A Comprehensive Study Employing Computational Analysis and Mendelian Randomization Has Revealed the Impact of Key Genes on Liver Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Data Acquisition
2.2. Differential Analysis
2.3. Gene Set Enrichment Analysis
2.4. Inference of Infiltrating Immune Cells
2.5. Instrumental Variable Selection
2.6. Instrumental Variable Analysis
2.7. Animals
2.8. Design of Rat HCC Experiment
2.9. Determination of Serological Indicators
2.10. Quantitative Real-Time PCR (qRT-PCR)
2.11. Liver Transcriptome Sequencing
2.12. Total Protein Extraction
2.13. Protein Expression Detection
2.14. Clinical Samples
2.15. Histology Staining Analysis
2.16. Immunohistochemistry (IHC) Analysis
2.17. Immunofluorescent (IF) Staining
3. Results
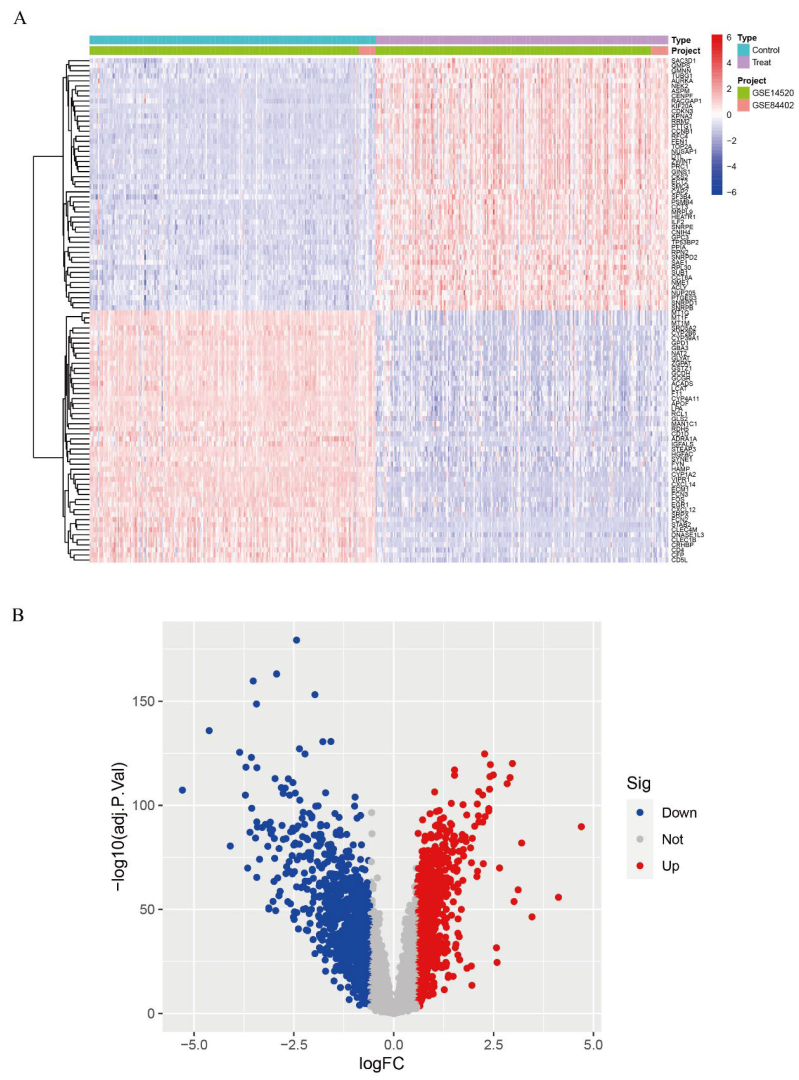
3.1. Differentially Expressed Gene (DEG) Analysis
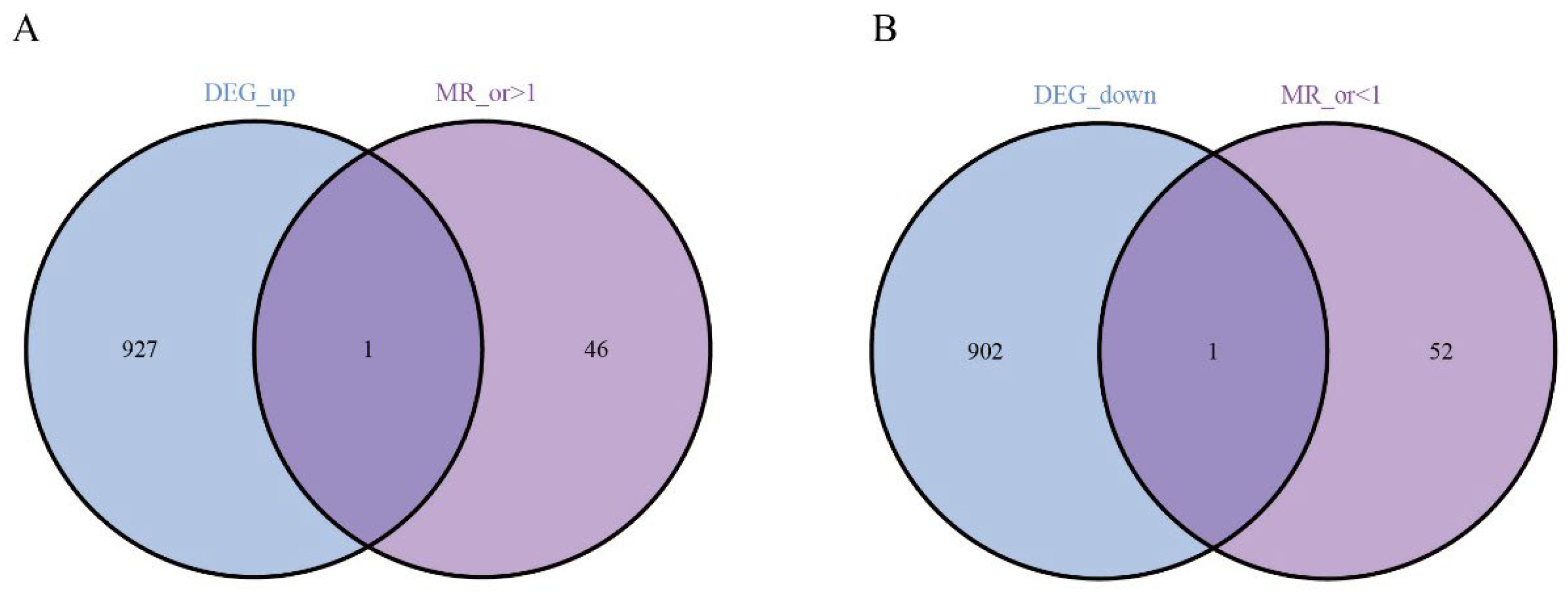
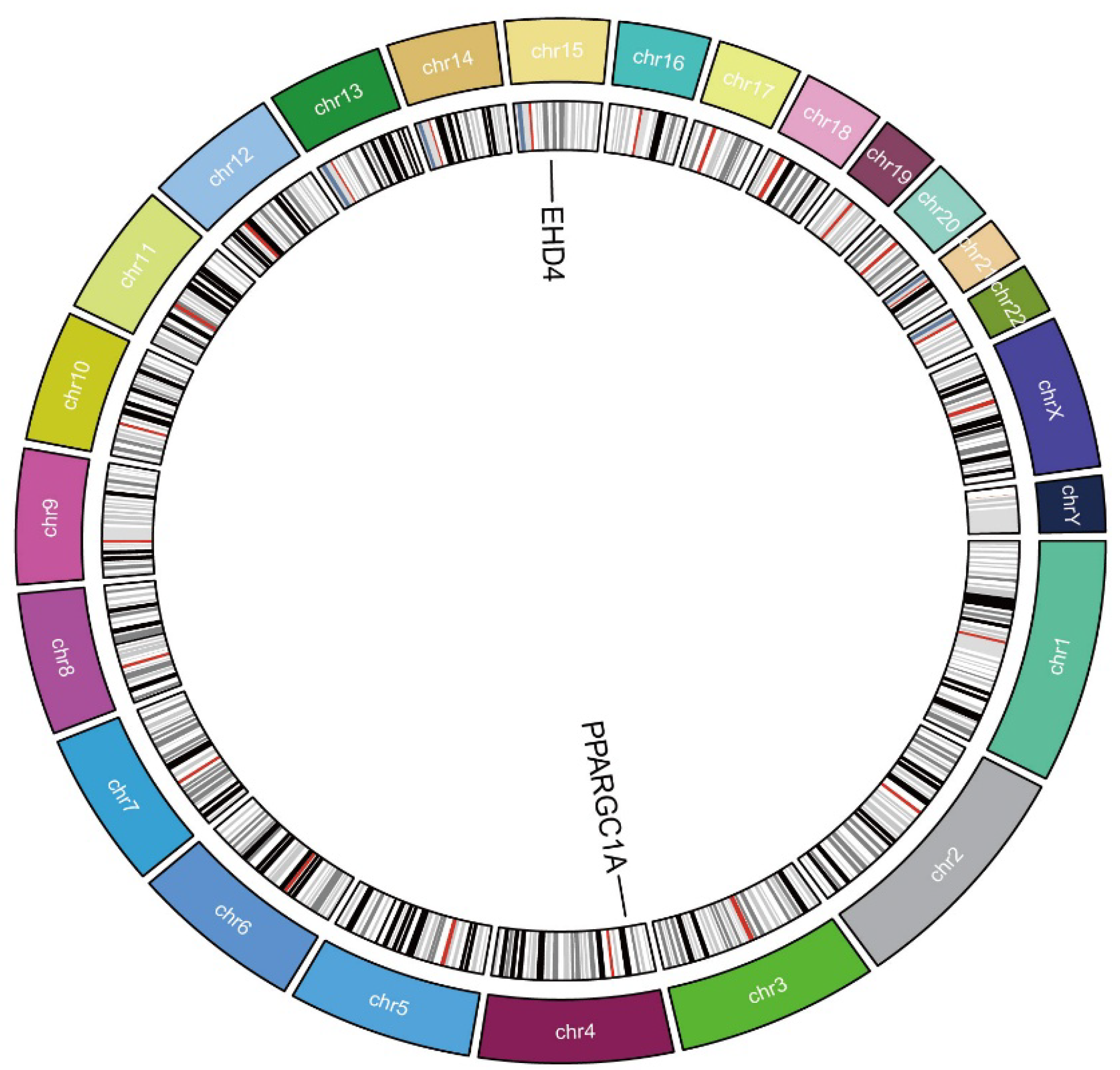
3.2. The Hub Genes Analysis
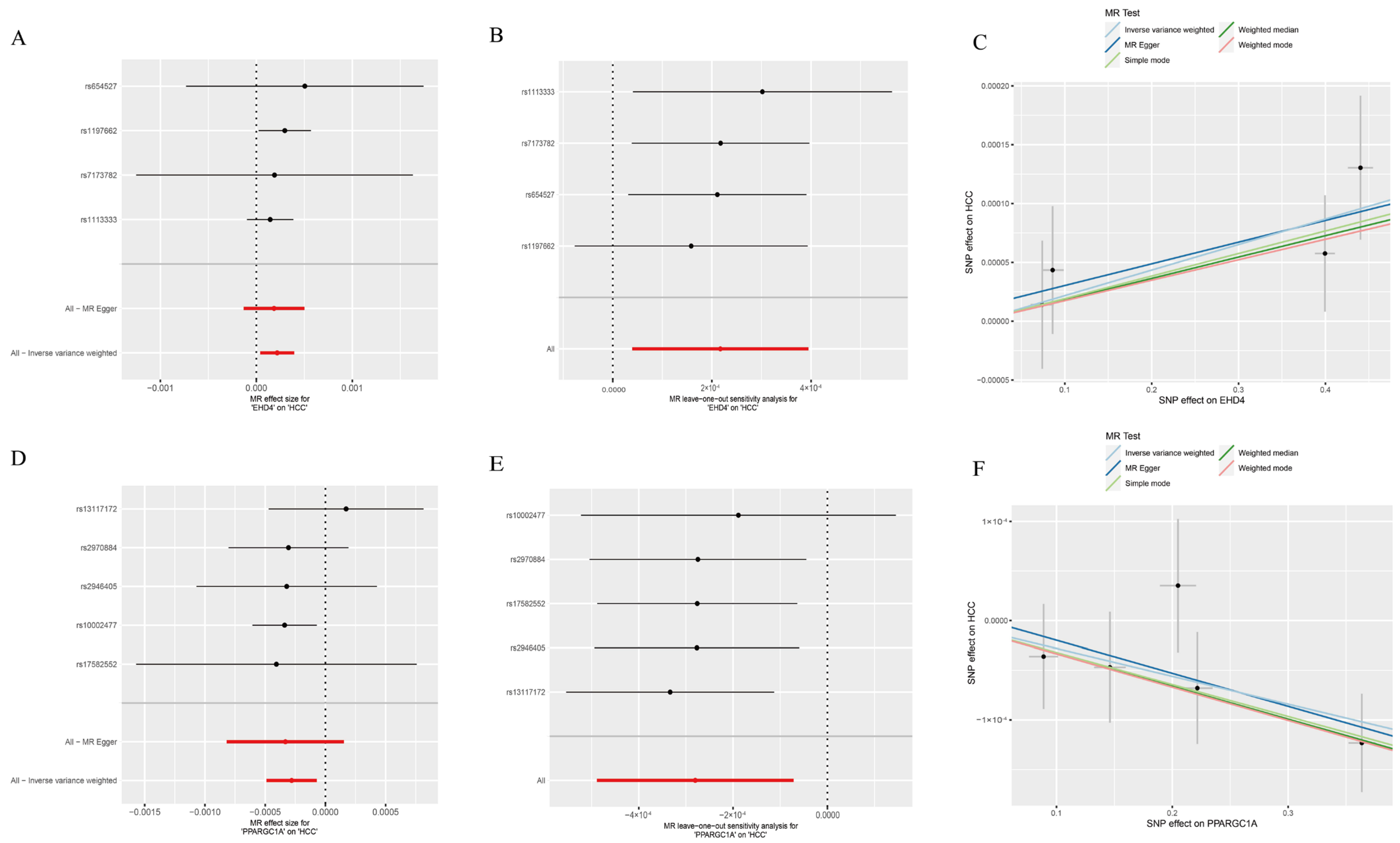
3.3. Causal Relationship Between the Hub Genes and HCC
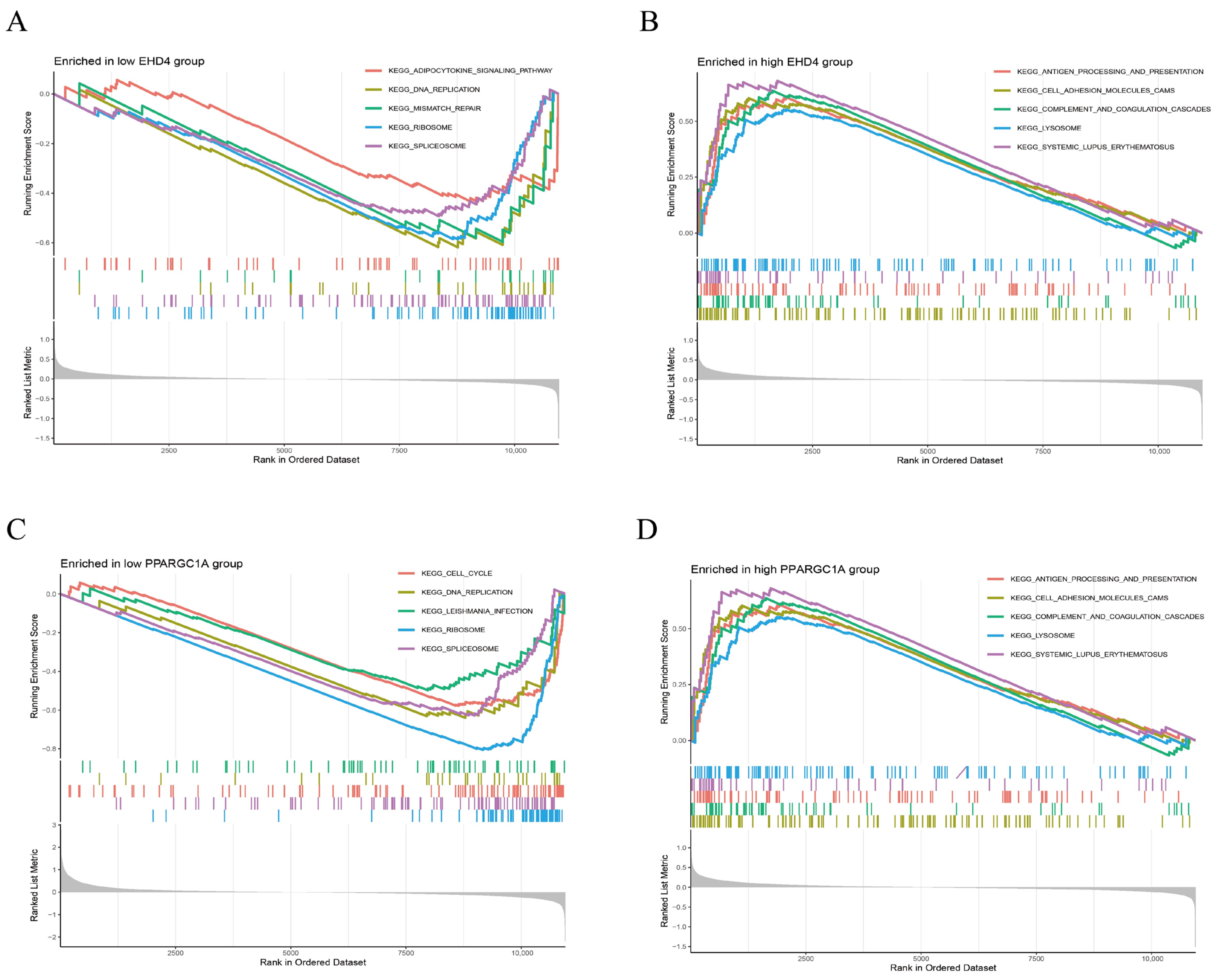
3.4. Gene Set Enrichment Analysis (GSEA) of the Hub Genes
3.5. Assessment of Immune Cell Infiltration in HCC
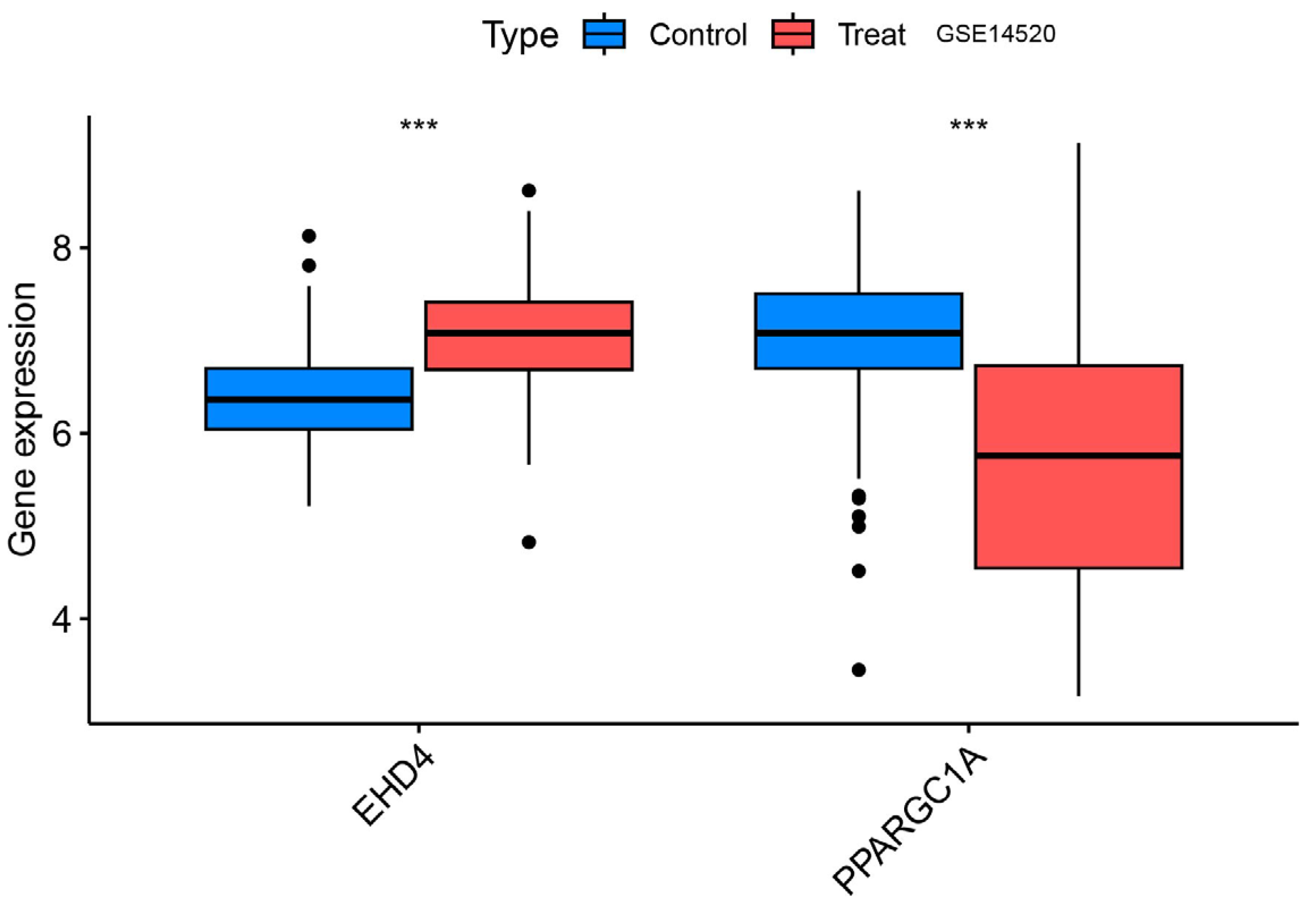
3.6. Validation Group Differential Analysis of the Hub Genes
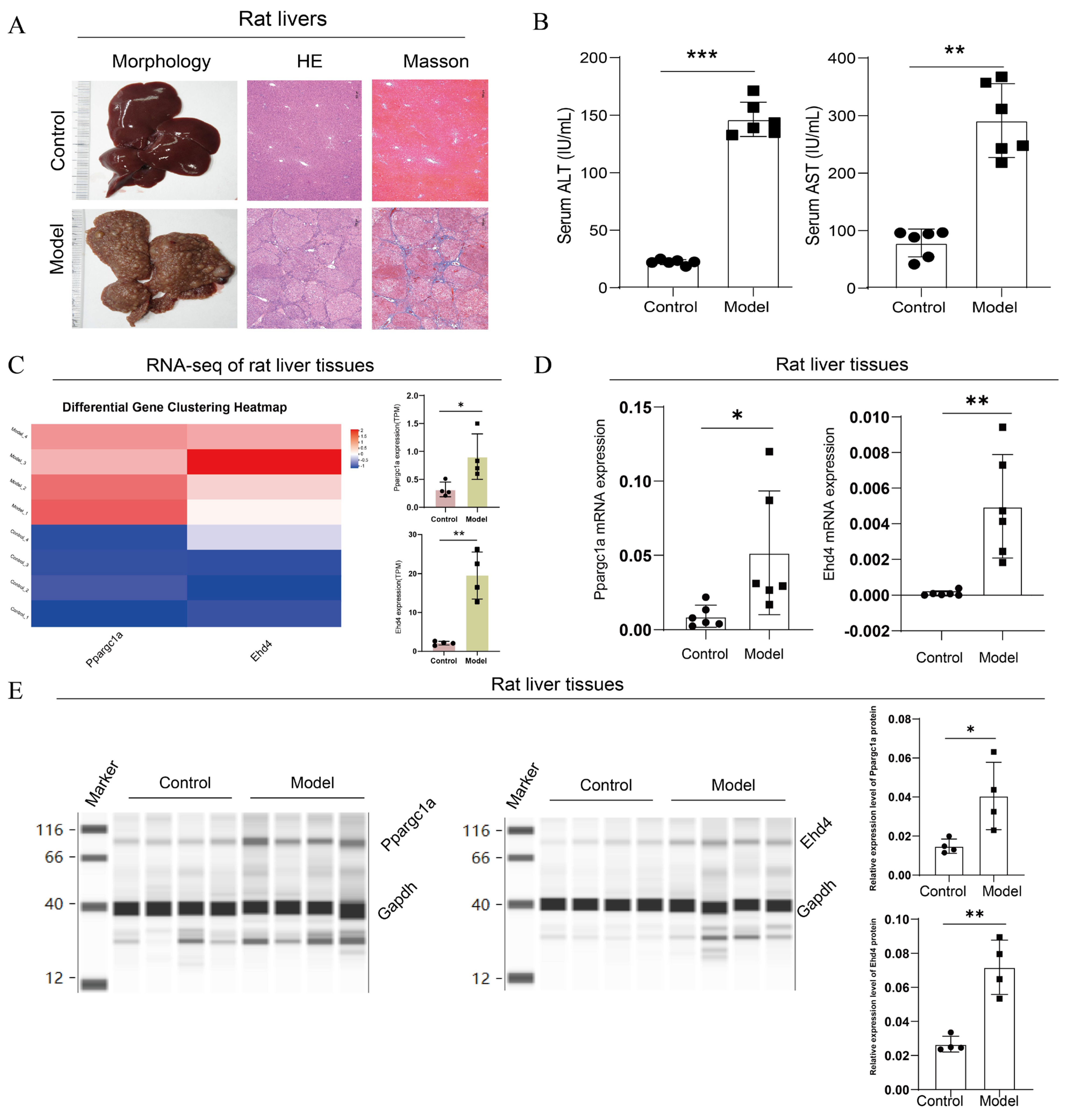
3.7. Expression Analysis of PPARGC1A and EHD4 in Rat HCC Model
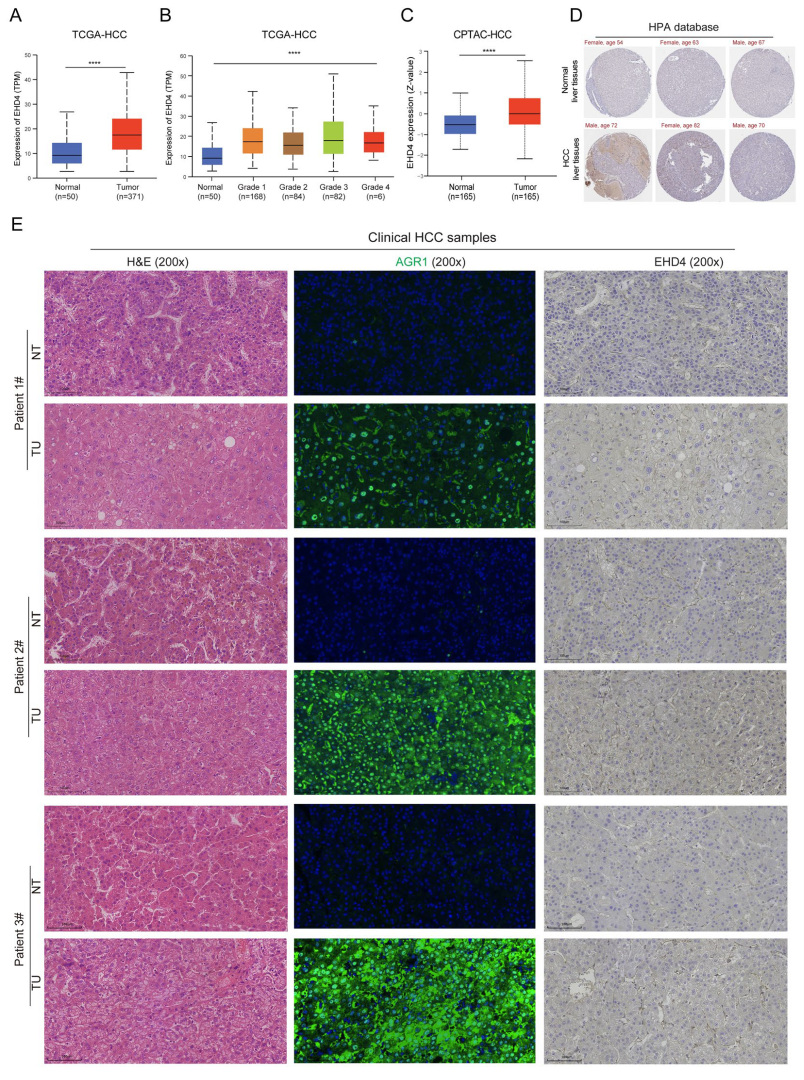
3.8. Expression Analysis of EHD4 in Clinical Samples
4. Discussion
Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Kinsey, E.; Lee, H.M. Management of Hepatocellular Carcinoma in 2024: The Multidisciplinary Paradigm in an Evolving Treatment Landscape. Cancers 2024, 16, 666. [Google Scholar] [CrossRef]
- Toh, M.R.; Wong, E.Y.T.; Wong, S.H.; Ng, A.W.T.; Loo, L.-H.; Chow, P.K.-H.; Ngeow, J. Global Epidemiology and Genetics of Hepatocellular Carcinoma. Gastroenterology 2023, 164, 766–782. [Google Scholar] [CrossRef]
- Joliat, G.R.; Allemann, P.; Labgaa, I.; Demartines, N.; Halkic, N. Treatment and outcomes of recurrent hepatocellular carcinomas. Langenbeck’s Arch. Surg. 2017, 402, 737–744. [Google Scholar] [CrossRef]
- Dagogo-Jack, I.; Shaw, A.T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 2018, 15, 81–94. [Google Scholar] [CrossRef]
- Hemminki, K.; Sundquist, K.; Sundquist, J.; Försti, A.; Liska, V.; Hemminki, A.; Li, X. Familial risks for liver, gallbladder and bile duct cancers and for their risk factors in Sweden, a low-incidence country. Cancers 2022, 14, 1938. [Google Scholar] [CrossRef]
- Mucci, L.A.; Hjelmborg, J.B.; Harris, J.R.; Czene, K.; Havelick, D.J.; Scheike, T.; Graff, R.E.; Holst, K.; Möller, S.; Unger, R.H.; et al. Familial Risk and Heritability of Cancer Among Twins in Nordic Countries. JAMA 2016, 315, 68–76. [Google Scholar] [CrossRef]
- Hassan, M.M.; Li, D.; Han, Y.; Byun, J.; Hatia, R.I.; Long, E.; Choi, J.; Kelley, R.K.; Cleary, S.P.; Lok, A.S.; et al. Genome-wide association study identifies high-impact susceptibility loci for hepatocellular carcinoma in North America. Hepatology 2024, 80, 87–101. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, Q.; Hu, W.; Liang, Y.; Jiang, D.; Chen, H. A transcriptome-wide association study identified susceptibility genes for hepatocellular carcinoma in East Asia. Gastroenterol. Rep. 2024, 12, goae057. [Google Scholar] [CrossRef] [PubMed]
