Investigating the Diagnostic Utility of LncRNA GAS5 in NAFLD Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Subjects
2.2. Methods
2.2.1. Routine Laboratory Tests
2.2.2. RNA Extraction
2.2.3. cDNA Synthesis
2.2.4. Quantitative Real-Time PCR
2.3. Statistical Analysis
3. Results
3.1. Characterization of Subjects
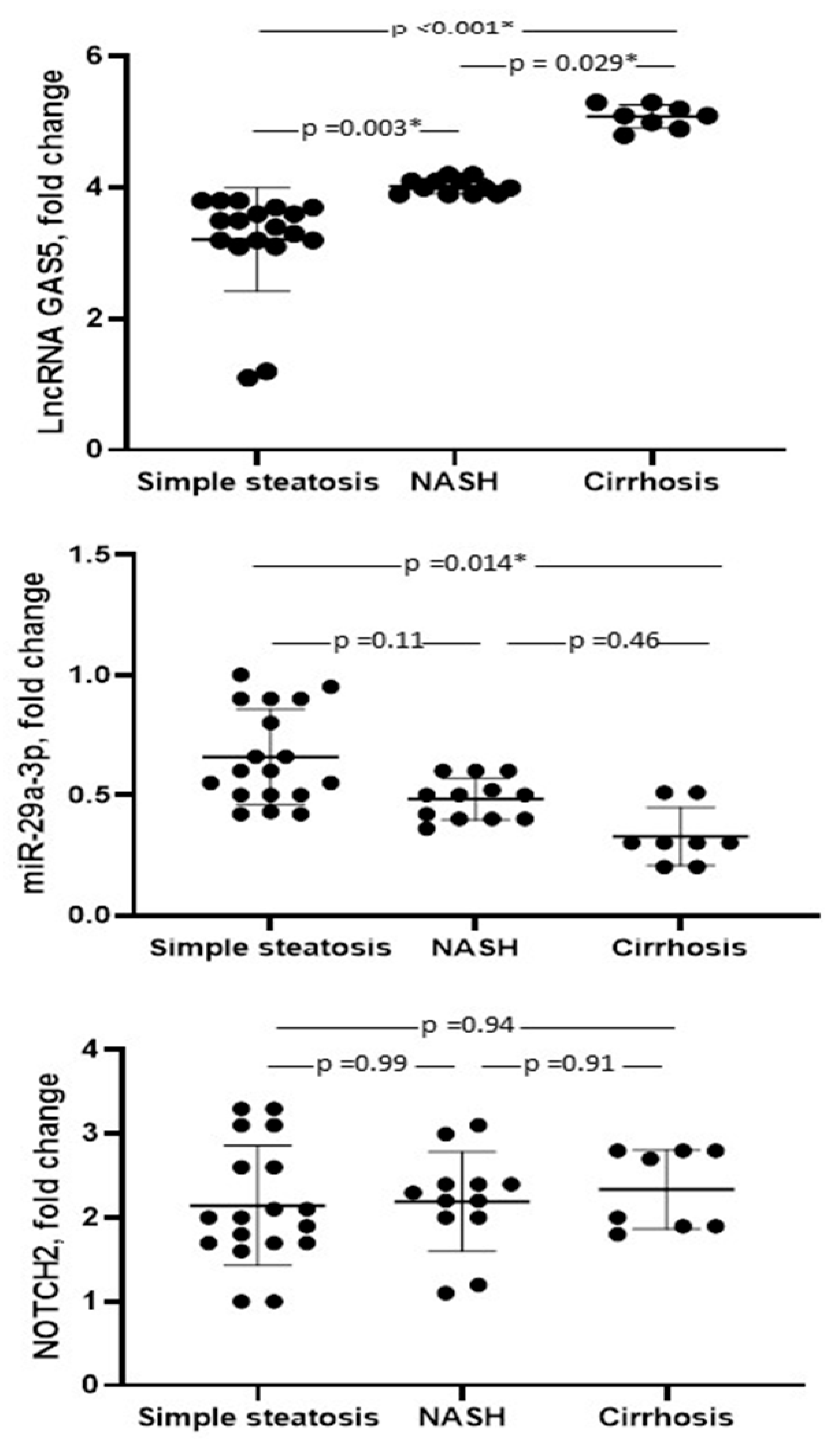
3.2. Marker Levels
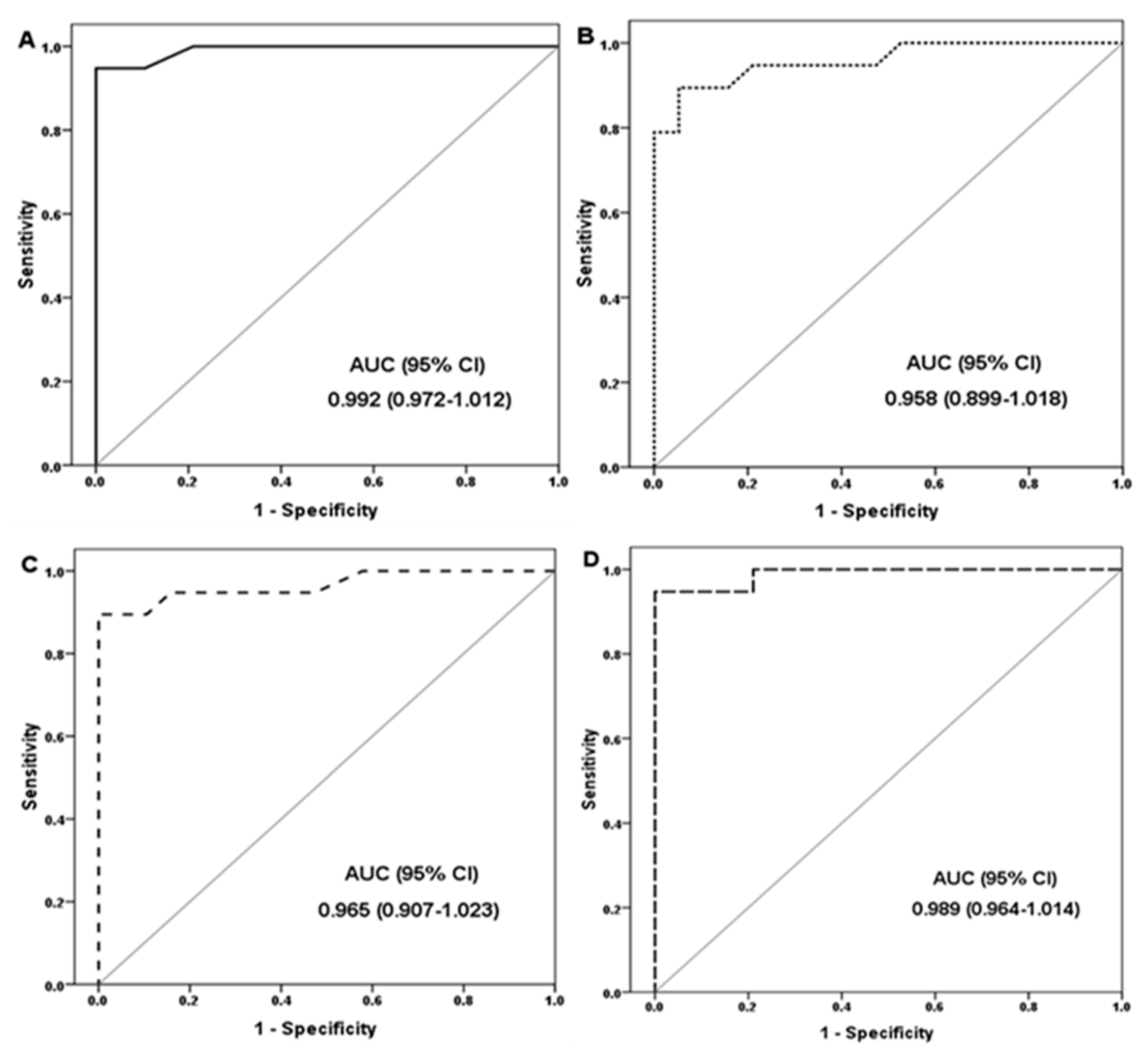
3.3. Diagnostic Performance of the Marker
3.4. Correlation Analysis of the Studied Markers
3.5. Logistic Regression Analysis of MVC Risk Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NAFLD | Non-alcoholic fatty liver disease |
| NASH | Non-alcoholic steatohepatitis |
| HCC | Hepatocellular carcinoma |
| lncRNAs | Long non-coding RNAs |
| TC | Total cholesterol |
| TG | Triglycerides |
| LDL-C | Low-density lipoprotein cholesterol |
| HDL-C | High-density lipoprotein cholesterol |
| ALT | Alanine aminotransferase |
| AST | Aspartate aminotransferase |
| cDNA | Complementary DNA |
| BMI | Body mass index |
| GAS | Growth arrest-specific |
| NOTCH2 | Neurogenic locus notch homolog protein 2. |
| miRNAs | microRNAs |
References
- Neuschwander-Tetri, B.A. Non-alcoholic fatty liver disease. BMC Med. 2017, 15, 45. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.G.; Kim, S.U.; Wong, V.W. New trends on obesity and NAFLD in Asia. J. Hepatol. 2017, 67, 862–873. [Google Scholar] [CrossRef]
- Maurice, J.; Manousou, P. Non-alcoholic fatty liver disease. Clin. Med. 2018, 18, 245–250. [Google Scholar] [CrossRef]
- Byrne, C.D.; Targher, G. NAFLD: A multisystem disease. J. Hepatol. 2015, 62 (Suppl. S1), S47–S64. [Google Scholar] [CrossRef] [PubMed]
- Ter Horst, K.W.; Serlie, M.J. Fructose Consumption, Lipogenesis, and Non-Alcoholic Fatty Liver Disease. Nutrients 2017, 9, 981. [Google Scholar] [CrossRef]
