Investigation of Novel Therapeutic Targets for Rheumatoid Arthritis Through Human Plasma Proteome
Abstract
1. Introduction
2. Materials and Methods
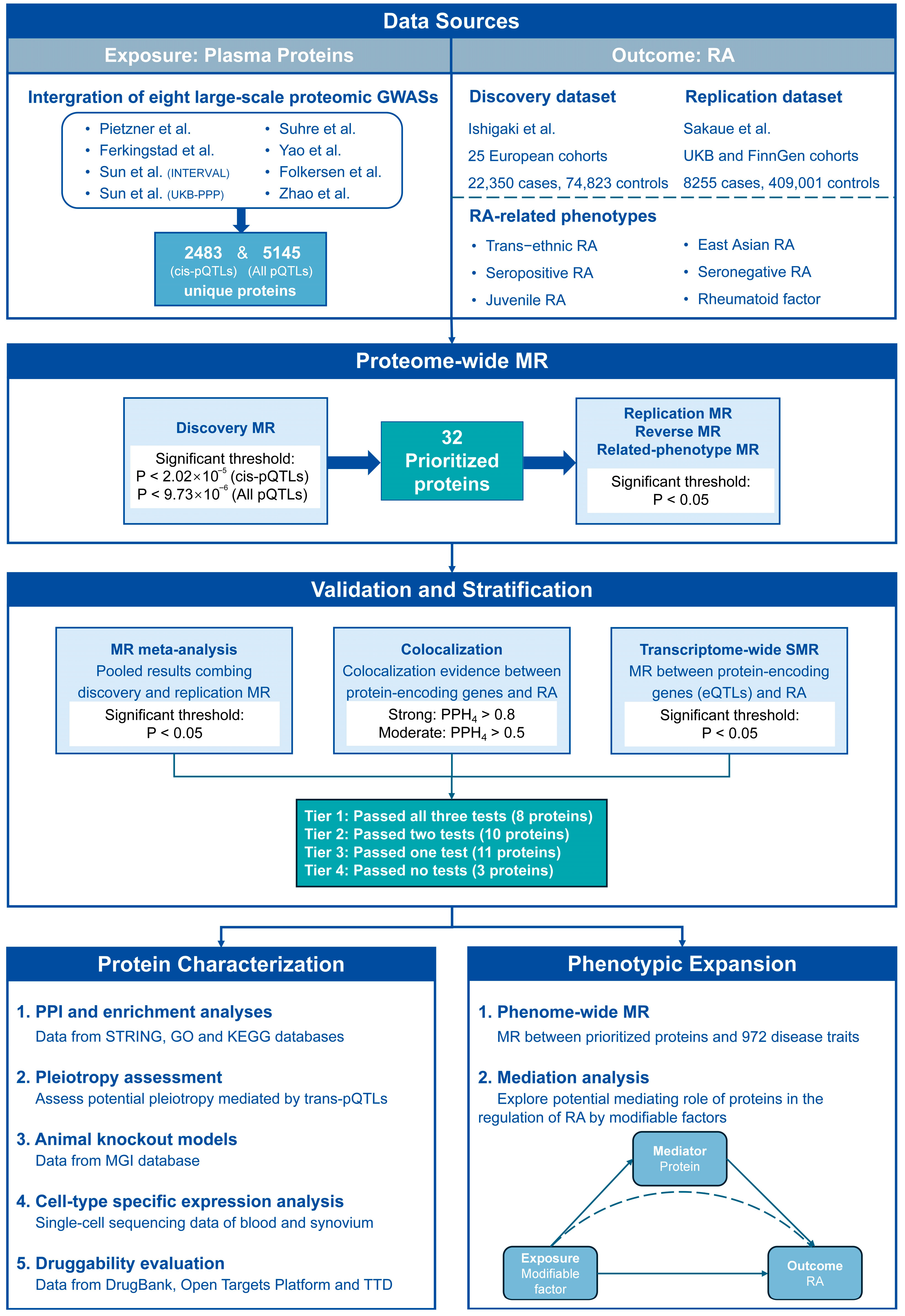
2.1. Data Sources of Plasma Proteins and RA
2.2. Pro-MR Analysis
2.3. Colocalization Analysis
2.4. Transcriptome-Wide Summary-Data-Based MR (SMR)
2.5. Reliability Stratification of Prioritized Proteins
2.6. Protein–Protein Interaction and Enrichment Analyses
2.7. Pleiotropy Assessment
2.8. Genetically Engineered Mouse Models
2.9. Cell-Type-Specific Expression Analysis
2.10. Druggability Evaluation
2.11. Phenome-Wide MR (Phe-MR) Analysis
2.12. Mediation Analysis
3. Results
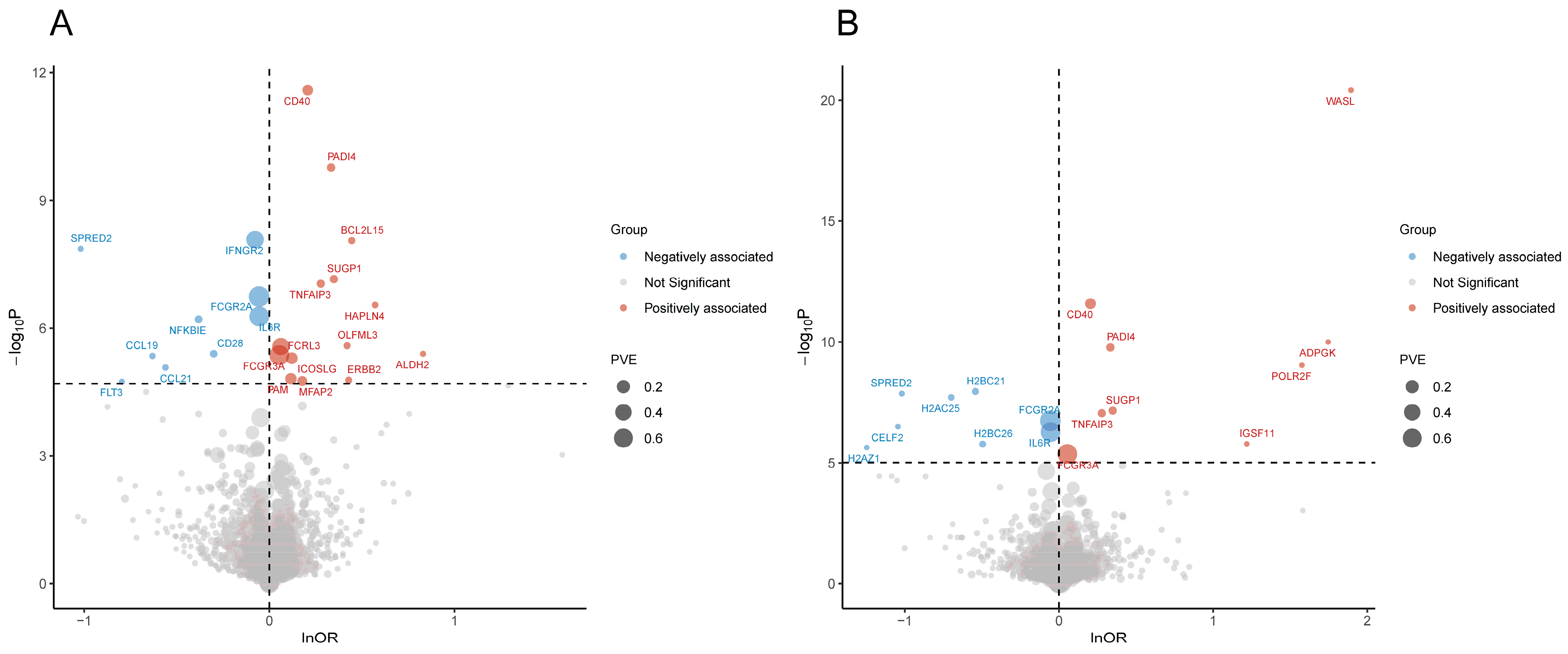
3.1. Pro-MR Prioritized 32 Plasma Proteins for RA
3.2. Colocalization Analysis Supported Causations Between 14 Prioritized Proteins and RA
3.3. SMR Validated 18 Prioritized Proteins of RA from Transcriptive Perspective
3.4. Four Reliability Tiers of Prioritized Proteins
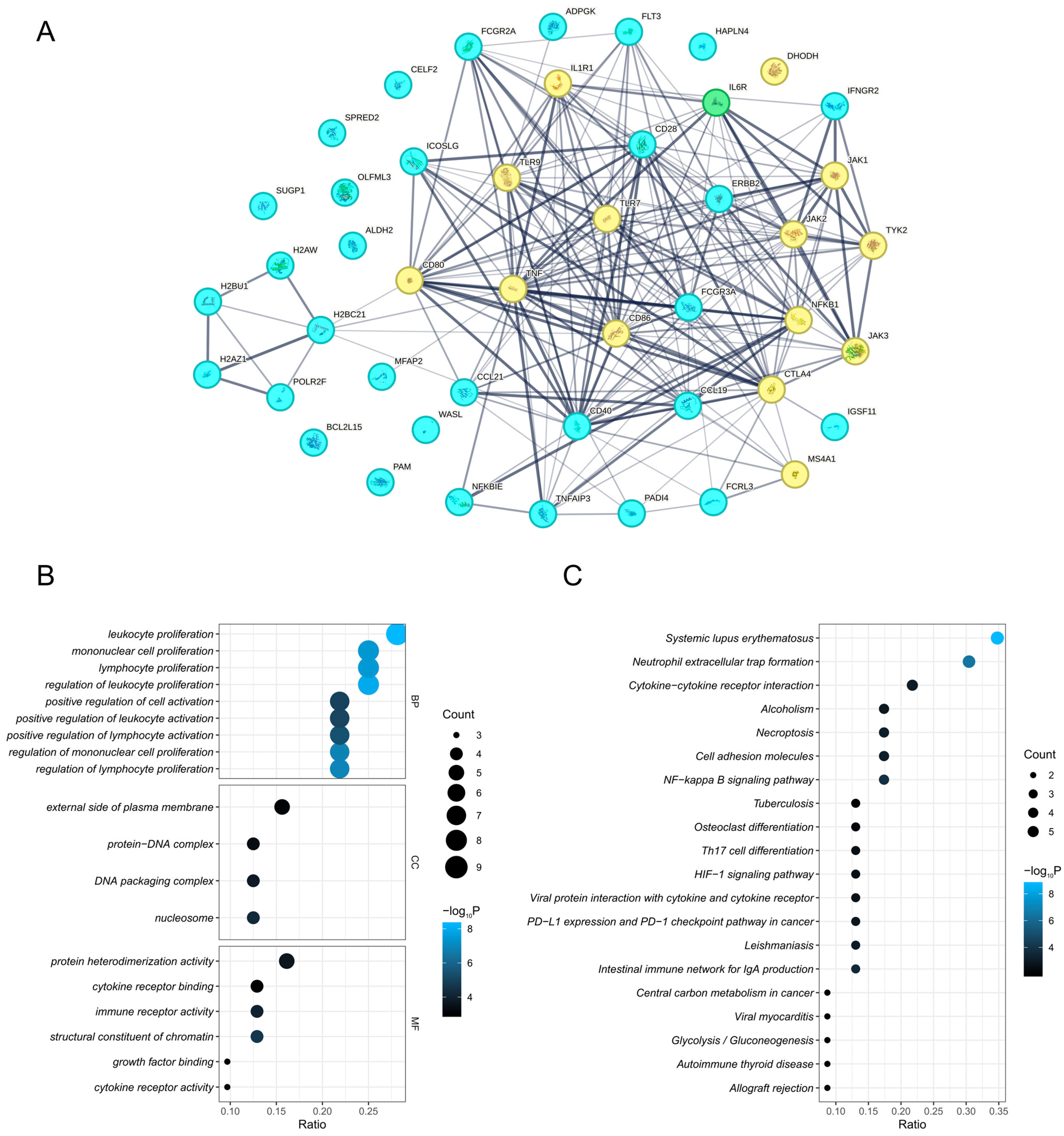
3.5. PPI Network and Enriched Pathways of Prioritized Proteins
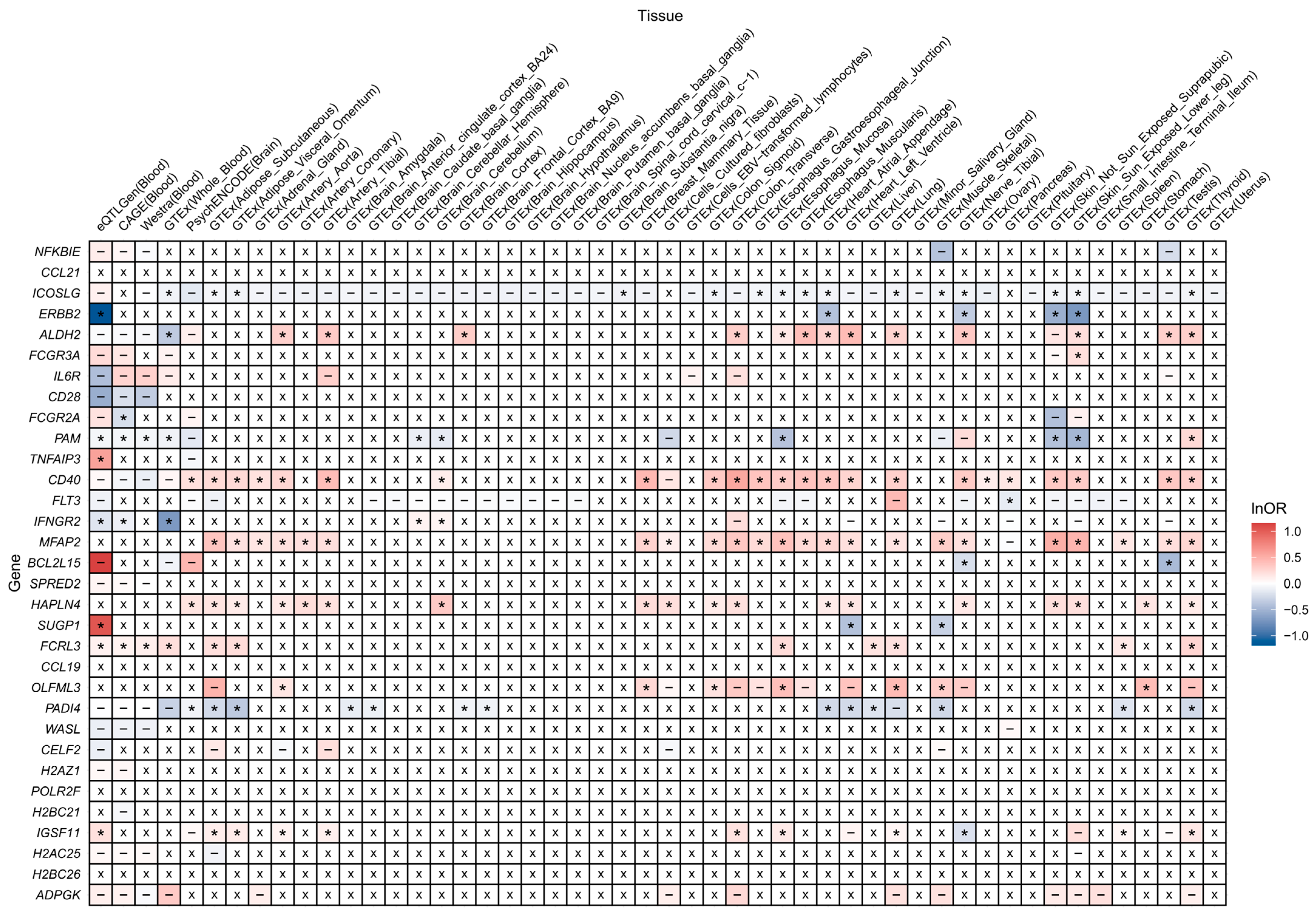
3.6. Four Trans-Associated Proteins Exhibited Vertical or Horizontal Pleiotropy
3.7. Genetically Engineered Mouse Models of Prioritized Protein-Coding Genes
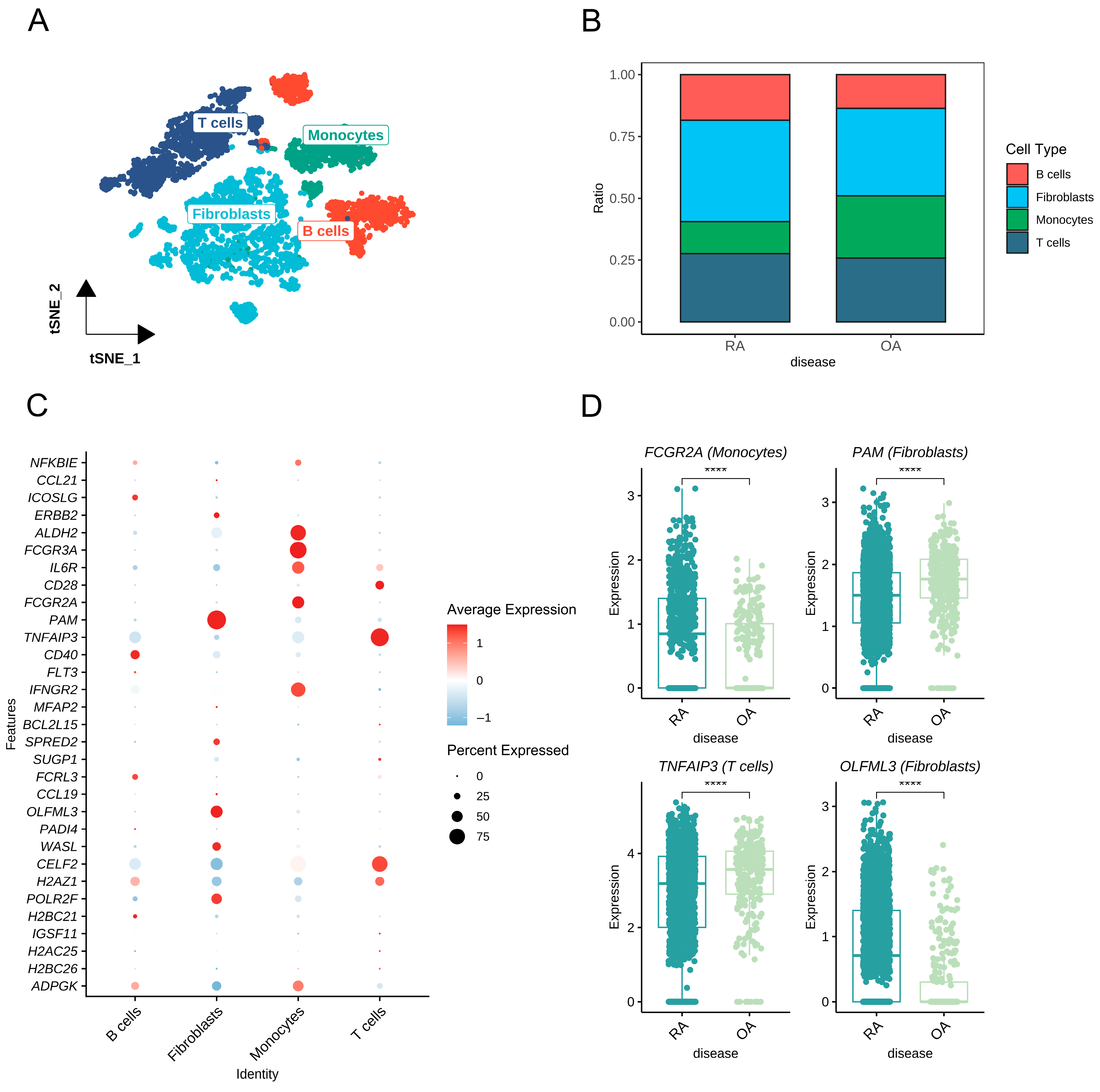
3.8. Cell-Type-Specific Expression of Prioritized Proteins in Synovium
3.9. Druggable Evidence for Nine Prioritized Proteins
3.10. Phe-MR Identified Potential Side Effects of Nine Prioritized Proteins
3.11. Six Prioritized Proteins Partially Mediate the Effects of Modifiable Factors on RA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RA | rheumatoid arthritis |
| DMARD | disease-modifying antirheumatic drug |
| SNP | single nucleotide polymorphism |
| GWAS | genome-wide association study |
| pQTL | protein quantitative trait locus |
| MR | Mendelian randomization |
| IVs | instrumental variables |
| Pro-MR | proteome-wide Mendelian randomization |
| LD | linkage disequilibrium |
| PPH4 | posterior probability for hypothesis 4 |
| SMR | summary-data-based Mendelian randomization |
| eQTL | expression quantitative trait locus |
| HEIDI | Heterogeneity in Dependent Instruments |
| PPI | protein–protein interaction |
| STRING | Search Tool for Recurring Instances of Neighboring Genes |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MGI | Mouse Genomics Informatics |
| scRNA-seq | single-cell RNA sequencing |
| TTD | Therapeutic Target Database |
| Phe-MR | phenome-wide Mendelian randomization |
| MVMR | multivariate Mendelian randomization |
| NFKB | nuclear factor kappa-B |
| ECM | extracellular matrix |
References
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- GBD 2021 Rheumatoid Arthritis Collaborators. Global, regional, and national burden of rheumatoid arthritis, 1990–2020, and projections to 2050: A systematic analysis of the global burden of disease study 2021. Lancet Rheumatol. 2023, 5, e594–e610. [Google Scholar] [CrossRef]
