Duodenal Adenocarcinoma Is Characterized by Acidity, High Infiltration of Macrophage, and Activated Linc01559–GRSF1 Axis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Cell Culture
2.2. RNA-Seq
2.3. Enrichment Analysis of Functional Pathways
2.4. Immune Infiltration Analysis
2.5. RNA Pulldown
2.6. RNA-Binding Protein Immunoprecipitation (RIP)
2.7. RT-qPCR
2.8. Cell Proliferation
2.9. Colony Formation
2.10. Cell Migration and Invasion Assays
2.11. Apoptosis
2.12. Western Blotting
2.13. Spheroid Formation Assay
2.14. Construction of GRSF1 Overexpression Cell
3. Results
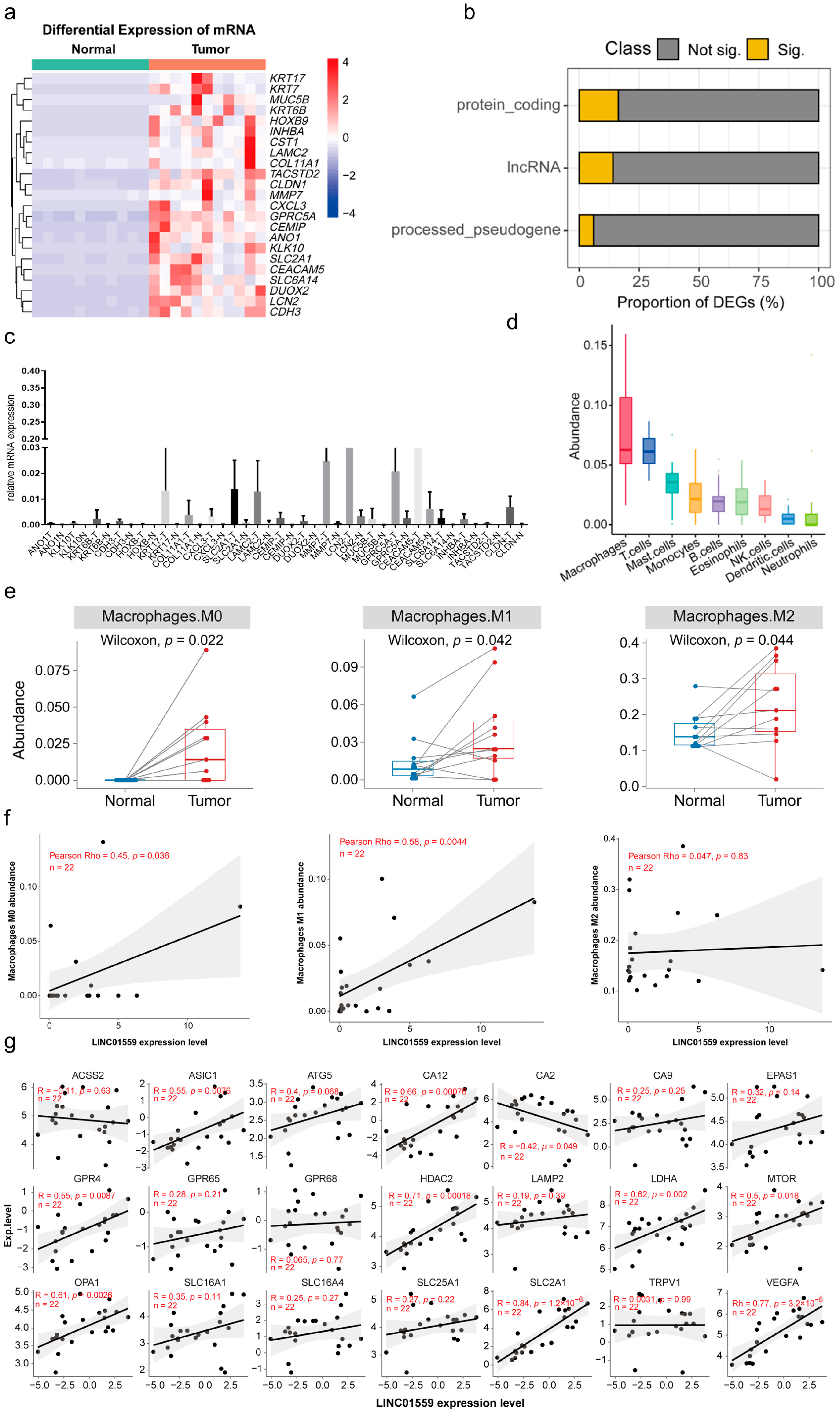
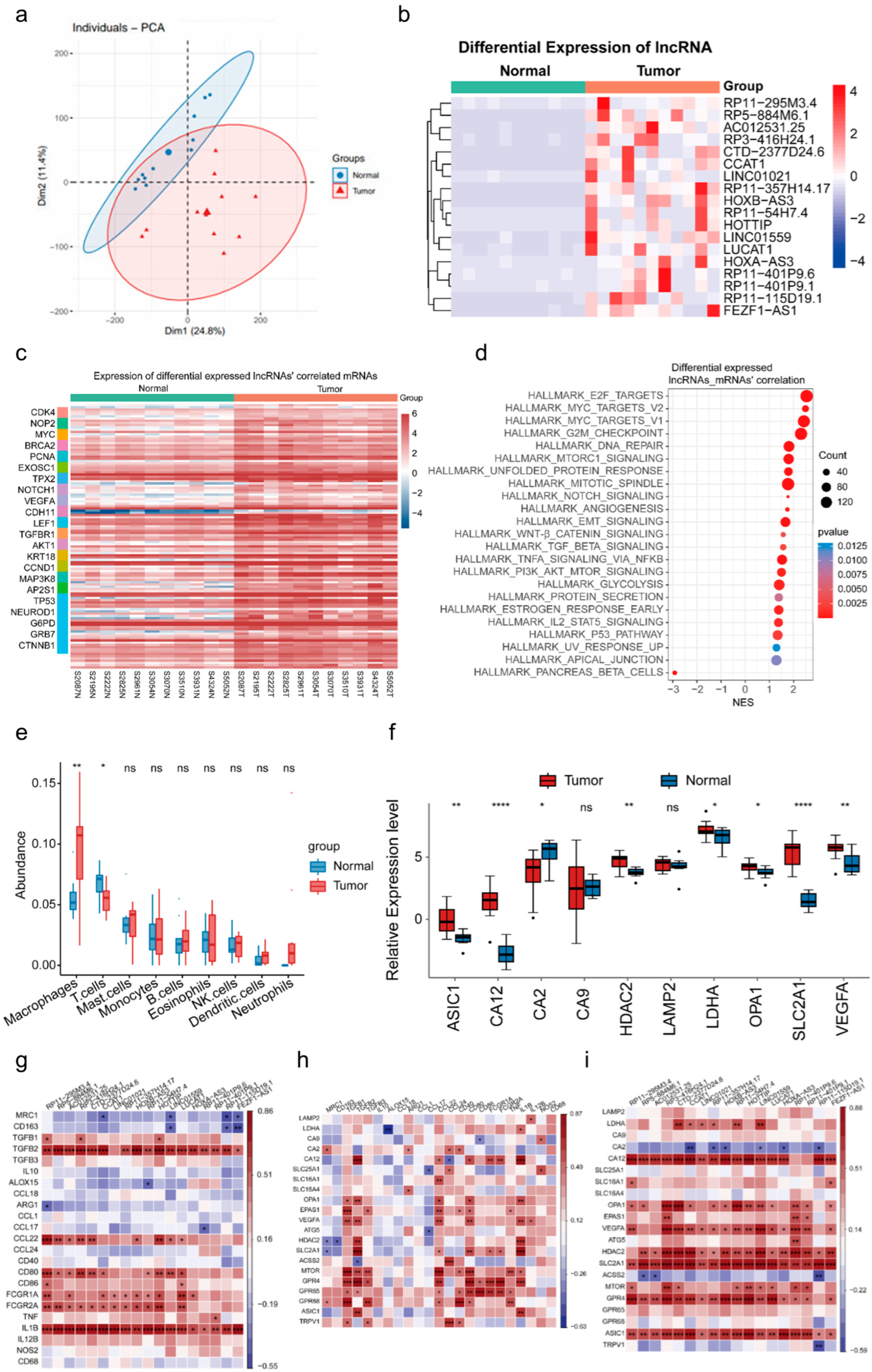
3.1. Macrophage Infiltration and Significant lncRNA Alterations in Duodenal Adenocarcinoma Under an Acidic Environment
3.2. Linc01559 Is Upregulated in Duodenal Adenocarcinoma and Is Correlated with Acidic Environment Markers
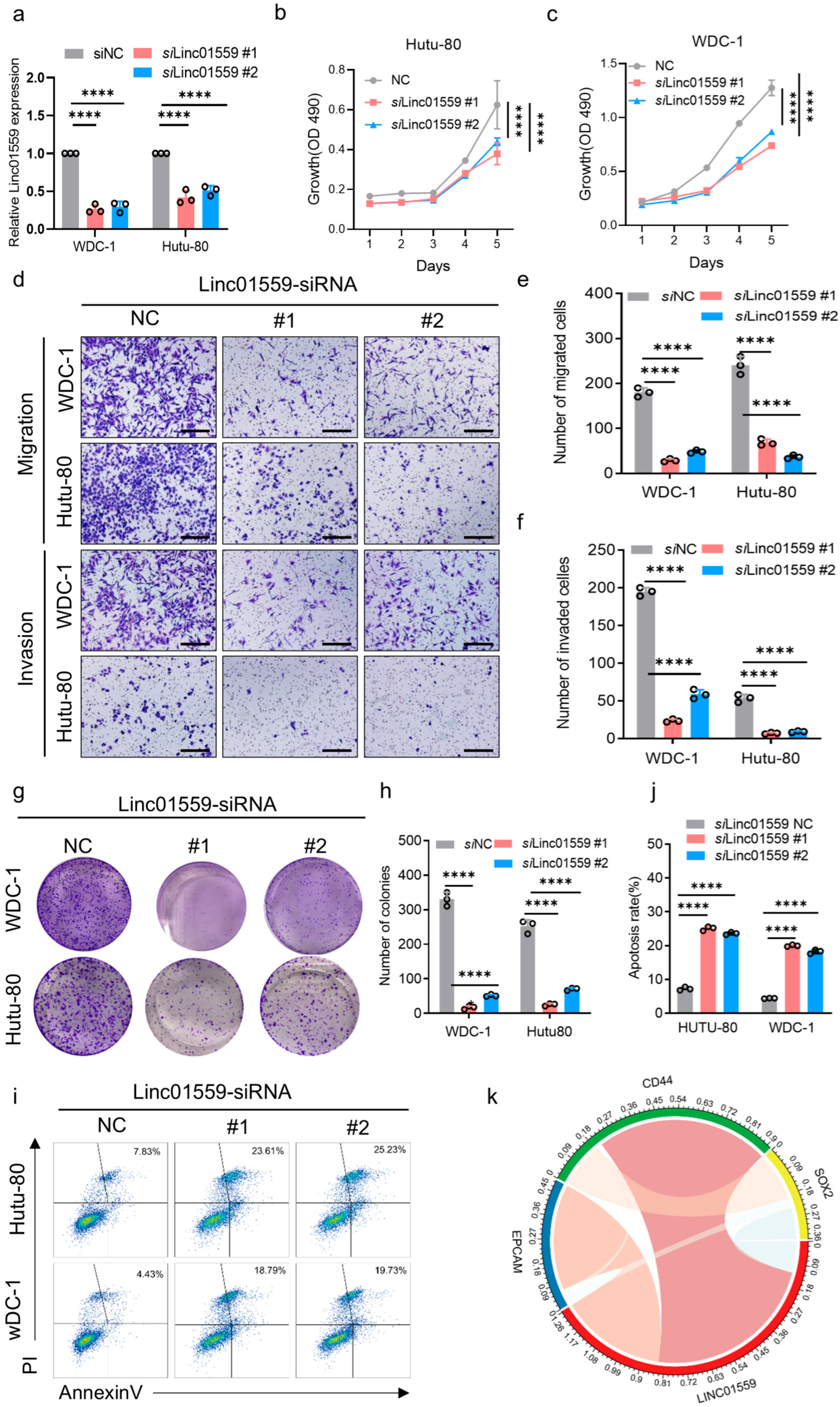
3.3. Linc01559 Knockdown Inhibits the Invasion, Migration, and Tumor Stem Cell Phenotype of Duodenal Adenocarcinoma
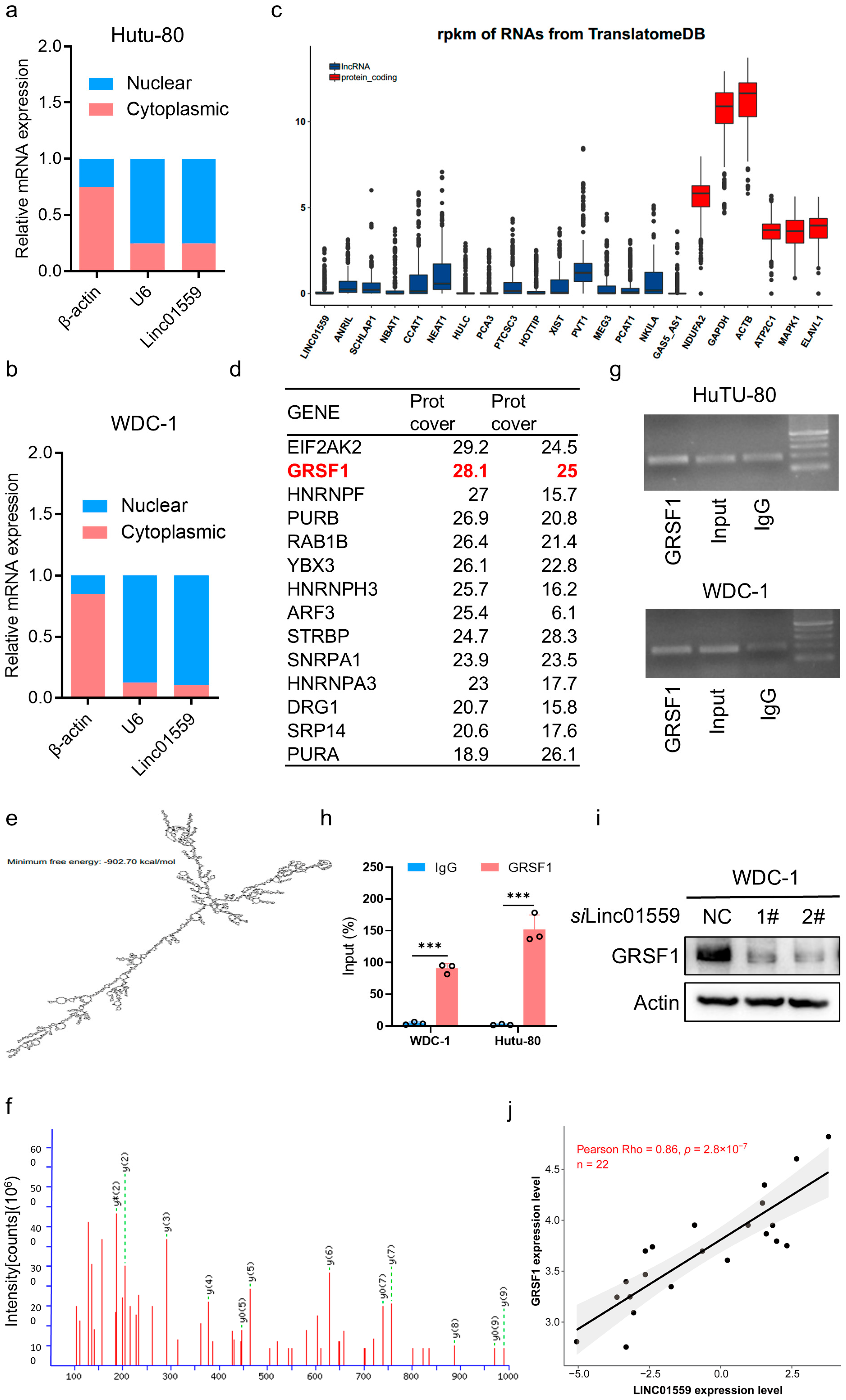
3.4. GRSF1 Is a Target Protein That Interacts with Linc01559
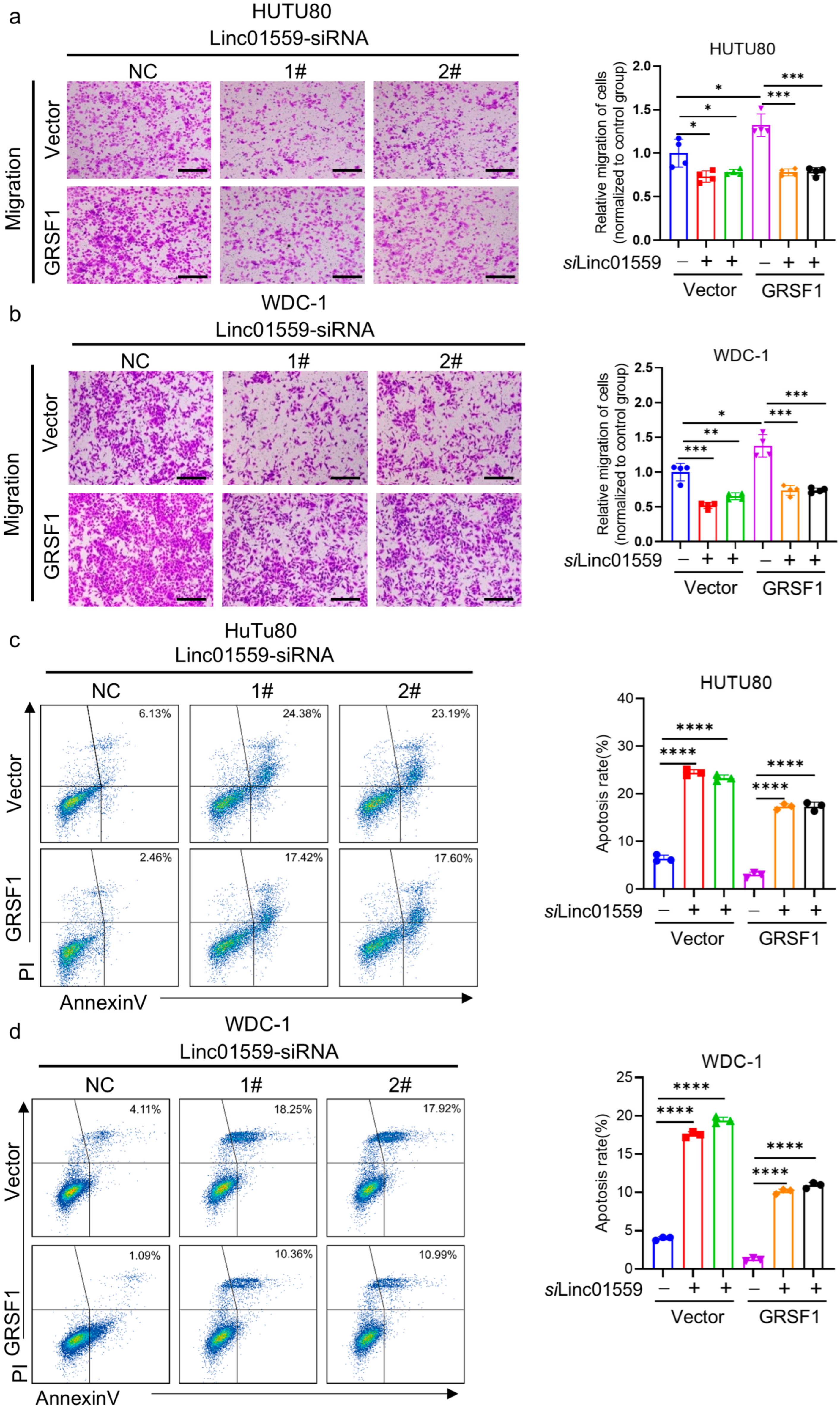
3.5. GRSF1 Is Highly Expressed in Acidic Environments and the Linc01559–GRSF1 Axis Regulates the Tumor Stem Cell Phenotype and Metastasis
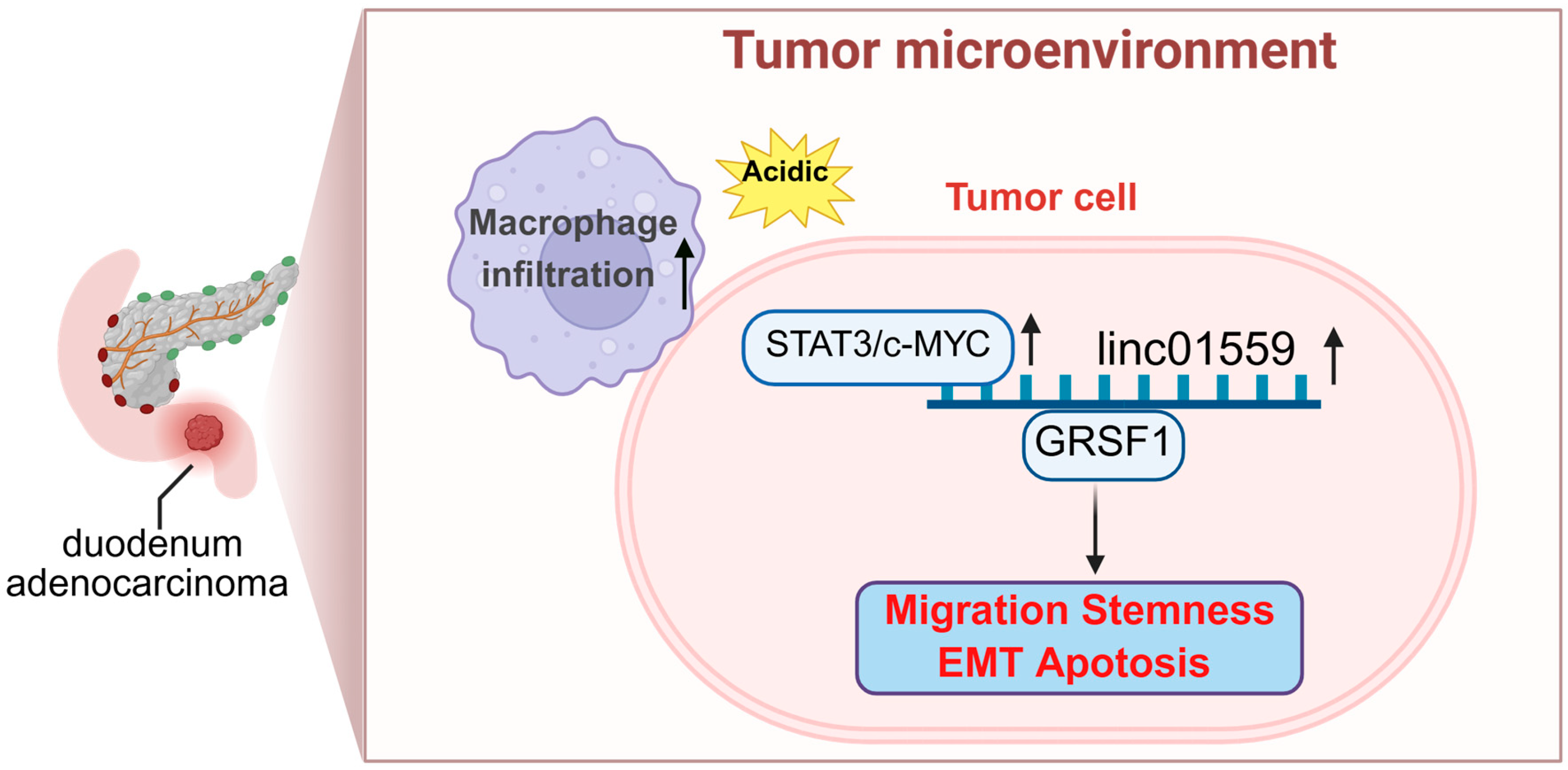
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DA | Duodenal adenocarcinoma |
| TME | tumor microenvironment |
| RNA-seq | RNA sequencing |
| LncRNA | long non-coding RNAs |
| GRSF1 | G-rich sequence binding factor 1 |
| STAT3 | Signal transducer and activator of transcription 3 |
| c-MYC | bHLH transcription factor |
| TAMs | tumor-associated macrophages |
| NSCLC | non-small cell lung cancer |
| EMT | epithelial-mesenchymal transition |
| PCA | principal component analysis |
| GSEA | Gene Set Enrichment Analysis |
| HIF1α | hypoxia inducible factor 1 subunit α |
| EPCAM | epithelial cell adhesion molecule |
| KRT | keratin |
| MUC | mucin |
| UBE2K | ubiquitin-conjugating enzyme E2 K |
| XRCC5 | X-ray repair cross complementing 5 |