- Cataldo, I.; Sarcognato, S.; Sacchi, D.; Cacciatore, M.; Baciorri, F.; Mangia, A.; Cazzagon, N.; Guido, M. Pathology of non-alcoholic fatty liver disease. Pathologica 2021, 113, 194–202. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Wu, H.; Zhong, Z.; Wang, A.; Yuan, C.; Ning, K.; Hu, H.; Wang, C.; Yin, X. LncRNA FTX represses the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via regulating the M1/M2 polarization of Kupffer cells. Cancer Cell Int. 2020, 24, 266. [Google Scholar] [CrossRef]
- Shabgah, A.G.; Norouzi, F.; Hedayati-Moghadam, M.; Soleimani, D.; Pahlavani, N.; Navashenaq, J.G. A comprehensive review of long non-coding RNAs in the pathogenesis and development of non-alcoholic fatty liver disease. Nutr. Metab. 2021, 18, 22. [Google Scholar] [CrossRef]
- Fernández-Tussy, P.; Fernández-Ramos, D.; Lopitz-Otsoa, F.; Simón, J.; Barbier-Torres, L.; Gomez-Santos, B.; Nuñez-Garcia, M.; Azkargorta, M.; Gutiérrez-de Juan, V.; Serrano-Macia, M.; et al. miR-873-5p targets mitochondrial GNMT-Complex II interface contributing to non-alcoholic fatty liver disease. Mol. Metab. 2019, 29, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Jampoka, K.; Muangpaisarn, P.; Khongnomnan, K.; Treeprasertsuk, S.; Tangkijvanich, P.; Payungporn, S. Serum miR-29a and miR-122 as Potential Biomarkers for Non-Alcoholic Fatty Liver Disease (NAFLD). Microrna 2018, 7, 215–222. [Google Scholar] [CrossRef]
- Sun, C.; Huang, F.; Liu, X.; Xiao, X.; Yang, M.; Hu, G.; Liu, H.; Liao, L. miR-21 regulates triglyceride and cholesterol metabolism in non-alcoholic fatty liver disease by targeting HMGCR. Int. J. Mol. Med. 2015, 35, 847–853. [Google Scholar] [CrossRef]
- Argentou, N.; Germanidis, G.; Hytiroglou, P.; Apostolou, E.; Vassiliadis, T.; Patsiaoura, K.; Sideras, P.; Germenis, A.E.; Speletas, M. TGF-beta signaling is activated in patients with chronic HBV infection and repressed by SMAD7 overexpression after successful antiviral treatment. Inflamm. Res. 2016, 65, 355–365. [Google Scholar] [CrossRef]
