Gut Microbiota Dysbiosis in Japanese Female Patients with Nontuberculous Mycobacteria-Associated Lung Disease: An Observational Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
Inclusion and Exclusion Criteria
2.2. Selection of Participants
2.3. 16S rRNA Data Analysis
2.4. Gut Microbiota Analysis
2.5. Statistical Analysis
3. Results
3.1. Demographic Characteristics
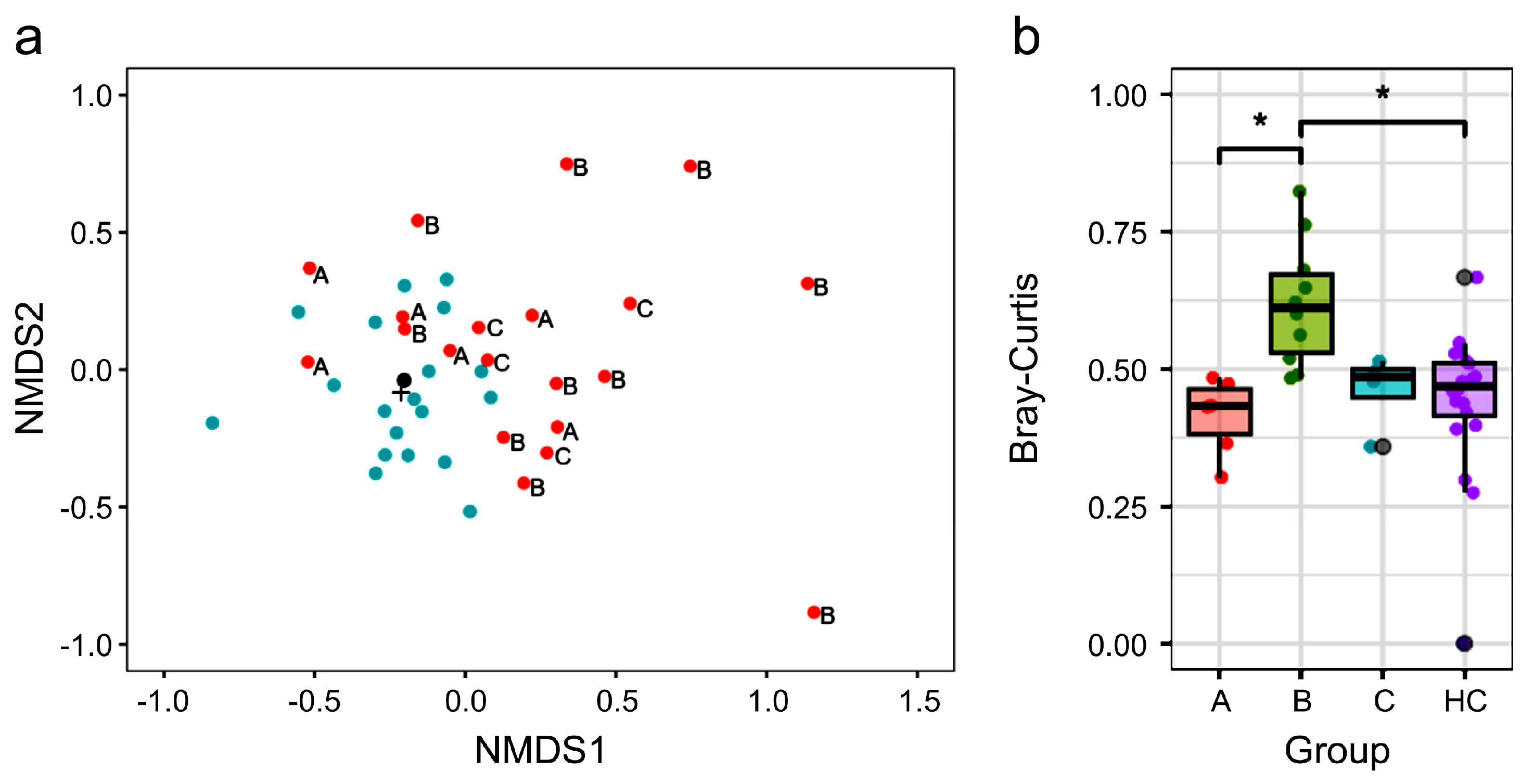
3.2. Different Gut Microbiota Composition Between NTM and HC Groups
3.3. Treatment Status and β Diversity of Gut Microbiota in Participants
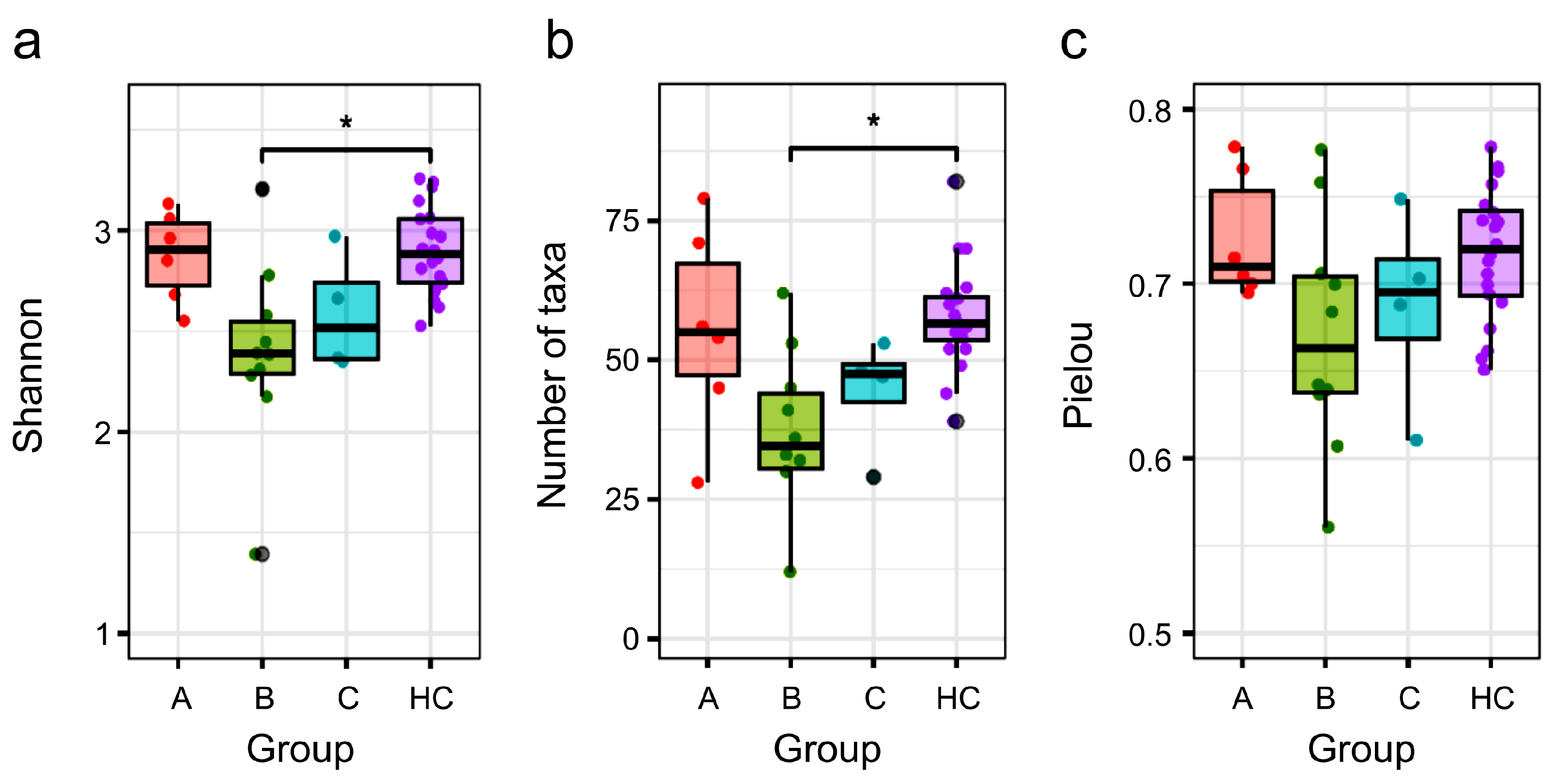
3.4. Comparison of α Diversity Between the HC Group and NTM Subgroups
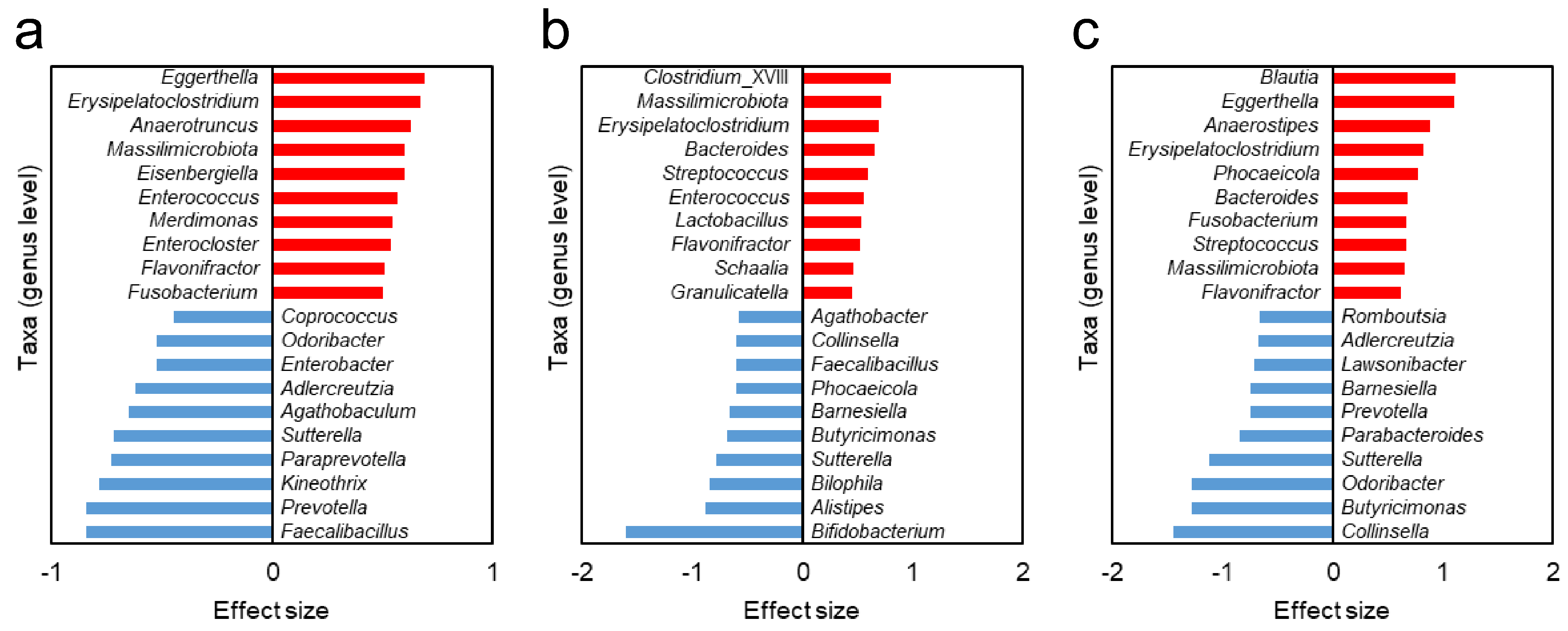
3.5. Different Gut Bacteria Between HC Group and NTM Subgroups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALDEx2 | ANOVA-like differential expression tool |
| BMI | Body mass index |
| FMT | Fecal Microbiota Transplantation |
| HC | Healthy control |
| NTM | Nontuberculous mycobacteria |
| NTM-LD | Nontuberculous mycobacterial lung disease |
| NTM-PD | Nontuberculous mycobacterial pulmonary disease NTM-PD |
| NMDS | Nonmetric multidimensional scaling |
| PERMANOVA | Permutational Multivariate Analysis of Variance |
| PERMDISP | Permutational Multivariate Analysis of Dispersion |
| TLR2 | Toll-like receptor 2 |
References
- Cowman, S.; van Ingen, J.; Griffith, D.E.; Loebinger, M.R. Non-tuberculous mycobacterial pulmonary disease. Eur. Respir. J. 2019, 54, 1900250. [Google Scholar] [CrossRef]