- Fazal, S.A.; Khan, M.; Nishi, S.E.; Alam, F.; Zarin, N.; Bari, M.T.; Ashraf, G.M. A clinical update and global economic burden of rheumatoid arthritis. Endocr. Metab. Immune Disord. Drug Targets 2018, 18, 98–109. [Google Scholar] [CrossRef]
- Brown, P.; Pratt, A.G.; Hyrich, K.L. Therapeutic advances in rheumatoid arthritis. BMJ 2024, 384, e070856. [Google Scholar] [CrossRef]
- Aletaha, D.; Smolen, J.S. Diagnosis and Management of Rheumatoid Arthritis: A Review. JAMA 2018, 320, 1360. [Google Scholar] [CrossRef]
- Suhre, K.; McCarthy, M.I.; Schwenk, J.M. Genetics meets proteomics: Perspectives for large population-based studies. Nat. Rev. Genet. 2021, 22, 19–37. [Google Scholar] [CrossRef]
- Zeng, T.; Tan, L.; Wu, Y.; Yu, J. 14-3-3η protein in rheumatoid arthritis: Promising diagnostic marker and independent risk factor for osteoporosis. Lab. Med. 2020, 51, 529–539. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, P.; Song, J.; Tang, Y.; Xie, Y.; Jin, X.; Zhu, D.; Fang, X.; Wei, C.; Li, R.; et al. Soluble CD24 is an inflammatory biomarker in early and seronegative rheumatoid arthritis. Ann. Med. 2023, 55, 2246370. [Google Scholar] [CrossRef]
- Ying, L.; Gong, L.; Meng, S.; Wu, X.; Li, M.; Li, Y. Circulating interleukin-39 as a potential biomarker for rheumatoid arthritis diagnosis. Clin. Biochem. 2023, 119, 110616. [Google Scholar] [CrossRef] [PubMed]
- Birney, E. Mendelian Randomization. Cold Spring Harb. Perspect. Med. 2022, 12, a041302. [Google Scholar] [CrossRef]
- Yuan, S.; Xu, F.; Li, X.; Chen, J.; Zheng, J.; Mantzoros, C.S.; Larsson, S.C. Plasma proteins and onset of type 2 diabetes and diabetic complications: Proteome-wide Mendelian randomization and colocalization analyses. Cell Rep. Med. 2023, 4, 101174. [Google Scholar] [CrossRef]
- Sun, J.; Zhao, J.; Jiang, F.; Wang, L.; Xiao, Q.; Han, F.; Chen, J.; Yuan, S.; Wei, J.; Larsson, S.C.; et al. Identification of novel protein biomarkers and drug targets for colorectal cancer by integrating human plasma proteome with genome. Genome Med. 2023, 15, 75. [Google Scholar] [CrossRef]
- Lin, J.; Zhou, J.; Xu, Y. Potential drug targets for multiple sclerosis identified through Mendelian randomization analysis. Brain J. Neurol. 2023, 146, 3364–3372. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Ren, F.; Song, P. Innovate therapeutic targets for autoimmune diseases: Insights from proteome-wide mendelian randomization and Bayesian colocalization. Autoimmunity 2024, 57, 2330392. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Liu, S.; Gong, W.; Guo, P.; Xue, F.; Zhou, X.; Wang, S.; Yuan, Z. Protein-centric omics integration analysis identifies candidate plasma proteins for multiple autoimmune diseases. Hum. Genet. 2023, 143, 1035–1048. [Google Scholar] [CrossRef]
- Pietzner, M.; Wheeler, E.; Carrasco-Zanini, J.; Cortes, A.; Koprulu, M.; Wörheide, M.A.; Oerton, E.; Cook, J.; Stewart, I.D.; Kerrison, N.D.; et al. Mapping the proteo-genomic convergence of human diseases. Science 2021, 374, eabj1541. [Google Scholar] [CrossRef]
