Detection of High-Risk Human Papillomavirus in Bladder Cancer: An Exploratory Study from a UK-Based Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Recruitment of Patients and Bladder Tissue Specimen Collection
2.2. DNA Extraction and Purification
2.3. Detection and Genotyping of HPV DNA
2.4. HPV DNA Sequencing
2.5. Immunohistochemistry
3. Results
3.1. Clinicopathological Characteristics of the Patients
3.2. Detection of HR-HPV DNA in Bladder Cancer Specimens
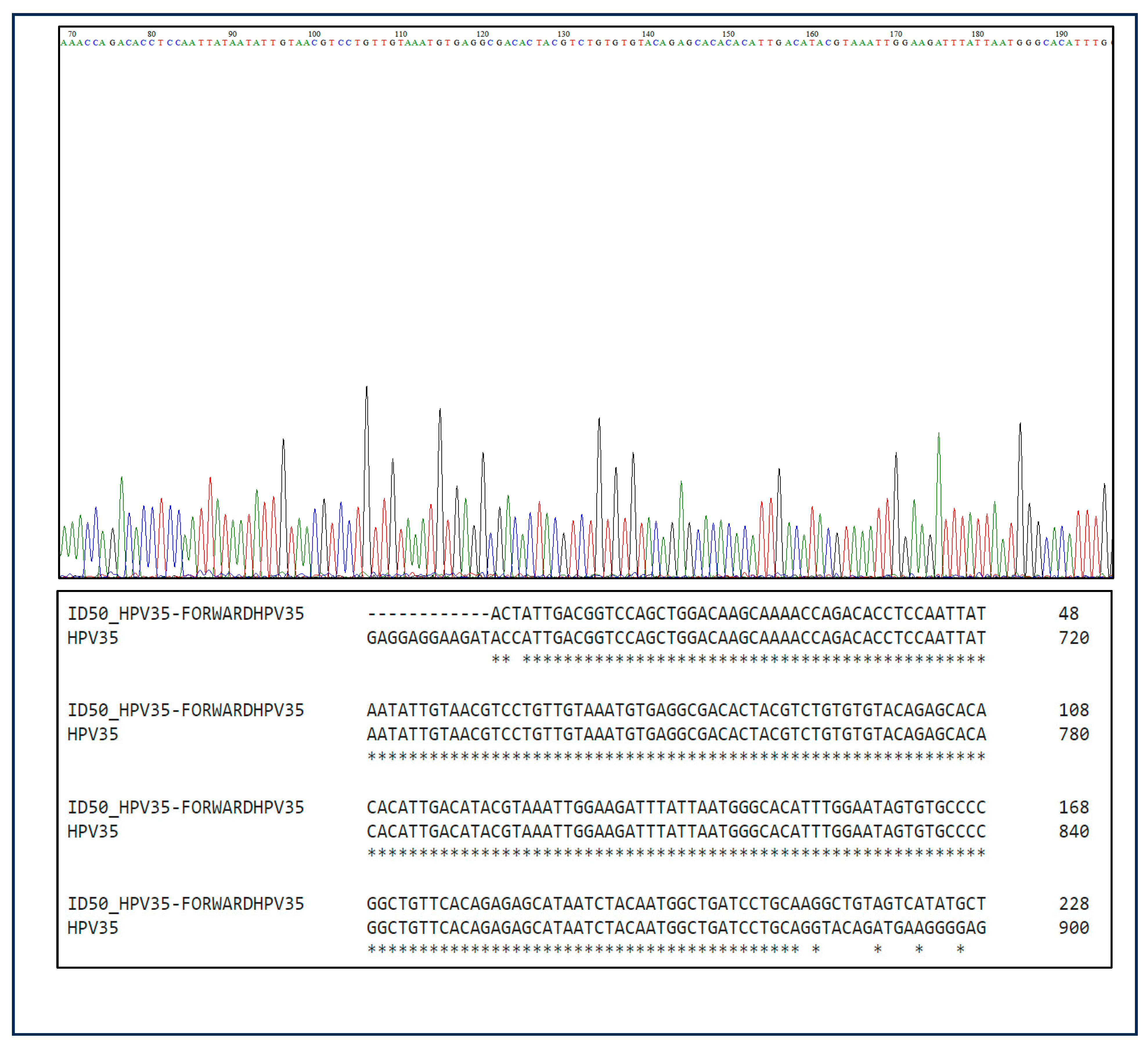
3.3. Sanger Sequencing Results
3.4. Prevalence and Distribution of HR-HPV Genotypes in Bladder Cancer
3.5. Frequency and Grade-Based Analysis of HPV Co-Infection
3.6. HPV Co-Infection Patterns and Combinatorial Genotypes
3.7. The Expression of HPV Protein in Samples Positive for HPV
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research, U.K. Bladder Cancer Statistics. [Online]. Cancer Research UK. 2022. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bladder-cancer#BladderCS0 (accessed on 20 July 2023).