- Erzurumluoglu, A.M.; Liu, M.; Jackson, V.E.; Barnes, D.R.; Datta, G.; Melbourne, C.A.; Young, R.; Batini, C.; Surendran, P.; Jiang, T.; et al. Meta-analysis of up to 622,409 individuals identifies 40 novel smoking behaviour associated genetic loci. Mol. Psychiatry 2020, 25, 2392–2409. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.P.; Colaço, A.A.; Oliveira, P.A. Animal models as a tool in hepatocellular carcinoma research: A Review. Tumor Biol. 2017, 39, 1010428317695923. [Google Scholar] [CrossRef]
- Uehara, T.; Pogribny, I.P.; Rusyn, I. The DEN and CCl4-Induced Mouse Model of Fibrosis and Inflammation-Associated Hepatocellular Carcinoma. Curr. Protoc. Pharmacol. 2014, 66, 14.30.1–14.30.10. [Google Scholar] [CrossRef]
- Han, B.; Zheng, R.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 2024, 4, 47–53. [Google Scholar] [CrossRef]
- Wu, Z.; Yu, Y.; Xie, F.; Chen, Q.; Cao, Z.; Chen, S.; Liu, G.G. Economic burden of patients with leading cancers in China: A cost-of-illness study. BMC Health Serv. Res. 2024, 24, 1135. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Andersen, J.B.; Thorgeirsson, S.S. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer. 2015, 15, 653–667. [Google Scholar] [CrossRef]
- Schulze, K.; Nault, J.C.; Villanueva, A. Genetic profiling of hepatocellular carcinoma using next-generation sequencing. J. Hepatol. 2016, 65, 1031–1042. [Google Scholar] [CrossRef]