- Ferkingstad, E.; Sulem, P.; Atlason, B.A.; Sveinbjornsson, G.; Magnusson, M.I.; Styrmisdottir, E.L.; Gunnarsdottir, K.; Helgason, A.; Oddsson, A.; Halldorsson, B.V.; et al. Large-scale integration of the plasma proteome with genetics and disease. Nat. Genet. 2021, 53, 1712–1721. [Google Scholar] [CrossRef]
- Sun, B.B.; Maranville, J.C.; Peters, J.E.; Stacey, D.; Staley, J.R.; Blackshaw, J.; Burgess, S.; Jiang, T.; Paige, E.; Surendran, P.; et al. Genomic atlas of the human plasma proteome. Nature 2018, 558, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.B.; Chiou, J.; Traylor, M.; Benner, C.; Hsu, Y.-H.; Richardson, T.G.; Surendran, P.; Mahajan, A.; Robins, C.; Vasquez-Grinnell, S.G.; et al. Plasma proteomic associations with genetics and health in the UK Biobank. Nature 2023, 622, 329–338. [Google Scholar] [CrossRef]
- Suhre, K.; Arnold, M.; Bhagwat, A.M.; Cotton, R.J.; Engelke, R.; Raffler, J.; Sarwath, H.; Thareja, G.; Wahl, A.; DeLisle, R.K.; et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017, 8, 14357. [Google Scholar] [CrossRef]
- Yao, C.; Chen, G.; Song, C.; Keefe, J.; Mendelson, M.; Huan, T.; Sun, B.B.; Laser, A.; Maranville, J.C.; Wu, H.; et al. Genome-wide mapping of plasma protein QTLs identifies putatively causal genes and pathways for cardiovascular disease. Nat. Commun. 2018, 9, 3268. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Gustafsson, S.; Wang, Q.; Hansen, D.H.; Hedman, Å.K.; Schork, A.; Page, K.; Zhernakova, D.V.; Wu, Y.; Peters, J.; et al. Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat. Metab. 2020, 2, 1135–1148. [Google Scholar] [CrossRef]
- Zhao, J.H.; Stacey, D.; Eriksson, N.; Macdonald-Dunlop, E.; Hedman, Å.K.; Kalnapenkis, A.; Enroth, S.; Cozzetto, D.; Digby-Bell, J.; Marten, J.; et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat. Immunol. 2023, 24, 1540–1551. [Google Scholar] [CrossRef]
- Ishigaki, K.; Sakaue, S.; Terao, C.; Luo, Y.; Sonehara, K.; Yamaguchi, K.; Amariuta, T.; Too, C.L.; Laufer, V.A.; Scott, I.C.; et al. Multi-ancestry genome-wide association analyses identify novel genetic mechanisms in rheumatoid arthritis. Nat. Genet. 2022, 54, 1640–1651. [Google Scholar] [CrossRef]
- Sakaue, S.; Kanai, M.; Tanigawa, Y.; Karjalainen, J.; Kurki, M.; Koshiba, S.; Narita, A.; Konuma, T.; Yamamoto, K.; Akiyama, M.; et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021, 53, 1415–1424. [Google Scholar] [CrossRef]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef]
- Giambartolomei, C.; Vukcevic, D.; Schadt, E.E.; Franke, L.; Hingorani, A.D.; Wallace, C.; Plagnol, V. Bayesian Test for Colocalisation between Pairs of Genetic Association Studies Using Summary Statistics. PLoS Genet. 2014, 10, e1004383. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, F.; Hu, H.; Bakshi, A.; Robinson, M.R.; Powell, J.E.; Montgomery, G.W.; Goddard, M.E.; Wray, N.R.; Visscher, P.M.; et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016, 48, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Rasooly, D.; Peloso, G.M.; Pereira, A.C.; Dashti, H.; Giambartolomei, C.; Wheeler, E.; Aung, N.; Ferolito, B.R.; Pietzner, M.; Farber-Eger, E.H.; et al. Genome-wide association analysis and Mendelian randomization proteomics identify drug targets for heart failure. Nat. Commun. 2023, 14, 3826. [Google Scholar] [CrossRef]