| SNRPD3 | small nuclear ribonucleoprotein D3 polypeptide |
| CDC123 | cell division cycle 123 |
Appendix A
| Name | Forward | Reverse |
|---|---|---|
| Primer Sequences for qRT-PCR | ||
| ANO1 | 5′-CTGATGCCGAGTGCAAGTATG-3 | 5′-AGGGCCTCTTGTGATGGTACA-3′ |
| KLK10 | 5′-CAAGGCGAACGGATGAGCA-3′ | 5′-GAGCACAGCGGTAGGGAAG-3′ |
| KRT6B | 5′-AGATTGCTCAGAGGAGCAGG-3′ | 5′-TGTAGGTTGGCACACTGCTT-3′ |
| CDH3 | 5′-ATCATCGTGACCGACCAGAAT-3′ | 5′-GACTCCCTCTAAGACACTCCC-3′ |
| HOXB | 5′-AGGAGACGGAGGCTATTTTCA-3′ | 5′-GTCTGCTCGTTCCCATAAGGG-3 |
| KRT17 | 5′-GGTGGGTGGTGAGATCAATGT -3′ | 5′-CGCGGTTCAGTTCCTCTGTC -3′ |
| COL11A1 | 5′-ACCCTCGCATTGACCTTCC -3′ | 5′-TTTGTGCAAAATCCCGTTGTTT-3′ |
| CXCL3 | 5′-CGCCCAAACCGAAGTCATAG-3′ | 5′-GCTCCCCTTGTTCAGTATCTTTT-3′ |
| SLC2A1 | 5′-GGCCAAGAGTGTGCTAAAGAA-3′ | 5′-ACAGCGTTGATGCCAGACAG-3′ |
| LAMC2 | 5′-GACAAACTGGTAATGGATTCCGC-3′ | 5′-TTCTCTGTGCCGGTAAAAGCC-3′ |
| CEMIP | 5′-CACGGTCTATTCCATCCACATC-3′ | 5′-GGTTCGCAAAACAATCGGCT-3′ |
| DUOX2 | 5′-CTGGGTCCATCGGGCAATC-3′ | 5′-GTCGGCGTAATTGGCTGGTA-3′ |
| MMP7 | 5′-GAGTGAGCTACAGTGGGAACA-3′ | 5′-CTATGACGCGGGAGTTTAACAT-3′ |
| LCN2 | 5′-CCACCTCAGACCTGATCCCA-3′ | 5′-CCCCTGGAATTGGTTGTCCTG-3′ |
| MUC5B | 5′-GCCTACGAGGACTTCAACGTC-3′ | 5′-CCTTGATGACAACACGGGTGA-3′ |
| GPRC5A | 5′-ATGGCTACAACAGTCCCTGAT-3′ | 5′-CCACCGTTTCTAGGACGATGC-3′ |
| CEACAM5 | 5′-CTGTCCAATGACAACAGGACC-3′ | 5′-ACGGTAATAGGTGTATGAGGGG-3′ |
| SLC6A14 | 5′-ACCGTGGTAACTGGTCCAAAA-3′ | 5′-CGCCTCCACCATTGCTGTAG-3′ |
| INHBA | 5′-CCTCCCAAAGGATGTACCCAA-3′ | 5′-CTCTATCTCCACATACCCGTTCT-3′ |
| TACSTD2 | 5′-ACAACGATGGCCTCTACGAC-3′ | 5′-GTCCAGGTCTGAGTGGTTGAA-3′ |
| CLDN | 5′-AGCTGCAAAATGTACGACTCG-3′ | 5′-GGAGACCACCATTAGGGCTC-3′ |
| LNC CCAT1 | 5′-CCATAATGTAGAATCAGTGGAAGC-3′ | 5′-TCTCATAGCAGCACAAACCCT-3′ |
| LNC RP11-357H14.17 | 5′-CCCACTCCCTTTCTTCCTTGA-3′ | 5′-CGCCTGTAATGAACCCTGTGA-3′ |
| LNC LUCAT1 | 5′-TACCTGTCCTGCGTGTTGAA-3′ | 5′-TCTTTGGGTAATTTTTGGGATCT-3′ |
| LNC HOTTIP | 5′-GTATCGGGCAAAGGTGGAAAA-3′ | 5′-ATGAAAAGGGAGCAAGGTCGT-3′ |
| LNC RP3-416H24.1 | 5′-TTGCCTTCAATGAGATGACCTTC-3′ | 5′-GCTTTCCCCTCTGGAGACTAA-3′ |
| LNC 01559 | 5′-TCTCCTTTTCTCACTCCTCCC-3′ | 5′-TTCCTCCTCTGGTTTCTCATG-3′ |
| LNC CTD-2377D24.6 | 5′-GAGGCAGCAGTCAATACCCAC-3′ | 5′-ATCTCAATGGAAGAATGCGACA-3′ |