- Kwon, Y.S.; Koh, W.J.; Daley, C.L. Treatment of Mycobacterium Avium Complex Pulmonary Disease. Tuberc. Respir. Dis. 2019, 82, 15–26. [Google Scholar] [CrossRef]
- Stout, J.E.; Koh, W.J.; Yew, W.W. Update on Pulmonary Disease Due to Non-Tuberculous Mycobacteria. Int. J. Infect. Dis. 2016, 45, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, H.; Kurashima, A.; Morimoto, K.; Hoshino, Y.; Hasegawa, N.; Ato, M.; Mitarai, S. Epidemiology of Pulmonary Nontuberculous Mycobacterial Disease, Japan. Emerg. Infect. Dis. 2016, 22, 1116–1117. [Google Scholar] [CrossRef]
- Morimoto, K.; Hasegawa, N.; Izumi, K.; Namkoong, H.; Uchimura, K.; Yoshiyama, T.; Hoshino, Y.; Kurashima, A.; Sokunaga, J.; Shibuya, S.; et al. A Laboratory-Based Analysis of Nontuberculous Mycobacterial Lung Disease in Japan from 2012 to 2013. Ann. Am. Thorac. Soc. 2017, 14, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Izumi, K.; Morimoto, K.; Hasegawa, N.; Uchimura, K.; Kawatsu, L.; Ato, M.; Mitarai, S. Epidemiology of Adults and Children Treated for Nontuberculous Mycobacterial Pulmonary Disease in Japan. Ann. Am. Thorac. Soc. 2019, 16, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Guglielmetti, L.; Mougari, F.; Lopes, A.; Gaillard, J.L.; Cambau, E. Non-tuberculous mycobacterial infections: Management challenges and future prospects. Clin. Microbiol. Infect. 2021, 27, 472–480. [Google Scholar] [CrossRef]