- Zhang, F.; Wei, K.; Slowikowski, K.; Fonseka, C.Y.; Rao, D.A.; Kelly, S.; Goodman, S.M.; Tabechian, D.; Hughes, L.B.; Salomon-Escoto, K.; et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019, 20, 928–942. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Della-Valle, V.; Roos-Weil, D.; Scourzic, L.; Mouly, E.; Aid, Z.; Darwiche, W.; Lecluse, Y.; Damm, F.; Mémet, S.; Mercher, T.; et al. Nfkbie-deficiency leads to increased susceptibility to develop B-cell lymphoproliferative disorders in aged mice. Blood Cancer J. 2020, 10, 38. [Google Scholar] [CrossRef]
- Myouzen, K.; Kochi, Y.; Okada, Y.; Terao, C.; Suzuki, A.; Ikari, K.; Tsunoda, T.; Takahashi, A.; Kubo, M.; Taniguchi, A.; et al. Functional variants in NFKBIE and RTKN2 involved in activation of the NF-κB pathway are associated with rheumatoid arthritis in Japanese. PLoS Genet. 2012, 8, e1002949. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.-C.; Song, G.G. FCGR2A, FCGR3A, FCGR3B polymorphisms and susceptibility to rheumatoid arthritis: A meta-analysis. Clin. Exp. Rheumatol. 2015, 33, 647–654. [Google Scholar] [PubMed]
- Umemura, T.; Ota, M.; Hamano, H.; Katsuyama, Y.; Kiyosawa, K.; Kawa, S. Genetic association of Fc receptor-like 3 polymorphisms with autoimmune pancreatitis in Japanese patients. Gut 2006, 55, 1367–1368. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Sun, Z.; Jiang, J.; Chang, X.; Shen, Y.; Gu, Y.; Liu, C. Inducible costimulator ligand (ICOSL) on CD19+ B cells is involved in immunopathological damage of rheumatoid arthritis (RA). Front. Immunol. 2022, 13, 1015831. [Google Scholar] [CrossRef] [PubMed]
- Román-Fernández, I.V.; García-Chagollán, M.; Cerpa-Cruz, S.; Jave-Suárez, L.F.; Palafox-Sánchez, C.A.; García-Arellano, S.; Sánchez-Zuno, G.A.; Muñoz-Valle, J.F. Assessment of CD40 and CD40L expression in rheumatoid arthritis patients, association with clinical features and DAS28. Clin. Exp. Med. 2019, 19, 427–437. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Chistiakov, A.P. Is FCRL3 a New General Autoimmunity Gene? Hum. Immunol. 2007, 68, 375–383. [Google Scholar] [CrossRef]
- Hamel, K.M.; Cao, Y.; Olalekan, S.A.; Finnegan, A. B cell-specific expression of inducible costimulator ligand is necessary for the induction of arthritis in mice. Arthritis Rheumatol. 2014, 66, 60–67. [Google Scholar] [CrossRef]
- Durie, F.H.; Fava, R.A.; Foy, T.M.; Aruffo, A.; Ledbetter, J.A.; Noelle, R.J. Prevention of collagen-induced arthritis with an antibody to gp39, the ligand for CD40. Science 1993, 261, 1328–1330. [Google Scholar] [CrossRef]