- Kuo, H.J.; Tran, N.T.; Clary, S.A.; Morris, N.P.; Glanville, R.W. Characterization of EHD4, an EH domain-containing protein expressed in the extracellular matrix. J. Biol. Chem. 2001, 276, 43103–43110. [Google Scholar] [CrossRef]
- Lu, Q.; Insinna, C.; Ott, C.; Stauffer, J.; Pintado, P.; Rahajeng, J.; Baxa, U.; Walia, V.; Cuenca, A.; Hwang, Y.-S.; et al. Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 2015, 17, 531. [Google Scholar] [CrossRef]
- Naslavsky, N.; Caplan, S. EHD proteins: Key conductors of endocytic transport. Trends Cell Biol. 2011, 21, 122–131. [Google Scholar] [CrossRef]
- Cai, B.; Giridharan, S.S.P.; Zhang, J.; Saxena, S.; Bahl, K.; Schmidt, J.A.; Sorgen, P.L.; Guo, W.; Naslavsky, N.; Caplan, S. Differential roles of C-terminal Eps15 homology domain proteins as vesiculators and tubulators of recycling endosomes. J. Biol. Chem. 2013, 288, 30172–30180. [Google Scholar] [CrossRef]
- Bahl, K.; Naslavsky, N.; Caplan, S. Role of the EHD2 unstructured loop in dimerization, protein binding and subcellular localization. PLoS ONE 2015, 10, e0123710. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Y.; Lin, Y.; Chen, R.; Xu, B.; Deng, J. SHU00238 promotes colorectal cancer cell apoptosis through miR-4701-3p and miR-4793-3p. Front. Genet. 2020, 10, 1320. [Google Scholar] [CrossRef]
- Liu, J.; Ni, W.; Qu, L.; Cui, X.; Lin, Z.; Liu, Q.; Zhao, H.; Ni, R. Decreased expression of EHD2 promotes tumor metastasis and indicates poor prognosis in hepatocellular carcinoma. Dig. Dis. Sci. 2016, 61, 2554–2567. [Google Scholar] [CrossRef]