| LNC FEZF1 | 5′-TTCAGTCAAGAAGGCAGGTAA-3′ | 5′-GATGTCTAACAGAAAGGCAGTG-3′ |
| LINC01021 | 5′-CGAGACCATCTTGGCTAACACT-3′ | 5′-TCGGCTCACTACAAGCTCTGC-3′ |
| LNC HOXA-AS3 | 5′-CTGGAAAGGTCGGTTGTAAAG-3′ | 5′-ATAGCGACTTTTGGGATAGTTTGC-3′ |
| LNC HOXB-AS3 | 5′-TTACTGGACTTGGAGGGAGGG-3′ | 5′-ATAGCGACTTTTGGGATAGTTTGC-3′ |
| STAT3 | 5′-CAGCAGCTTGACACACGGTA-3′ | 5′-AAACACCAAAGTGGCATGTGA-3′ |
| c-MYC | 5′-GGCTCCTGGCAAAAGGTCA-3′ | 5′-CTGCGTAGTTGTGCTGATGT-3′ |
| No | Gebder | Age | Location | Differentiation Grade | Therapy | CA199 u/mL | CEA ug/L |
|---|---|---|---|---|---|---|---|
| 1 | Female | 66 | Ampullae | Moderately differentiated | None | 157.2 | 2.1 |
| 2 | Male | 69 | descending part | Moderately differentiated | None | 157.5 | 5.5 |
| 3 | Male | 52 | Superior par | Moderately differentiated | None | 12.27 | 2.41 |
| 4 | Female | 62 | Papilla | Moderately differentiated | None | 313.9 | 1.58 |
| 5 | Male | 62 | horizontal part | Moderately differentiated | None | 10.43 | 1.3 |
| 6 | Male | 70 | Ampullae | Moderately differentiated | None | 17.71 | 9.59 |
| 7 | Male | 55 | descending part | Poorly differentiated | None | 0.69 | 2.85 |
| 8 | Male | 46 | Ampullae | Moderately differentiated | None | 21.56 | 3.19 |
| 9 | Male | 57 | Ampullae | Moderately differentiated | None | 24.15 | 3.14 |
| 10 | Male | 66 | Ampullae | Moderately differentiated | None | 58.6 | 6.8 |
| 11 | Female | 46 | Papilla | Moderately differentiated | None | 112.5 | 4.73 |
| Antibody | Source | Identifier |
|---|---|---|
| Anti-GRSF1 antibody | Signalway | 54507 |
| Anti-EpCAM antibody [E144] | abcam | ab32392 |
| Anti-Actin antibody | Proteintech | HRP-6609 |
| Anti-STAT3 antibody | SCBT | SC-8019 |
| Anti-GAPDH antibody | Proteintech | 60004-1-Ig |
| Anti-CD44 antibody [EPR1013Y] | abcam | ab51037 |
| Anti-c-MYC antibody | proteintech | 10828-1-AP |
| HRP-conjugated Goat Anti-Mouse IgG(H + L) HRP-conjugated Goat Anti-Rabbit IgG(H + L) | Proteintech Proteintech | SA00001-1 SA00001-2 |
| Reagents | Source | Identifier |
|---|---|---|
| EGF | Peprotech | AF-100-15 |
| FGF-basic | Peprotech | 100-18B |