- Cui, J.; Wang, Y.; Xue, H. Long non-coding RNA GAS5 contributes to the progression of non-alcoholic fatty liver disease by targeting the microRNA-29a-3p/NOTCH2 axis. Bioengineered 2022, 13, 8370–8381. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natt, A.M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for non-alcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef]
- Zhao, Y.; Xia, J.; He, H.; Liang, S.; Zhang, H.; Gan, W. Diagnostic performance of novel inflammatory biomarkers based on ratios of laboratory indicators for non-alcoholic fatty liver disease. Front. Endocrinol. 2022, 13, 981196. [Google Scholar] [CrossRef]
- Amor, A.J.; Pinyol, M.; Solà, E.; Catalan, M.; Cofán, M.; Herreras, Z.; Amigó, N.; Gilabert, R.; Sala-Vila, A.; Ros, E.; et al. Relationship between noninvasive scores of nonalcoholic fatty liver disease and nuclear magnetic resonance lipoprotein abnormalities: A focus on atherogenic dyslipidemia. J. Clin. Lipidol. 2017, 11, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Deprince, A.; Haas, J.T.; Staels, B. Dysregulated lipid metabolism links NAFLD to cardiovascular disease. Mol. Metab. 2020, 42, 101092. [Google Scholar] [CrossRef]
- Zhang, Q.Q.; Lu, L.G. Non-alcoholic Fatty Liver Disease: Dyslipidemia, Risk for Cardiovascular Complications, and Treatment Strategy. J. Clin. Transl. Hepatol. 2015, 3, 78–84. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chan, C.; Lee, H.W.; Huang, C.; Chen, Y.J.; Liu, P.C.; Lu, S.N.; Chuang, W.L.; Huang, J.F.; Yu, M.L.; et al. Influence of Non-alcoholic Fatty Liver Disease with Increased Liver Enzyme Levels on the Risk of Cirrhosis and Hepatocellular Carcinoma. Clin. Gastroenterol. Hepatol. 2023, 21, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Men, X.; Liu, Y.; Jiang, H.; Bi, C.; Qu, Y.; Wang, K.; Wang, X.; Jing, J.; Liu, Y. Association with the plasma atherogenic index with hepatic steatosis and fibrosis in the US population. Medicine 2024, 103, e37152. [Google Scholar] [CrossRef]