- Yu, J.; Yan, Y.; Hua, C.; Song, H. EHD3 promotes gastric cancer progression via Wnt/β-catenin/EMT pathway and associates with clinical prognosis and immune infiltration. Am. J. Cancer Res. 2023, 13, 4401–4417. [Google Scholar]
- Gao, J.; Meng, Q.; Zhao, Y.; Chen, X.; Cai, L. EHD1 confers resistance to cisplatin in non-small cell lung cancer by regulating intracellular cisplatin concentrations. BMC Cancer 2016, 16, 470. [Google Scholar] [CrossRef]
- Di Meo, A.; Batruch, I.; Brown, M.D.; Diamandis, E.P.; Yang, C.; Finelli, A.; Jewett, M.A.; Yousef, G.M. Searching for prognostic biomarkers for small renal masses in the urinary proteome. Int. J. Cancer 2020, 146, 2315–2325. [Google Scholar] [CrossRef]
- Taghvaei, S.; Saremi, L.; Babaniamansour, S. Computational analysis of Gly482Ser single-nucleotide polymorphism in PPARGC1A gene associated with CAD, NAFLD, T2DM, obesity, hypertension, and metabolic diseases. PPAR Res. 2021, 2021, 5544233. [Google Scholar] [CrossRef]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in metabolism, immunity, and cancer: Unified and diverse mechanisms of action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef]
- Huang, A.; Lv, B.; Zhang, Y.; Yang, J.; Li, J.; Li, C.; Yu, Z.; Xia, J. Construction of a tumor immune infiltration macrophage signature for predicting prognosis and immunotherapy response in liver cancer. Front. Mol. Biosci. 2022, 9, 983840. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiong, L.; Wei, T.; Liu, Q.; Yan, L.; Chen, J.; Dai, L.; Shi, L.; Zhang, W.; Yang, J.; et al. Hypoxia-responsive PPARGC1A/BAMBI/ACSL5 axis promotes progression and resistance to lenvatinib in hepatocellular carcinoma. Oncogene 2023, 42, 1509–1523. [Google Scholar] [CrossRef]