| HEPES | Gibco | 15630080 |
| PIPES | Sigma | P1851 |
| Falcon® 40 µm Cell Strainer | Corning | 352340 |
| B-27™ Supplement (50×), minus vitamin A | Gibco | 12587010 |
| Collagenase, Type I, powder | Gibco | 17100017 |
| StemPro™ Accutase™ Cell Dissociation | Gibco | A1110501 |
| ViaFect™ Transfection Reagent | promega | E4981 |
Appendix B

References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Gelsomino, F.; Balsano, R.; De Lorenzo, S.; Garajová, I. Small Bowel Adenocarcinoma: From Molecular Insights to Clinical Management. Curr. Oncol. 2022, 29, 1223–1236. [Google Scholar] [CrossRef]
- Zhong, L.; Shanahan, E.R.; Raj, A.; Koloski, N.A.; Fletcher, L.; Morrison, M.; Walker, M.M.; Talley, N.J.; Holtmann, G. Dyspepsia and the microbiome: Time to focus on the small intestine. Gut 2017, 66, 1168–1169. [Google Scholar] [CrossRef] [PubMed]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.A.; Cooper, H.S.; Deming, D.A.; Garrido-Laguna, I.; et al. Small Bowel Adenocarcinoma, Version 1.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2019, 17, 1109–1133. [Google Scholar] [CrossRef]
- Hamad, A.; Singhi, A.D.; Bahary, N.; McGrath, K.; Amarin, R.; Zeh, H.J.; Zureikat, A.H. Neoadjuvant Treatment with Trastuzumab and FOLFOX Induces a Complete Pathologic Response in a Metastatic ERBB2 (HER2)-Amplified Duodenal Cancer. J. Natl. Compr. Cancer Netw. 2017, 15, 983–988. [Google Scholar] [CrossRef]
- Laforest, A.; Aparicio, T.; Zaanan, A.; Silva, F.P.; Didelot, A.; Desbeaux, A.; Le Corre, D.; Benhaim, L.; Pallier, K.; Aust, D.; et al. ERBB2 gene as a potential therapeutic target in small bowel adenocarcinoma. Eur. J. Cancer 2014, 50, 1740–1746. [Google Scholar] [CrossRef]
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Li, S.; Kong, F.; Ruan, H.; Xu, X.; Zhang, X.; Wu, Z.; Zhang, L.; Xu, Y.; Yuan, H.; et al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics 2020, 10, 265–280. [Google Scholar] [CrossRef]