- Han, M.H.; Lee, J.H.; Kim, G.; Lee, E.; Lee, Y.R.; Jang, S.Y.; Lee, H.W.; Chun, J.M.; Han, Y.S.; Yoon, J.S.; et al. Expression of the Long Noncoding RNA GAS5 Correlates with Liver Fibrosis in Patients with Non-alcoholic Fatty Liver Disease. Genes. 2020, 11, 545. [Google Scholar] [CrossRef]
- Xu, S.; Wang, Y.; Li, Z.; Hua, Q.; Jiang, M.; Fan, X. LncRNA GAS5 Knockdown Mitigates Hepatic Lipid Accumulation via Regulating MiR-26a-5p/PDE4B to Activate cAMP/CREB Pathway. Front. Endocrinol. 2022, 13, 889858. [Google Scholar] [CrossRef]
- Yang, Y.L.; Wang, P.W.; Wang, F.S.; Lin, H.Y.; Huang, Y.H. miR-29a Modulates GSK3β/SIRT1-Linked Mitochondrial Proteostatic Stress to Ameliorate Mouse Non-Alcoholic Steatohepatitis. Int. J. Mol. Sci. 2020, 21, 6884. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; Li, C.; Li, J.; Chen, X.; Zhong, X. Analysis of non-alcoholic fatty liver disease microRNA expression spectra in rat liver tissues. Mol. Med. Rep. 2018, 18, 2669–2680. [Google Scholar] [CrossRef] [PubMed]
- Rusu, I.; Pirlog, R.; Chiroi, P.; Nutu, A.; Puia, V.R.; Fetti, A.C.; Rusu, D.R.; Berindan-Neagoe, I.; Al Hajjar, N. The Implications of Noncoding RNAs in the Evolution and Progression of Non-alcoholic Fatty Liver Disease (NAFLD)-Related HCC. Int. J. Mol. Sci. 2022, 23, 12370. [Google Scholar] [CrossRef]
- Chen, T.; Meng, Y.; Zhou, Z.; Li, H.; Wan, L.; Kang, A.; Guo, W.; Ren, K.; Song, X.; Chen, Y.; et al. GAS5 protects against nonalcoholic fatty liver disease via miR-28a-5p/MARCH7/NLRP3 axis-mediated pyroptosis. Cell Death Differ. 2023, 30, 1829–1848. [Google Scholar] [CrossRef] [PubMed]
| Target | Forward Primer | Reverse Primer | Tm |
|---|---|---|---|
| GAS5 | 5′-GTGTCTCTCTCTCTCTCTCTCTT-3′ | 5′-CCTCTTCAGCAGTAGCATAGTT-3′ | 57.9 |