- Aisyah, R.; Sadewa, A.H.; Patria, S.Y.; Wahab, A. The PPARGC1A is the gene responsible for thrifty metabolism related metabolic diseases: A scoping review. Genes 2022, 13, 1894. [Google Scholar] [CrossRef]
- Zuo, Z.; Chen, T.; Zhang, Y.; Han, L.; Liu, B.; Yang, B.; Han, T.; Zheng, Z. Construction of a ceRNA network in hepatocellular carcinoma and comprehensive analysis of immune infiltration patterns. Am. J. Transl. Res. 2021, 13, 13356–13379. [Google Scholar]
- Tanouti, I.A.; Fellah, H.; El Fihry, R.; Zerrad, C.; Abounouh, K.; Tahiri, M.; Belkouchi, A.; Badre, W.; Pineau, P.; Benjelloun, S.; et al. Association of peroxisome proliferator-activated receptor gamma coactivator 1 alpha coding variants with hepatocellular carcinoma risk in the Moroccan population: A case-control study. Asian Pac. J. Cancer Prev. 2023, 24, 3689–3696. [Google Scholar] [CrossRef]
- Qian, C.N.; Mei, Y.; Zhang, J. Cancer metastasis: Issues and challenges. Chin. J. Cancer 2017, 36, 38. [Google Scholar] [CrossRef]
- Li, J.D.; Feng, Q.C.; Qi, Y.; Cui, G.; Zhao, S. PPARGC1A is up-regulated and facilitates lung cancer metastasis. Exp. Cell Res. 2017, 359, 356–360. [Google Scholar] [CrossRef]
- Deblois, G.; St-Pierre, J.; Giguère, V. The PGC-1/ERR signalling axis in cancer. Oncogene 2013, 32, 3483–3490. [Google Scholar] [CrossRef]
- Hirayama, D.; Iida, T.; Nakase, H. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef]