- Zhou, Y.; Shao, Y.; Hu, W.; Zhang, J.; Shi, Y.; Kong, X.; Jiang, J. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ. 2023, 30, 111–124. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, H.; Zhao, J.; Li, J.; Chen, Y.; Cui, K.; Tian, L.; Zhang, J.; Li, C.; Sun, S.; et al. Long noncoding RNA DLGAP1-AS2 promotes tumorigenesis and metastasis by regulating the Trim21/ELOA/LHPP axis in colorectal cancer. Mol. Cancer 2022, 21, 210. [Google Scholar] [CrossRef]
- Lan, Y.; Xiao, X.; He, Z.; Luo, Y.; Wu, C.; Li, L.; Song, X. Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer. Nucleic Acids Res. 2018, 46, 5809–5821. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Pan, B.; Xu, X.; Xu, T.; Hu, X.; Sun, L.; et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer 2018, 17, 141. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Meng, Q.; Li, X.; Yang, H.; Xu, J.; Gao, N.; Sun, H.; Wu, S.; Familiari, G.; Relucenti, M.; et al. Long Noncoding RNA MIR17HG Promotes Colorectal Cancer Progression via miR-17-5p. Cancer Res. 2019, 79, 4882–4895. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wan, J.; Guo, M.; Yang, Z.; Li, Z.; Wang, Y.; Ming, L. Long non-coding RNA LINC01559 exerts oncogenic role via enhancing autophagy in lung adenocarcinoma. Cancer Cell Int. 2021, 21, 624. [Google Scholar] [CrossRef]
- Li, H.; Liu, J.; Lai, Y.; Huang, S.; Zheng, L.; Fan, N. LINC01559 promotes colorectal cancer via sponging miR-1343-3p to modulate PARP1/PTEN/AKT pathway. Pathol. Res. Pract. 2021, 224, 153521. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Qian, X.; Xu, X.; Lv, P. Long non-coding RNA LINC01559 serves as a competing endogenous RNA accelerating triple-negative breast cancer progression. Biomed. J. 2022, 45, 512–521. [Google Scholar] [CrossRef]
- Shen, H.; Zhu, H.; Chen, Y.; Shen, Z.; Qiu, W.; Qian, C.; Zhang, J. ZEB1-induced LINC01559 expedites cell proliferation, migration and EMT process in gastric cancer through recruiting IGF2BP2 to stabilize ZEB1 expression. Cell Death Dis. 2021, 12, 349. [Google Scholar] [CrossRef]
- Liu, D.; Xu, C.; Gong, Z.; Zhao, Y.; Fang, Z.; Rao, X.; Chen, Q.; Li, G.; Kong, W.; Chen, J. GRSF1 antagonizes age-associated hypercoagulability via modulation of fibrinogen mRNA stability. Cell Death Dis. 2023, 14, 717. [Google Scholar] [CrossRef]