| NOTCH2 | 5′-GTGGCATACTGGGAGGAGAA -3′ | 5′-GATGGAGAAACCAGGGAACA-3′ | 57.9 |
| miR-29a-3p | 5′-GCACCGTCAAGGCTGAGAAC-3′ | 5′-CAGCCCATCGACTGGTG-3′ | 59 |
| GAPDH | 5′-ACCAGGAAATGAGCTTGACA-3′ | 5′-GACCACAGTCCATGCCATC-3′ | 57.6 |
| U6 | 5′-ACACGCACAAACGAGAAAGG-3′ | 5′-AGTGCAGGGTCCGAGGTATT-3′ | 59.5 |
| Parameters | NAFLD (No.: 38) | Controls (No.: 38) | p |
|---|---|---|---|
| Age, years | 45 ± 6.4 | 45.4 ± 6.8 | 0.83 |
| Sex (Female/Male) | 30/8 (78.9/21.1) | 28/10 (73.7/26.3) | 0.70 |
| BMI, Kg/m2 | 29.3 ± 2.6 | 25.8 ± 2.1 | <0.001 * |
| Total cholesterol, mg/dL | 169.3 ± 53.9 | 130.2 ± 35 | 0.012 * |
| Triglyceride, mg/dL | 98.7 ± 24.5 | 92.2 ± 20.5 | 0.44 |
| HDL-C, mg/dL | 46.8 ± 6.1 | 43.7 ± 5.4 | 0.10 |
| LDL-C, mg/dL | 103.1 ± 52.6 | 69.7 ± 31.8 | 0.023 * |
| AST (IU/L) | 52.7 ± 16.4 | 23.4 ±5.6 | <0.001 * |
| ALT (IU/L) | 69.5 ± 17 | 23.9 ± 7.5 | <0.001 * |
| LncRNA GAS5, fold change | 3.9 ± 0.95 | 1.04 ± 0.07 | <0.001 * |
| miR-29a-3p, fold change | 0.53 ± 0.21 | 0.99 ± 0.09 | <0.001 * |
| NOTCH2, fold change | 2.2 ± 0.63 | 1.03 ± 0.13 | <0.001 * |
| Marker | Cutoff (Fold Change) | Youden’s Index | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| LncRNA GAS5 | ≥1.16 | 0.95 | 100 | 94.7 | 95 | 100 | 97.4 |
| miR-29a-3p | ≤0.91 | 0.89 | 89.4 | 94.7 | 94.4 | 90 | 92.1 |
| NOTCH2 | ≥1.4 | 0.84 | 89.4 | 100 | 100 | 90.5 | 94.7 |
| Combined lncRNA GAS5, microRNA-29a-3p, and NOTCH2 | Same cutoffs | 0.95 | 94.7 | 100 | 100 | 95 | 97.4 |
| Parameters | LncRNA GAS5 | miR-29a-3p | NOTCH2 | |||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| Age | −0.02 | 0.89 | −0.01 | 0.94 | −0.03 | 0.86 |
| BMI | 0.62 | <0.001 * | −0.55 | <0.001 * | 0.42 | 0.009 * |
| Total cholesterol | 0.39 | 0.014 * | −0.17 | 0.28 | 0.31 | 0.06 |
| Triglyceride | 0.17 | 0.29 | −0.05 | 0.75 | 0.15 | 0.37 |
| HDL-C | 0.16 | 0.34 | −0.06 | 0.74 | −0.04 | 0.98 |
| LDL-C | 0.35 | 0.031 * | −0.14 | 0.41 | 0.28 | 0.09 |