- Kochi, Y.; Myouzen, K.; Yamada, R.; Suzuki, A.; Kurosaki, T.; Nakamura, Y.; Yamamoto, K. FCRL3, an autoimmune susceptibility gene, has inhibitory potential on B-cell receptor-mediated signaling. J. Immunol. Baltim. 2009, 183, 5502–5510. [Google Scholar] [CrossRef]
- Espié, P.; He, Y.; Koo, P.; Sickert, D.; Dupuy, C.; Chokoté, E.; Schuler, R.; Mergentaler, H.; Ristov, J.; Milojevic, J.; et al. First-in-human clinical trial to assess pharmacokinetics, pharmacodynamics, safety, and tolerability of iscalimab, an anti-CD40 monoclonal antibody. Am. J. Transplant. 2020, 20, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Visvanathan, S.; Daniluk, S.; Ptaszyński, R.; Müller-Ladner, U.; Ramanujam, M.; Rosenstock, B.; Eleftheraki, A.G.; Vinisko, R.; Petříková, A.; Kellner, H.; et al. Effects of BI 655064, an antagonistic anti-CD40 antibody, on clinical and biomarker variables in patients with active rheumatoid arthritis: A randomised, double-blind, placebo-controlled, phase IIa study. Ann. Rheum. Dis. 2019, 78, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.E.; Amoura, Z.; Cheah, B.; Hiepe, F.; Sullivan, B.A.; Zhou, L.; Arnold, G.E.; Tsuji, W.H.; Merrill, J.T.; Chung, J.B. Brief Report: A Randomized, Double-Blind, Parallel-Group, Placebo-Controlled, Multiple-Dose Study to Evaluate AMG 557 in Patients with Systemic Lupus Erythematosus and Active Lupus Arthritis. Arthritis Rheumatol. 2018, 70, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Yamada, R.; Chang, X.; Tokuhiro, S.; Sawada, T.; Suzuki, M.; Nagasaki, M.; Nakayama-Hamada, M.; Kawaida, R.; Ono, M.; et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat. Genet. 2003, 34, 395–402. [Google Scholar] [CrossRef]
- Lee, Y.H.; Bae, S.-C. Association between susceptibility to rheumatoid arthritis and PADI4 polymorphisms: A meta-analysis. Clin. Rheumatol. 2016, 35, 961–971. [Google Scholar] [CrossRef]
- Wang, L.-S.; Wu, Z.-X. ALDH2 and Cancer Therapy. Adv. Exp. Med. Biol. 2019, 1193, 221–228. [Google Scholar] [CrossRef]
- Matsuo, K.; Xiang, Y.; Nakamura, H.; Masuko, K.; Yudoh, K.; Noyori, K.; Nishioka, K.; Saito, T.; Kato, T. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res. Ther. 2006, 8, R175. [Google Scholar] [CrossRef]
- Cheng, K.-C.; Wu, Y.-J.; Cheng, K.-H.; Cheng, K.-Y.; Chen, K.-J.; Wu, W.-C.; Lee, P.-Y.; Chang, C.-H. Autoantibody against aldehyde dehydrogenase 2 could be a biomarker to monitor progression of Graves’ orbitopathy. Graefe’s Arch. Clin. Exp. Ophthalmol. 2018, 256, 1195–1201. [Google Scholar] [CrossRef]
- Chen, C.-H.; Budas, G.R.; Churchill, E.N.; Disatnik, M.-H.; Hurley, T.D.; Mochly-Rosen, D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science 2008, 321, 1493–1495. [Google Scholar] [CrossRef]
- Correale, M.; Brunetti, N.D. Alcohol consumption and subclinical and clinical coronary heart disease: New insight into potential causal mechanisms. Eur. J. Prev. Cardiol. 2022, 29, 2003–2005. [Google Scholar] [CrossRef]