- Yang, Z.; Sun, Q.; Guo, J.; Wang, S.; Song, G.; Liu, W.; Liu, M.; Tang, H. GRSF1-mediated MIR-G-1 promotes malignant behavior and nuclear autophagy by directly upregulating TMED5 and LMNB1 in cervical cancer cells. Autophagy 2019, 15, 668–685. [Google Scholar] [CrossRef]
- Huang, J.; Liu, J.; Lan, J.; Sun, J.; Zhou, K.; Deng, Y.; Liang, L.; Liu, L.; Liu, X. Guanine-Rich RNA Sequence Binding Factor 1 Deficiency Promotes Colorectal Cancer Progression by Regulating PI3K/AKT Signaling Pathway. Cancer Manag. Res. 2024, 16, 629–638. [Google Scholar] [CrossRef]
- Han, L.; Huang, C.; Wang, X.; Tong, D. The RNA-binding protein GRSF1 promotes hepatocarcinogenesis via competitively binding to YY1 mRNA with miR-30e-5p. J. Exp. Clin. Cancer Res. 2022, 41, 17. [Google Scholar] [CrossRef]
- Zhong, X.; He, X.; Wang, Y.; Hu, Z.; Huang, H.; Zhao, S.; Wei, P.; Li, D. Warburg effect in colorectal cancer: The emerging roles in tumor microenvironment and therapeutic implications. J. Hematol. Oncol. 2022, 15, 160. [Google Scholar] [CrossRef]
- Liu, S.; Zhao, H.; Hu, Y.; Yan, C.; Mi, Y.; Li, X.; Tao, D.; Qin, J. Lactate promotes metastasis of normoxic colorectal cancer stem cells through PGC-1α-mediated oxidative phosphorylation. Cell Death Dis. 2022, 13, 651. [Google Scholar] [CrossRef] [PubMed]
- Ji, K.; Mayernik, L.; Moin, K.; Sloane, B.F. Acidosis and proteolysis in the tumor microenvironment. Cancer Metastasis Rev. 2019, 38, 103–112. [Google Scholar] [CrossRef]
- Wang, H.; Tian, T.; Zhang, J. Tumor-Associated Macrophages (TAMs) in Colorectal Cancer (CRC): From Mechanism to Therapy and Prognosis. Int. J. Mol. Sci. 2021, 22, 8470. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Xu, Y.; Xu, H.; Ren, J.; Meng, T.; Ni, Y.; Zhu, Q.; Zhang, W.B.; Pan, Y.B.; Jin, J.; et al. Lactate-induced M2 polarization of tumor-associated macrophages promotes the invasion of pituitary adenoma by secreting CCL17. Theranostics 2021, 11, 3839–3852. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Shi, W.; Xu, Y.; Xu, C.; Zhao, T.; Geng, B.; Yang, J.; Pan, J.; Hu, S.; Zhang, C.; et al. Tumor-derived lactate induces M2 macrophage polarization via the activation of the ERK/STAT3 signaling pathway in breast cancer. Cell Cycle 2018, 17, 428–438. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Liao, Q. Long noncoding RNA: A dazzling dancer in tumor immune microenvironment. J. Exp. Clin. Cancer Res. 2020, 39, 231. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wang, X.; Zhai, S.; Shi, M.; Peng, C.; Deng, X.; Fu, D.; Wang, J.; Shen, B. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 2022, 15, 128. [Google Scholar] [CrossRef]
- Nakagawa, K.; Sho, M.; Fujishiro, M.; Kakushima, N.; Horimatsu, T.; Okada, K.I.; Iguchi, M.; Uraoka, T.; Kato, M.; Yamamoto, Y.; et al. Clinical practice guidelines for duodenal cancer 2021. J. Gastroenterol. 2022, 57, 927–941. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Devoe, C.E.; McWilliams, R.; Sun, J.; Aparicio, T.; Stephens, P.J.; Ross, J.S.; Wilson, R.; Miller, V.A.; Ali, S.M.; et al. Genomic Profiling of Small-Bowel Adenocarcinoma. JAMA Oncol. 2017, 3, 1546–1553. [Google Scholar] [CrossRef]
- Yuan, W.; Zhang, Z.; Dai, B.; Wei, Q.; Liu, J.; Liu, Y.; Liu, Y.; He, L.; Zhou, D. Whole-exome sequencing of duodenal adenocarcinoma identifies recurrent Wnt/β-catenin signaling pathway mutations. Cancer 2016, 122, 1689–1696. [Google Scholar] [CrossRef]
- Boedtkjer, E.; Pedersen, S.F. The Acidic Tumor Microenvironment as a Driver of Cancer. Annu. Rev. Physiol. 2020, 82, 103–126. [Google Scholar] [CrossRef]
- Longhitano, L.; Vicario, N.; Forte, S.; Giallongo, C.; Broggi, G.; Caltabiano, R.; Barbagallo, G.M.V.; Altieri, R.; Raciti, G.; Di Rosa, M.; et al. Lactate modulates microglia polarization via IGFBP6 expression and remodels tumor microenvironment in glioblastoma. Cancer Immunol. Immunother. 2023, 72, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Song, X.; Zhang, H.; Liu, Q.; Wei, R.; Guo, L.; Yuan, C.; Chen, F.; Xue, K.; Lai, Y.; et al. Single-cell transcriptomic landscape deciphers olfactory neuroblastoma subtypes and intra-tumoral heterogeneity. Nat. Cancer 2024, 5, 1919–1939. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Yang, Z.; Chen, P.; Yeh, Y.; Sun, C.; Xie, T.; Huang, W.; Zhang, X. GPR65 sensing tumor-derived lactate induces HMGB1 release from TAM via the cAMP/PKA/CREB pathway to promote glioma progression. J. Exp. Clin. Cancer Res. 2024, 43, 105. [Google Scholar] [CrossRef]