- Farha, M.; Jairath, N.K.; Lawrence, T.S.; El Naqa, I. Characterization of the tumor immune microenvironment identifies M0 macrophage-enriched cluster as a poor prognostic factor in hepatocellular carcinoma. JCO Clin. Cancer Inform. 2020, 4, 1002–1013. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Z.; Ding, Y.; Qin, Y. Tumor microenvironment-mediated immune evasion in hepatocellular carcinoma. Front. Immunol. 2023, 10, 1133308. [Google Scholar] [CrossRef] [PubMed]
- Ishtiaq, S.M.; Arshad, M.I.; Khan, J.A. PPARγ signaling in hepatocarcinogenesis: Mechanistic insights for cellular reprogramming and therapeutic implications. Pharmacol. Ther. 2022, 240, 108298. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.H.; Lai, A.G. The pan-cancer mutational landscape of the PPAR pathway reveals universal patterns of dysregulated metabolism and interactions with tumor immunity and hypoxia. Ann. N. Y. Acad. Sci. 2019, 1448, 65–82. [Google Scholar] [CrossRef]
- Zao, X.; Cao, X.; Liang, Y.; Zhang, J.; Chen, H.; Zhang, N.; Liu, R.; Jin, Q.; Chen, Y.; Li, X.; et al. The Chinese herbal KangXianYiAi formula inhibits hepatocellular carcinoma by reducing glutathione and inducing ferroptosis. Pharmacol. Res.-Mod. Chin. Med. 2023, 8, 100276. [Google Scholar] [CrossRef]
- Wang, C.; Dong, L.; Li, X.; Zhou, J.; Zhang, X.; Li, Y.; Wang, L.; Xu, J.; Li, D.; Liu, L. The PGC1α/NRF1-MPC1 axis suppresses tumor progression and enhances the sensitivity to sorafenib/doxorubicin treatment in hepatocellular carcinoma. Free Radic. Biol. Med. 2021, 163, 141–152. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| Gapdh | F: ATGGGACGATGCTGGTACTGA R: TGCTGACAACCTTGAGTGAAAT |