- Abdollahzadeh, P.; Madani, S.H.; Khazaei, S.; Sajadimajd, S.; Izadi, B.; Najafi, F. Association between human Papillomavirus and transitional cell carcinoma of the bladder. Urol. J. 2017, 14, 5047–5050.4. [Google Scholar]
- Llewellyn, M.A.; Gordon, N.S.; Abbotts, B.; James, N.D.; Zeegers, M.P.; Cheng, K.K.; Macdonald, A.; Roberts, S.; Parish, J.L.; Ward, D.G.; et al. Defining the frequency of human papillomavirus and polyomavirus infection in urothelial bladder tumours. Sci. Rep. 2018, 8, 11290. [Google Scholar] [CrossRef]
- Park, S.; Reuter, V.E.; Hansel, D.E. Non-urothelial carcinomas of the bladder. Histopathology 2019, 74, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Szymonowicz, K.A.; Chen, J. Biological and clinical aspects of HPV-related cancers. Cancer Biol. Med. 2020, 17, 864. [Google Scholar] [CrossRef]
- Manini, I.; Montomoli, E. Epidemiology and prevention of Human Papillomavirus. Annali Di Igiene 2018, 30, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Stanley, M. Immune responses to human papillomavirus and the development of human papillomavirus vaccines. In Human Papillomavirus; Academic Press: Cambridge, MA, USA, 2020; pp. 283–297. [Google Scholar]
- Hatano, T.; Sano, D.; Takahashi, H.; Oridate, N. Pathogenic role of immune evasion and integration of human papillomavirus in oropharyngeal cancer. Microorganisms 2021, 9, 891. [Google Scholar] [CrossRef]
- Ashrafi, G.H.; Salman, N.A. Pathogenesis of Human Papillomavirus–Immunological Responses to HPV Infection. In Human Papillomavirus-Research in a Global Perspective; Rajkumar, R., Ed.; InTech: Rijeka, Croatia, 2016; pp. 243–253. ISBN 978-953-51-2439-9. [Google Scholar]
- Kusakabe, M.; Taguchi, A.; Sone, K.; Mori, M.; Osuga, Y. Carcinogenesis and management of human papillomavirus-associated cervical cancer. Int. J. Clin. Oncol. 2023, 28, 965–974. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Salman, N.A.; Sandhu, S.; Cakir, M.O.; Seddon, A.M.; Kuehne, C.; Ashrafi, G.H. Detection of high-risk Human Papillomavirus in prostate cancer from a UK based population. Sci. Rep. 2023, 13, 7633. [Google Scholar] [CrossRef]
- Salman, N.A.; Davies, G.; Majidy, F.; Shakir, F.; Akinrinade, H.; Perumal, D.; Ashrafi, G.H. Association of High Risk Human Papillomavirus and Breast cancer: A UK based Study. Sci. Rep. 2017, 7, 43591. [Google Scholar] [CrossRef]
- Kitamura, T.; Yogo, Y.; Ueki, T.; Murakami, S.; Aso, Y. Presence of human papillomavirus type 16 genome in bladder carcinoma in situ of a patient with mild immunodeficiency. Cancer Res. 1988, 48, 7207–7211. [Google Scholar] [PubMed]
- Li, N.; Yang, L.; Zhang, Y.; Zhao, P.; Zheng, T.; Dai, M. Human papillomavirus infection and bladder cancer risk: A meta-analysis. J. Infect. Dis. 2011, 204, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Schmid, S.C.; Thümer, L.; Schuster, T.; Horn, T.; Kurtz, F.; Slotta-Huspenina, J.; Seebach, J.; Straub, M.; Maurer, T.; Autenrieth, M.; et al. Human papilloma virus is not detectable in samples of urothelial bladder cancer in a central European population: A prospective translational study. Infect. Agents Cancer 2015, 10, 31. [Google Scholar] [CrossRef]
- Karaoğlan, B.B.; Ürün, Y. Unveiling the Role of Human Papillomavirus in Urogenital Carcinogenesis a Comprehensive Review. Viruses 2024, 16, 667. [Google Scholar] [CrossRef]
- Griffiths, T.R.L.; Mellon, J.K. Human papillomavirus and urological tumours: II. Role in bladder, prostate, renal and testicular cancer. BJU Int. 2000, 85, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Khatami, A.; Salavatiha, Z.; Razizadeh, M.H. Bladder cancer and human papillomavirus association: A systematic review and meta-analysis. Infect. Agents Cancer 2022, 17, 3. [Google Scholar] [CrossRef]