- Chen, Y.; Hong, C.; Qu, J.; Chen, J.; Qin, Z. Knockdown of lncRNA PCAT6 suppresses the growth of non-small cell lung cancer cells by inhibiting macrophages M2 polarization via miR-326/KLF1 axis. Bioengineered 2022, 13, 12834–12846. [Google Scholar] [CrossRef]
- Antonicka, H.; Sasarman, F.; Nishimura, T.; Paupe, V.; Shoubridge, E.A. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013, 17, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, L.; Lu, Y.; Liang, W.; Gao, Y.; Xi, H.; Chen, L. GRSF1 promotes tumorigenesis and EMT-mediated metastasis through PI3K/AKT pathway in gastric cancer. Biochem. Biophys. Res. Commun. 2021, 555, 61–66. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Shi, Y.; Liu, Z.; Wu, Y.; Luo, X.; Chen, D.; Wei, Z.; Chen, C.; Ju, H.; Wu, X.; et al. Duodenal Adenocarcinoma Is Characterized by Acidity, High Infiltration of Macrophage, and Activated Linc01559–GRSF1 Axis. Biomedicines 2025, 13, 1611. https://doi.org/10.3390/biomedicines13071611
Huang X, Shi Y, Liu Z, Wu Y, Luo X, Chen D, Wei Z, Chen C, Ju H, Wu X, et al. Duodenal Adenocarcinoma Is Characterized by Acidity, High Infiltration of Macrophage, and Activated Linc01559–GRSF1 Axis. Biomedicines. 2025; 13(7):1611. https://doi.org/10.3390/biomedicines13071611
Chicago/Turabian StyleHuang, Xinxin, Ying Shi, Zekun Liu, Yihang Wu, Xiaotong Luo, Dongwen Chen, Zhengyu Wei, Chong Chen, Huaiqiang Ju, Xiaojian Wu, and et al. 2025. "Duodenal Adenocarcinoma Is Characterized by Acidity, High Infiltration of Macrophage, and Activated Linc01559–GRSF1 Axis" Biomedicines 13, no. 7: 1611. https://doi.org/10.3390/biomedicines13071611
APA StyleHuang, X., Shi, Y., Liu, Z., Wu, Y., Luo, X., Chen, D., Wei, Z., Chen, C., Ju, H., Wu, X., Liu, X., Chen, Z., & Hu, P. (2025). Duodenal Adenocarcinoma Is Characterized by Acidity, High Infiltration of Macrophage, and Activated Linc01559–GRSF1 Axis. Biomedicines, 13(7), 1611. https://doi.org/10.3390/biomedicines13071611