- Zhang, Z.-D.; Shi, C.-R.; Li, F.-X.; Gan, H.; Wei, Y.; Zhang, Q.; Shuai, X.; Chen, M.; Lin, Y.-L.; Xiong, T.-C.; et al. Disulfiram ameliorates STING/MITA-dependent inflammation and autoimmunity by targeting RNF115. Cell. Mol. Immunol. 2024, 21, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Ye, L.; Bennett, S.; Xu, H.; He, D.; Xu, J. Molecular structure and function of microfibrillar-associated proteins in skeletal and metabolic disorders and cancers. J. Cell. Physiol. 2021, 236, 41–48. [Google Scholar] [CrossRef]
- Craft, C.S.; Pietka, T.A.; Schappe, T.; Coleman, T.; Combs, M.D.; Klein, S.; Abumrad, N.A.; Mecham, R.P. The extracellular matrix protein MAGP1 supports thermogenesis and protects against obesity and diabetes through regulation of TGF-β. Diabetes 2014, 63, 1920–1932. [Google Scholar] [CrossRef]
- Chmelova, M.; Androvic, P.; Kirdajova, D.; Tureckova, J.; Kriska, J.; Valihrach, L.; Anderova, M.; Vargova, L. A view of the genetic and proteomic profile of extracellular matrix molecules in aging and stroke. Front. Cell. Neurosci. 2023, 17, 1296455. [Google Scholar] [CrossRef]
- Bekku, Y.; Saito, M.; Moser, M.; Fuchigami, M.; Maehara, A.; Nakayama, M.; Kusachi, S.; Ninomiya, Y.; Oohashi, T. Bral2 is indispensable for the proper localization of brevican and the structural integrity of the perineuronal net in the brainstem and cerebellum. J. Comp. Neurol. 2012, 520, 1721–1736. [Google Scholar] [CrossRef]
- Li, X.; Shen, A.; Zhao, Y.; Xia, J. Mendelian Randomization Using the Druggable Genome Reveals Genetically Supported Drug Targets for Psychiatric Disorders. Schizophr. Bull. 2023, 49, 1305–1315. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Yu, C.-Y.; Theusch, E.; Naidoo, D.; Stevens, K.; Kuang, Y.-L.; Schuetz, E.; Chaudhry, A.S.; Medina, M.W. SUGP1 is a novel regulator of cholesterol metabolism. Hum. Mol. Genet. 2016, 25, 3106–3116. [Google Scholar] [CrossRef]
- Myasoedova, E.; Crowson, C.S.; Kremers, H.M.; Roger, V.L.; Fitz-Gibbon, P.D.; Therneau, T.M.; Gabriel, S.E. Lipid paradox in rheumatoid arthritis: The impact of serum lipid measures and systemic inflammation on the risk of cardiovascular disease. Ann. Rheum. Dis. 2011, 70, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Bruhns, P.; Iannascoli, B.; England, P.; Mancardi, D.A.; Fernandez, N.; Jorieux, S.; Daëron, M. Specificity and affinity of human fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009, 113, 3716–3725. [Google Scholar] [CrossRef] [PubMed]
- Rose-John, S.; Jenkins, B.J.; Garbers, C.; Moll, J.M.; Scheller, J. Targeting IL-6 trans-signalling: Past, present and future prospects. Nat. Rev. Immunol. 2023, 23, 666–681. [Google Scholar] [CrossRef] [PubMed]
- Traylor, M.; Knevel, R.; Cui, J.; Taylor, J.; Harm-Jan, W.; Conaghan, P.G.; Cope, A.P.; Curtis, C.; Emery, P.; Newhouse, S.; et al. Genetic associations with radiological damage in rheumatoid arthritis: Meta-analysis of seven genome-wide association studies of 2,775 cases. PLoS ONE 2019, 14, e0223246. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Raygoza Garay, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Chen, J.; Spracklen, C.N.; Marenne, G.; Varshney, A.; Corbin, L.J.; Luan, J.; Willems, S.M.; Wu, Y.; Zhang, X.; Horikoshi, M.; et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 2021, 53, 840–860. [Google Scholar] [CrossRef]