- Jørgensen, K.R.; Jensen, J.B. Human papillomavirus and urinary bladder cancer revisited. APMIS 2020, 128, 72–79. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, H.; Jiang, C.; Ma, X.; Zhou, X.; Tian, X.; Song, Y.; Chen, X.; Yu, L.; Li, R.; et al. Human papillomavirus prevalence and integration status in tissue samples of bladder cancer in the Chinese population. J. Infect. Dis. 2021, 224, 114–122. [Google Scholar] [CrossRef]
- Athanasiou, A.; Bowden, S.; Paraskevaidi, M.; Fotopoulou, C.; Martin-Hirsch, P.; Paraskevaidis, E.; Kyrgiou, M. HPV vaccination and cancer prevention. Best Prac. Res. Clin. Obstet. Gynaecol. 2020, 65, 109–124. [Google Scholar] [CrossRef]
- Ferreira, D.A.; Idris, A.; McMillan, N.A. Analysis of a hit-and-run tumor model by HPV in oropharyngeal cancers. J. Med. Virol. 2023, 95, e28260. [Google Scholar] [CrossRef] [PubMed]
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Padala, S.A.; Barsouk, A. Epidemiology of bladder cancer. Med. Sci. 2020, 8, 15. [Google Scholar] [CrossRef] [PubMed]


| Total Samples N (%) | |
|---|---|
| Total number of samples | 55 (100) |
| Female | 8/55 (14.5) |
| Male | 47/55 (85.5) |
| Age (Year) | |
| (45–96) | |
| <60 | 6/55 (10.9) |
| 61–70 | 10/55 (18.2) |
| 71–80 | 18/55 (32.7) |
| 81–90 | 16/55 (29.1) |
| >90 | 5/55 (9.1) |
| Pathological status | |
| Squamous cell carcinoma | 2/55 (3.6) |
| Transitional cell carcinoma | 49/55 (89.1) |
| No cancer | 4/55 (7.3) |
| Pathological Status | HPV Genotypes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HPV in TCC n = 17 17/49 (35%) | 16 | 31 | 33 | 35 | 18 | 39 | 45 | 59 | 52 | 56 | 58 | 66 |
| Grade 1 | 2/5 (40) | - | 1/1 (100) | 2/5 (40) | - | - | 1/4 (25) | - | 4/5 (80) | - | - | 2/2 (100) |
| Grade 2 | - | - | - | 1/5 (20) | - | - | 1/4 (25) | 1/2 (50) | - | - | - | - |
| Grade 3 | 3/5 (60) | - | - | 2/5 (40) | - | 1/1 (100) | 2/4 (50) | 1/2 (50) | 1/5 (20) | - | - | - |
| Total Prevalence of Specific HPV Genotype Cases n = | 5 | - | 1 | 5 | - | 1 | 4 | 2 | 5 | - | - | 2 |
| Pathological Status | Total Number of Cases | Total HPV + N (%) | HPV Co-Infections N (%) |
|---|---|---|---|
| Transitional Cell Carcinoma Cases | 49/49 (100) | 17/49 (34.7) | 6/49 (12.2) |
| Grade 1 | 16 | 8/16 (50) | 3/16 (19) |
| Grade 2 | 14 | 2/14 (14) | 1/14 (7) |
| Grade 3 | 19 | 7/19 (37) | 2/19 (11) |
| Total | 49 | 17 | 6 |
| HPV Co-Infection Pairing | Frequency |
|---|---|
| HPV16-HPV39 | 1 |
| HPV35-HPV66 | 1 |
| HPV52-HPV66 | 1 |
| HPV45-HPV59 | 1 |
| HPV16-HPV33-HPV52 | 1 |
| HPV35-HPV45-HPV59 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, M.Y.; Cakir, M.O.; Sandhu, S.; Ashrafi, G.H. Detection of High-Risk Human Papillomavirus in Bladder Cancer: An Exploratory Study from a UK-Based Population. Biomedicines 2025, 13, 1548. https://doi.org/10.3390/biomedicines13071548
Ahmed MY, Cakir MO, Sandhu S, Ashrafi GH. Detection of High-Risk Human Papillomavirus in Bladder Cancer: An Exploratory Study from a UK-Based Population. Biomedicines. 2025; 13(7):1548. https://doi.org/10.3390/biomedicines13071548
Chicago/Turabian StyleAhmed, Mohammed Yahya, Muharrem Okan Cakir, Sarbjinder Sandhu, and G. Hossein Ashrafi. 2025. "Detection of High-Risk Human Papillomavirus in Bladder Cancer: An Exploratory Study from a UK-Based Population" Biomedicines 13, no. 7: 1548. https://doi.org/10.3390/biomedicines13071548
APA StyleAhmed, M. Y., Cakir, M. O., Sandhu, S., & Ashrafi, G. H. (2025). Detection of High-Risk Human Papillomavirus in Bladder Cancer: An Exploratory Study from a UK-Based Population. Biomedicines, 13(7), 1548. https://doi.org/10.3390/biomedicines13071548