- Wallace, R.J.; Brown-Elliott, B.A.; McNulty, S.; Philley, J.V.; Killingley, J.; Wilson, R.W.; York, D.S.; Shepherd, S.; Griffith, D.E. Macrolide/Azalide Therapy for Nodular/Bronchiectatic Mycobacterium Avium Complex Lung Disease. Chest 2014, 146, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Shamaei, M.; Mirsaeidi, M. Nontuberculous Mycobacteria, Macrophages, and Host Innate Immune Response. Infect. Immun. 2021, 89, e0081220. [Google Scholar] [CrossRef]
- Kim, S.Y.; Han, S.A.; Kim, D.H.; Koh, W.-J. Nontuberculous Mycobacterial Lung Disease: Ecology, Microbiology, Pathogenesis, and Antibiotic Resistance Mechanisms. Precis. Future Med. 2017, 1, 99–114. [Google Scholar] [CrossRef]
- Prevots, D.R.; Marras, T.K. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: A review. Clin. Chest Med. 2015, 36, 13–34. [Google Scholar] [CrossRef]
- Choi, J.Y.; Shim, B.; Park, Y.; Kang, Y.A. Alterations in lung and gut microbiota reduce diversity in patients with nontuberculous mycobacterial pulmonary disease. Korean J. Intern. Med. 2023, 38, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.L.; Kuo, Y.-L.; Lai, J.-H.; Lu, C.-C.; Yuan, C.-T.; Hsu, C.-Y.; Yan, B.-S.; Wu, L.S.-H.; Wu, T.-S.; Wang, J.-Y.; et al. Gut Microbiota Dysbiosis-Related Susceptibility to Nontuberculous Mycobacterial Lung Disease. Gut Microbes 2024, 16, 2361490. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.