- Strawbridge, R.J.; Dupuis, J.; Prokopenko, I.; Barker, A.; Ahlqvist, E.; Rybin, D.; Petrie, J.R.; Travers, M.E.; Bouatia-Naji, N.; Dimas, A.S.; et al. Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 2011, 60, 2624–2634. [Google Scholar] [CrossRef]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Evidence for causal effects of lifetime smoking on risk for depression and schizophrenia: A mendelian randomisation study. Psychol. Med. 2020, 50, 2435–2443. [Google Scholar] [CrossRef]
- Jansen, P.R.; Watanabe, K.; Stringer, S.; Skene, N.; Bryois, J.; Hammerschlag, A.R.; de Leeuw, C.A.; Benjamins, J.S.; Muñoz-Manchado, A.B.; Nagel, M.; et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat. Genet. 2019, 51, 394–403. [Google Scholar] [CrossRef]
- van de Vegte, Y.J.; Said, M.A.; Rienstra, M.; van der Harst, P.; Verweij, N. Genome-wide association studies and mendelian randomization analyses for leisure sedentary behaviours. Nat. Commun. 2020, 11, 1770. [Google Scholar] [CrossRef] [PubMed]
- Merino, J.; Dashti, H.S.; Sarnowski, C.; Lane, J.M.; Todorov, P.V.; Udler, M.S.; Song, Y.; Wang, H.; Kim, J.; Tucker, C.; et al. Genetic analysis of dietary intake identifies new loci and functional links with metabolic traits. Nat. Hum. Behav. 2022, 6, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhao, J.; Zhou, S.; Li, X.; Li, T.; Wang, L.; Yuan, S.; Chen, D.; Law, P.J.; Larsson, S.C.; et al. Systematic investigation of genetically determined plasma and urinary metabolites to discover potential interventional targets for colorectal cancer. J. Natl. Cancer Inst. 2024, 116, 1303–1312. [Google Scholar] [CrossRef] [PubMed]

| Protein Short Name a | Uniprot ID | Full Name | MR Meta-Analysis | Colocalization | SMR | Tier | |
|---|---|---|---|---|---|---|---|
| OR | p Value | ||||||
| NFKBIE | O00221 | NF-kappa-B inhibitor epsilon | 0.747 | 3.560 × 10−17 | + | − | 2 |
| CCL21 | O00585 | C-C motif chemokine 21 | 0.654 | 9.605 × 10−6 | − | − | 3 |
| ICOSLG | O75144 | ICOS ligand | 1.095 | 1.286 × 10−5 | + | + | 1 |
| ERBB2 | P04626 | Receptor tyrosine-protein kinase erbB-2 | 1.320 | 5.989 × 10−2 | − | + | 3 |
| ALDH2 | P05091 | Aldehyde dehydrogenase, mitochondrial | 1.804 | 3.840 × 10−6 | + | + | 1 |
| FCGR3A | P08637 | Low-affinity immunoglobulin gamma Fc region receptor III-A | 1.052 | 7.118 × 10−10 | − | + | 2 |
| IL6R | P08887 | Interleukin-6 receptor subunit alpha | 0.958 | 2.113 × 10−8 | + | − | 2 |
| CD28 | P10747 | T cell-specific surface glycoprotein CD28 | 0.795 | 5.446 × 10−6 | − | − | 3 |
| FCGR2A | P12318 | Low-affinity immunoglobulin gamma Fc region receptor II-a | 0.955 | 2.933 × 10−9 | + | + | 1 |
| PAM | P19021 | Peptidyl-glycine alpha-amidating monooxygenase | 1.054 | 4.011 × 10−1 | − | + | 3 |