| AST | 0.78 | <0.001 * | −0.69 | <0.001 * | 0.67 | <0.001 * |
| ALT | 0.82 | <0.001 * | −0.71 | <0.001 * | 0.68 | <0.001 * |
| miR-29a-3p | −0.91 | <0.001 * | 1 | - | −0.78 | <0.001 * |
| NOTCH2 | 0.8 | <0.001 * | −0.78 | <0.001 * | 1 | - |
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| OR (95% CI) | p | AOR (95% CI) | p | |
| BMI | 1.8 (1.2–2.6) | 0.001 * | 1.93 (0.91–4.1) | 0.09 |
| Total cholesterol | 1.02 (1–1.04) | 0.026 * | 1.02 (0.78–1.32) | 0.90 |
| LDL-C | 1.02 (1–1.04) | 0.039 * | 0.99 (0.73–1.35) | 0.95 |
| LncRNA GAS5 | 324 (18.7–5588) | <0.001 * | 162 (5.7–4629) | 0.003 * |
| miR-29a-3p | 0.01 (0.001–011) | <0.001 * | 0.1 (0–8.3) | 0.25 |
| NOTCH2 | 72 (9–573) | <0.001 * | 1.9 (0.02–159) | 0.76 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mobasher, M.A.; Alsirhani, A.M.; Alkhodair, S.A.; Abd-elhameed, A.; Baioumy, S.A.; Esawy, M.M.; Shabana, M.A. Investigating the Diagnostic Utility of LncRNA GAS5 in NAFLD Patients. Biomedicines 2025, 13, 1873. https://doi.org/10.3390/biomedicines13081873
Mobasher MA, Alsirhani AM, Alkhodair SA, Abd-elhameed A, Baioumy SA, Esawy MM, Shabana MA. Investigating the Diagnostic Utility of LncRNA GAS5 in NAFLD Patients. Biomedicines. 2025; 13(8):1873. https://doi.org/10.3390/biomedicines13081873
Chicago/Turabian StyleMobasher, Maysa A., Alaa Muqbil Alsirhani, Sahar Abdulrahman Alkhodair, Amir Abd-elhameed, Shereen A. Baioumy, Marwa M. Esawy, and Marwa A. Shabana. 2025. "Investigating the Diagnostic Utility of LncRNA GAS5 in NAFLD Patients" Biomedicines 13, no. 8: 1873. https://doi.org/10.3390/biomedicines13081873
APA StyleMobasher, M. A., Alsirhani, A. M., Alkhodair, S. A., Abd-elhameed, A., Baioumy, S. A., Esawy, M. M., & Shabana, M. A. (2025). Investigating the Diagnostic Utility of LncRNA GAS5 in NAFLD Patients. Biomedicines, 13(8), 1873. https://doi.org/10.3390/biomedicines13081873