-W.; Hirose, Y.; Morita, H.; Hattori, M. The Gut Microbiome of Healthy Japanese and Its Microbial and Functional Uniqueness. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2016, 23, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Wang, X.; Su, L.; Pan, S.; Liu, N.; Li, D.; Liu, L.; Cui, J.; Zhao, H.; Yang, F. Intestinal microbiome dysbiosis increases Mycobacteria pulmonary colonization in mice by regulating the Nos2-associated pathways. eLife 2024, 13, RP99282. [Google Scholar] [CrossRef]
- Hatayama, K.; Ebara, A.; Okuma, K.; Tokuno, H.; Hasuko, K.; Masuyama, H.; Ashikari, I.; Shirasawa, T. Characteristics of Intestinal Microbiota in Japanese Patients with Mild Cognitive Impairment and a Risk-Estimating Method for the Disorder. Biomedicines 2023, 11, 1789. [Google Scholar] [CrossRef]
- Hatayama, K.; Kono, K.; Okuma, K.; Hasuko, K.; Masuyama, H.; Benno, Y. Sex Differences in Intestinal Microbiota and Their Association with Some Diseases in a Japanese Population Observed by Analysis Using a Large Dataset. Biomedicines 2023, 11, 376. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and Tools for High Throughput RRNA Analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Chao, A.; Jost, L. Coverage-Based Rarefaction and Extrapolation: Standardizing Samples by Completeness Rather than Size. Ecology 2012, 93, 2533–2547. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5-7. 2020. Available online: https://github.com/vegandevs/vegan (accessed on 18 February 2025).