| TNFAIP3 | P21580 | Tumor necrosis factor alpha-induced protein 3 | 1.275 | 2.751 × 10−10 | − | + | 2 |
| CD40 | P25942 | Tumor necrosis factor receptor superfamily member 5 | 1.174 (cis-pQTL) 1.191 (all pQTLs) | 7.047 × 10−4 (cis-pQTL) 1.007 × 10−12 (all pQTLs) | + | + | 1 |
| FLT3 | P36888 | Receptor-type tyrosine-protein kinase FLT3 | 0.602 | 8.046 × 10−2 | − | + | 3 |
| IFNGR2 | P38484 | Interferon gamma receptor 2 | 0.943 | 9.349 × 10−4 | + | + | 1 |
| MFAP2 | P55001 | Microfibrillar-associated protein 2 | 1.172 | 2.050 × 10−9 | − | + | 2 |
| BCL2L15 | Q5TBC7 | Bcl-2-like protein 15 | 1.290 | 1.829 × 10−1 | − | + | 3 |
| SPRED2 | Q7Z698 | Sprouty-related, EVH1 domain-containing protein 2 | 0.474 | 7.533 × 10−3 | − | − | 3 |
| HAPLN4 | Q86UW8 | Hyaluronan and proteoglycan link protein 4 | 1.478 | 4.386 × 10−2 | + | + | 1 |
| SUGP1 | Q8IWZ8 | SURP and G-patch domain-containing protein 1 | 1.252 | 7.287 × 10−2 | + | + | 2 |
| FCRL3 | Q96P31 | Fc receptor-like protein 3 | 1.055 | 1.256 × 10−8 | + | + | 1 |
| CCL19 | Q99731 | C-C motif chemokine 19 | 0.618 | 3.105 × 10−6 | − | − | 3 |
| OLFML3 | Q9NRN5 | Olfactomedin-like protein 3 | 1.230 | 3.293 × 10−1 | − | + | 3 |
| PADI4 | Q9UM07 | Protein-arginine deiminase type-4 | 1.298 | 2.813 × 10−4 | + | + | 1 |
| WASL | O00401 | Actin nucleation-promoting factor WASL | 4.424 | 2.875 × 10−4 | + | − | 2 |
| CELF2 | O95319 | CUGBP Elav-like family member 2 | 0.565 | 2.405 × 10−1 | − | − | 4 |
| H2AZ1 | P0C0S5 | Histone H2A.Z | 0.491 | 1.975 × 10−1 | − | − | 4 |
| POLR2F | P61218 | DNA-directed RNA polymerases I, II, and III subunit RPABC2 | 4.250 | 1.693 × 10−13 | + | − | 2 |
| H2BC21 | Q16778 | Histone H2B type 2-E | 0.635 | 3.379 × 10−8 | − | − | 3 |
| IGSF11 | Q5DX21 | Immunoglobulin superfamily member 11 | 2.812 | 1.081 × 10−7 | − | + | 2 |
| H2AC25 | Q7L7L0 | Histone H2A type 3 | 0.646 | 1.039 × 10−1 | − | − | 4 |
| H2BC26 | Q8N257 | Histone H2B type 3-B | 0.654 | 3.602 × 10−6 | − | − | 3 |
| ADPGK | Q9BRR6 | ADP-dependent glucokinase | 4.840 | 3.853 × 10−14 | + | − | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Huang, C.; Huang, K.; Wu, T.; Liu, H. Investigation of Novel Therapeutic Targets for Rheumatoid Arthritis Through Human Plasma Proteome. Biomedicines 2025, 13, 1841. https://doi.org/10.3390/biomedicines13081841
Wang H, Huang C, Huang K, Wu T, Liu H. Investigation of Novel Therapeutic Targets for Rheumatoid Arthritis Through Human Plasma Proteome. Biomedicines. 2025; 13(8):1841. https://doi.org/10.3390/biomedicines13081841
Chicago/Turabian StyleWang, Hong, Chengyi Huang, Kangkang Huang, Tingkui Wu, and Hao Liu. 2025. "Investigation of Novel Therapeutic Targets for Rheumatoid Arthritis Through Human Plasma Proteome" Biomedicines 13, no. 8: 1841. https://doi.org/10.3390/biomedicines13081841
APA StyleWang, H., Huang, C., Huang, K., Wu, T., & Liu, H. (2025). Investigation of Novel Therapeutic Targets for Rheumatoid Arthritis Through Human Plasma Proteome. Biomedicines, 13(8), 1841. https://doi.org/10.3390/biomedicines13081841