| Ehd4 | F: CCTGCGCTCTCTGTACCAG R: TCCCCATACATCACAGCAATGA |
| Ppargc1a | F: TATGGAGTGACATAGAGTGTGCT R: CCACTTCAATCCACCCAGAAAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Qi, W.; Wu, J.; Luo, C.; Zheng, S.; Cao, X.; Wang, W.; Liu, Q.; Du, H.; Li, X.; et al. A Comprehensive Study Employing Computational Analysis and Mendelian Randomization Has Revealed the Impact of Key Genes on Liver Cancer. Biomedicines 2025, 13, 1313. https://doi.org/10.3390/biomedicines13061313
Li S, Qi W, Wu J, Luo C, Zheng S, Cao X, Wang W, Liu Q, Du H, Li X, et al. A Comprehensive Study Employing Computational Analysis and Mendelian Randomization Has Revealed the Impact of Key Genes on Liver Cancer. Biomedicines. 2025; 13(6):1313. https://doi.org/10.3390/biomedicines13061313
Chicago/Turabian StyleLi, Size, Wenying Qi, Junzheng Wu, Chunhua Luo, Shihao Zheng, Xu Cao, Wei Wang, Qiyao Liu, Hongbo Du, Xiaoke Li, and et al. 2025. "A Comprehensive Study Employing Computational Analysis and Mendelian Randomization Has Revealed the Impact of Key Genes on Liver Cancer" Biomedicines 13, no. 6: 1313. https://doi.org/10.3390/biomedicines13061313
APA StyleLi, S., Qi, W., Wu, J., Luo, C., Zheng, S., Cao, X., Wang, W., Liu, Q., Du, H., Li, X., Zao, X., & Ye, Y. (2025). A Comprehensive Study Employing Computational Analysis and Mendelian Randomization Has Revealed the Impact of Key Genes on Liver Cancer. Biomedicines, 13(6), 1313. https://doi.org/10.3390/biomedicines13061313