- Fernandes, A.D.; Macklaim, J.M.; Linn, T.G.; Reid, G.; Gloor, G.B. ANOVA-like Differential Expression (ALDEx) Analysis for Mixed Population RNA-Seq. PLoS ONE 2013, 8, e67019. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.D.; Reid, J.N.; Macklaim, J.M.; McMurrough, T.A.; Edgell, D.R.; Gloor, G.B. Unifying the Analysis of High-Throughput Sequencing Datasets: Characterizing RNA-Seq, 16S RRNA Gene Sequencing and Selective Growth Experiments by Compositional Data Analysis. Microbiome 2014, 2, 15. [Google Scholar] [CrossRef]
- Gloor, G.B.; Macklaim, J.M.; Fernandes, A.D. Displaying Variation in Large Datasets: Plotting a Visual Summary of Effect Sizes. J. Comput. Graph. Stat. 2016, 25, 971–979. [Google Scholar] [CrossRef]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate Dispersion as a Measure of Beta Diversity. Ecol. Lett. 2006, 9, 683–693. [Google Scholar] [CrossRef]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef] [PubMed]
- Flórez, A.B.; Vázquez, L.; Rodríguez, J.; Redruello, B.; Mayo, B. Transcriptional Regulation of the Equol Biosynthesis Gene Cluster in Adlercreutzia Equolifaciens DSM19450T. Nutrients 2019, 11, 993. [Google Scholar] [CrossRef]
- Oñate, F.P.; Chamignon, C.; Burz, S.D.; Lapaque, N.; Monnoye, M.; Philippe, C.; Bredel, M.; Chêne, L.; Farin, W.; Paillarse, J.-M.; et al. Adlercreutzia Equolifaciens Is an Anti-Inflammatory Commensal Bacterium with Decreased Abundance in Gut Microbiota of Patients with Metabolic Liver Disease. Int. J. Mol. Sci. 2023, 24, 12232. [Google Scholar] [CrossRef]
- Jackson, R.L.; Greiwe, J.S.; Schwen, R.J. Emerging Evidence of the Health Benefits of S-Equol, an Estrogen Receptor β Agonist. Nutr. Rev. 2011, 69, 432–448. [Google Scholar] [CrossRef]
- Hiippala, K.; Barreto, G.; Burrello, C.; Diaz-Basabe, A.; Suutarinen, M.; Kainulainen, V.; Bowers, J.R.; Lemmer, D.; Engelthaler, D.M.; Eklund, K.K.; et al. Novel Odoribacter Splanchnicus Strain and Its Outer Membrane Vesicles Exert Immunoregulatory Effects in Vitro. Front. Microbiol. 2020, 11, 575455. [Google Scholar] [CrossRef]
- Lawson, P.A.; Saavedra Perez, L.; Sankaranarayanan, K. Reclassification of Clostridium cocleatum, Clostridium ramosum, Clostridium spiroforme and Clostridium saccharogumia as Thomasclavelia cocleata Gen. Nov., Comb. Nov., Thomasclavelia ramosa Comb. Nov., Gen. Nov., Thomasclavelia spiroformis Comb. Nov. and Thomasclavelia saccharogumia Comb. Nov. Int. J. Syst. Evol. Microbiol. 2023, 73, 5694. [Google Scholar] [CrossRef]
- Senda, S.; Fujiyama, Y.; Ushijima, T.; Hodohara, K.; Bamba, T.; Hosoda, S.; Kobayashi, K. Clostridium Ramosum, an IgA Protease-Producing Species and Its Ecology in the Human Intestinal Tract. Microbiol. Immunol. 1985, 29, 1019–1028. [Google Scholar] [CrossRef]
- Harris, S.C.; Devendran, S.; Méndez-García, C.; Mythen, S.M.; Wright, C.L.; Fields, C.J.; Hernandez, A.G.; Cann, I.; Hylemon, P.B.; Ridlon, J.M. Bile Acid Oxidation by Eggerthella Lenta Strains C592 and DSM 2243T. Gut Microbes 2018, 9, 523–539. [Google Scholar] [CrossRef]
- Hylemon, P.B.; Harris, S.C.; Ridlon, J.M. Metabolism of Hydrogen Gases and Bile Acids in the Gut Microbiome. FEBS Lett. 2018, 592, 2070–2082. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Calvo, J.M.; Rodríguez-Iglesias, M.A.; Molinillo, J.M.G. Bioavailability of Phytoestrogens: The Role of the Gut Microbiota. Pharmaceutics 2021, 13, 219. [Google Scholar] [CrossRef]
