgaris (dominant) | N.A. | T. rubrum |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Fu, Y.; Zhang, Q.; Chen, Q.; Yang, Y.; Xue, Y.; Ren, Y. Bullous Congenital Ichthyosiform Erythroderma with Tinea Capitis in Half-Siblings: Rare Phenomenon in Ichthyosis with Co-Existing Trichophyton rubrum Infection and Blocker Displacement Amplification for Mosaic Mutation Detection. Biomedicines 2025, 13, 2015. https://doi.org/10.3390/biomedicines13082015
Liu J, Fu Y, Zhang Q, Chen Q, Yang Y, Xue Y, Ren Y. Bullous Congenital Ichthyosiform Erythroderma with Tinea Capitis in Half-Siblings: Rare Phenomenon in Ichthyosis with Co-Existing Trichophyton rubrum Infection and Blocker Displacement Amplification for Mosaic Mutation Detection. Biomedicines. 2025; 13(8):2015. https://doi.org/10.3390/biomedicines13082015
Chicago/Turabian StyleLiu, Jipeng, Yujuan Fu, Qihao Zhang, Qi Chen, Yuxiang Yang, Yi Xue, and Yunqing Ren. 2025. "Bullous Congenital Ichthyosiform Erythroderma with Tinea Capitis in Half-Siblings: Rare Phenomenon in Ichthyosis with Co-Existing Trichophyton rubrum Infection and Blocker Displacement Amplification for Mosaic Mutation Detection" Biomedicines 13, no. 8: 2015. https://doi.org/10.3390/biomedicines13082015
APA StyleLiu, J., Fu, Y., Zhang, Q., Chen, Q., Yang, Y., Xue, Y., & Ren, Y. (2025). Bullous Congenital Ichthyosiform Erythroderma with Tinea Capitis in Half-Siblings: Rare Phenomenon in Ichthyosis with Co-Existing Trichophyton rubrum Infection and Blocker Displacement Amplification for Mosaic Mutation Detection. Biomedicines, 13(8), 2015. https://doi.org/10.3390/biomedicines13082015