- Gaulke, C.A.; Sharpton, T.J. The Influence of Ethnicity and Geography on Human Gut Microbiome Composition. Nat. Med. 2018, 24, 1495–1496. [Google Scholar] [CrossRef] [PubMed]
- Okuma, K.; Hatayama, K.; Tokuno, H.; Ebara, A.; Odachi, A.; Masuyama, H.; Hoshiko, N.; Tanaka, N. A Risk Estimation Method for Depression Based on the Dysbiosis of Intestinal Microbiota in Japanese Patients. Front. Psychiatry 2024, 15, 1382175. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium Nucleatum—Symbiont, Opportunist and Oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Borody, T.J.; Campbell, J. Fecal microbiota transplantation: Current status and future directions. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 653–655. [Google Scholar] [CrossRef]
| NTM (n = 20) | HC (n = 20) | p-Value | |
|---|---|---|---|
| Age (years) | 67.9 ± 8.4 | 67.6 ± 8.2 | 0.745 |
| BMI (kg/m2) | 18.1 ± 1.9 | 21.0 ± 2.9 | <0.001 |
| NTM Subgroups | |||
|---|---|---|---|
| A (n = 6) | B (n = 10) | C (n = 4) | |
| During NTM treatment | No | Yes | No |
| Previous treatment | No | Yes or No | Yes |
| Use of antibiotics within 3 months | No | Yes | No |
| NTM pathogens (n) | |||
| Mycobacterium avium | 3 | 5 | 2 |
| Mycobacterium intracellulare | 2 | 1 | 1 |
| Mycobacterium avium and Mycobacterium intracellulare | 0 | 2 | 1 |
| Mycobacterium abscesuss | 0 | 1 | 0 |
| Not identified | 1 | 1 | 0 |
| Age (years) | 65.7 ± 8.0 | 68.5 ± 9.4 | 69.5 ± 2.6 |
| BMI (kg/m2) | 17.1 ± 2.0 | 18.0 ± 1.4 | 19.7 ± 1.6 |
| Subgroup A and B | Subgroup B and C | Subgroup A and C | |
|---|---|---|---|
| Taxa with higher relative abundance | Erysipelatoclostridium | Erysipelatoclostridium | Erysipelatoclostridium |
| Massilimicrobiota | Massilimicrobiota | Massilimicrobiota | |
| Flavonifractor | Flavonifractor | Flavonifractor | |
| Enterococcus | Streptococcus | Eggerthella | |
| Bacteroides | Fusobacterium | ||
| Taxa with lower relative abundance | Sutterella | Sutterella | Sutterella |
| Faecalibacillus | Bamesiella | Adlercreutzia | |
| Collinsella | Odoribacter | ||
| Butyricimonas | Prevotella |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kono, K.; Kozu, Y.; Yokota, S.; Hatayama, K.; Mizumura, K.; Maruoka, S.; Masuyama, H.; Gon, Y. Gut Microbiota Dysbiosis in Japanese Female Patients with Nontuberculous Mycobacteria-Associated Lung Disease: An Observational Study. Biomedicines 2025, 13, 1264. https://doi.org/10.3390/biomedicines13051264
Kono K, Kozu Y, Yokota S, Hatayama K, Mizumura K, Maruoka S, Masuyama H, Gon Y. Gut Microbiota Dysbiosis in Japanese Female Patients with Nontuberculous Mycobacteria-Associated Lung Disease: An Observational Study. Biomedicines. 2025; 13(5):1264. https://doi.org/10.3390/biomedicines13051264
Chicago/Turabian StyleKono, Kanako, Yutaka Kozu, Shun Yokota, Kouta Hatayama, Kenji Mizumura, Shuichiro Maruoka, Hiroaki Masuyama, and Yasuhiro Gon. 2025. "Gut Microbiota Dysbiosis in Japanese Female Patients with Nontuberculous Mycobacteria-Associated Lung Disease: An Observational Study" Biomedicines 13, no. 5: 1264. https://doi.org/10.3390/biomedicines13051264
APA StyleKono, K., Kozu, Y., Yokota, S., Hatayama, K., Mizumura, K., Maruoka, S., Masuyama, H., & Gon, Y. (2025). Gut Microbiota Dysbiosis in Japanese Female Patients with Nontuberculous Mycobacteria-Associated Lung Disease: An Observational Study. Biomedicines, 13(5), 1264. https://doi.org/10.3390/biomedicines13051264







