Cardiac Phase-Resolved T2* Magnetic Resonance Imaging Reveals Differences Between Normal Hearts and a Humanized Mouse Model of Hypertrophic Cardiomyopathy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse Model Carrying Human HCM Gene Mutation
2.2. Free Breathing, Cardiac Phased-Resolved T2* Mapping
2.3. In Vivo Cardiac MRI
2.4. Cardiac Phase-Resolved T2* Mapping
2.5. Data Analysis
2.6. Statistical Analysis
3. Results
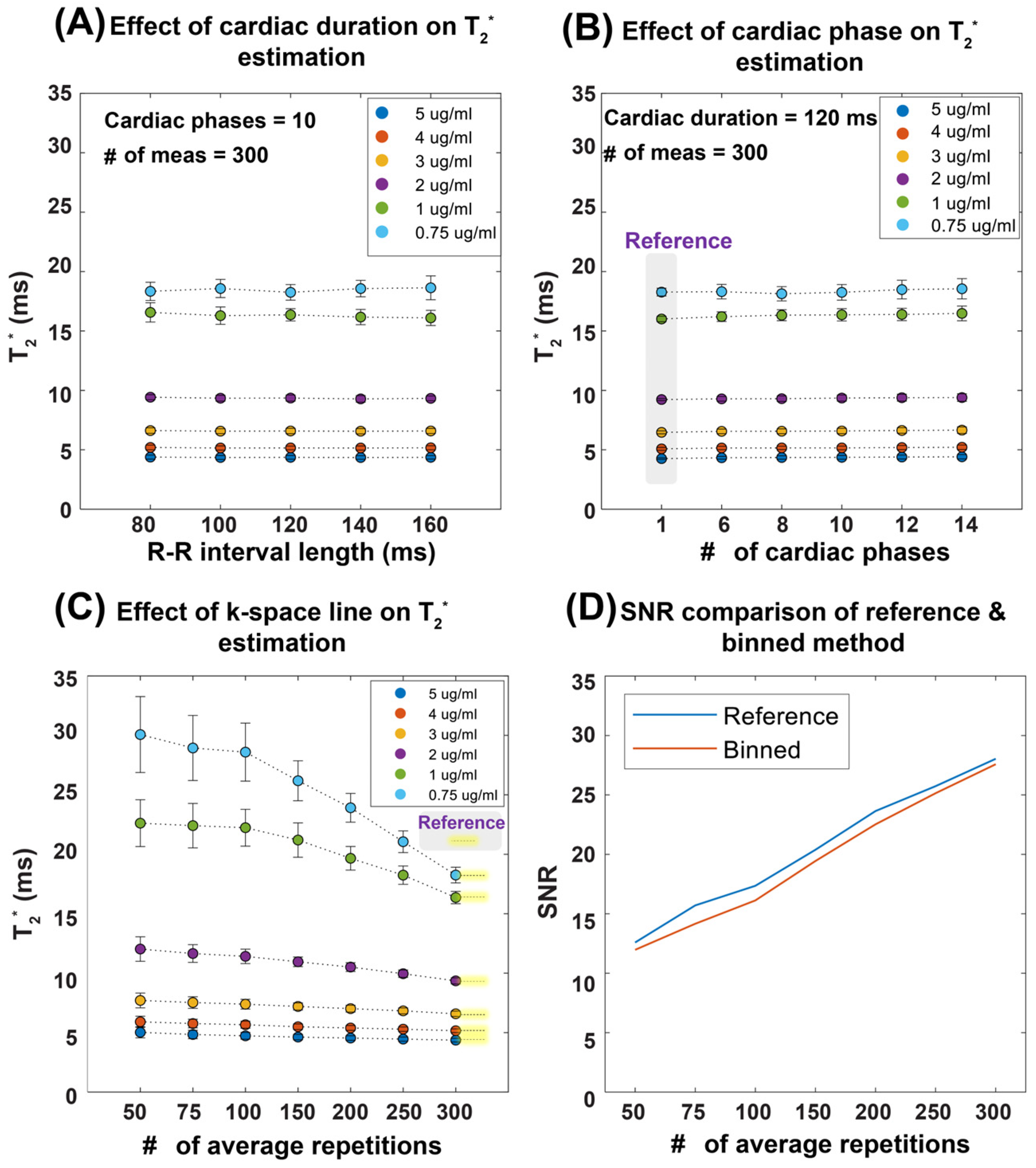
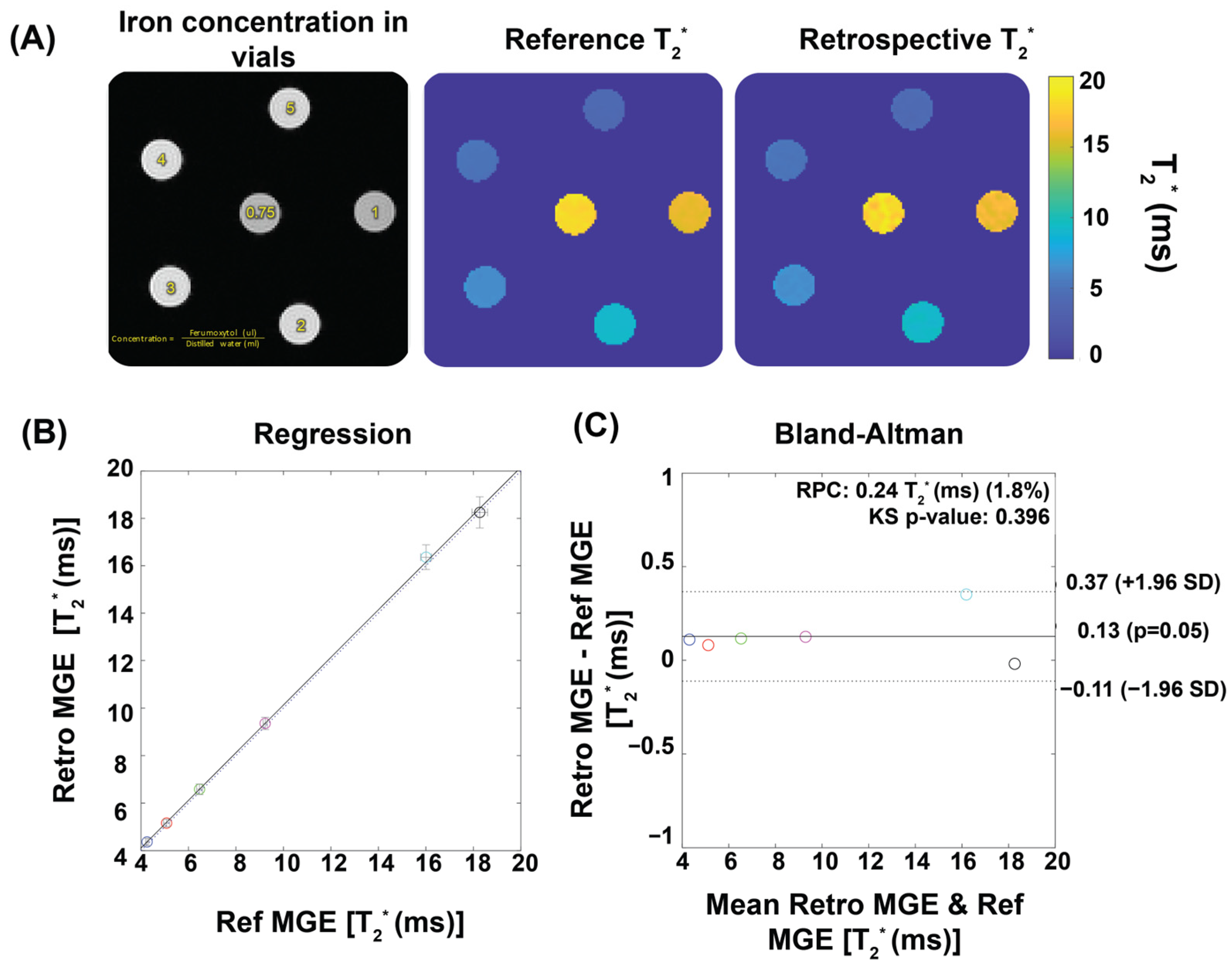
3.1. Validation in Phantom Study
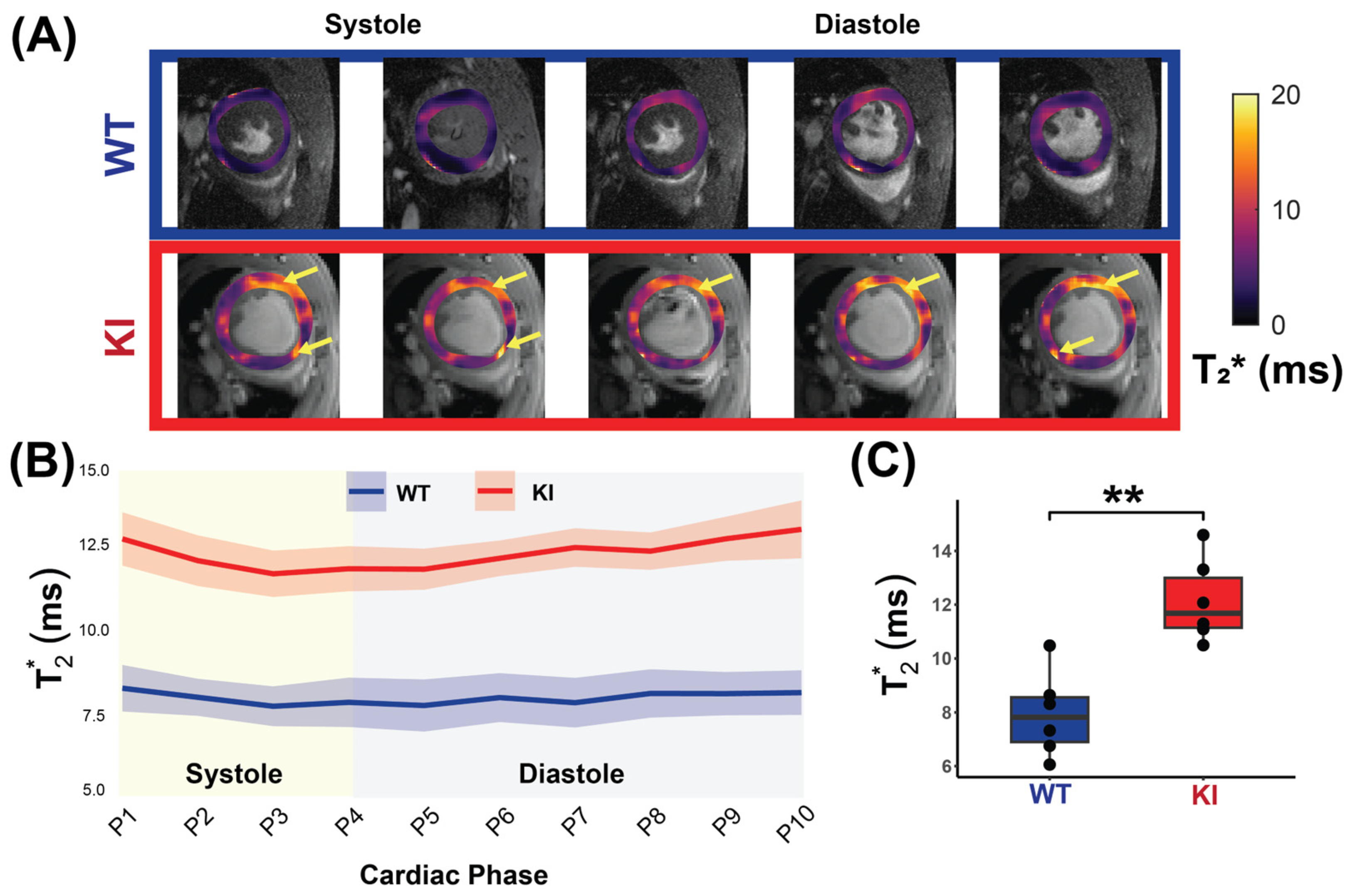
3.2. Mouse Model Carrying Mybpc3 Gene Mutation Shows HCM Phenotype Revealed by CMR
3.3. Cardiac Phase-Resolved T2* Mapping Detects Changes Across the Cardiac Phases
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MRI | Magnetic resonance imaging |
| CMR | Cardiovascular magnetic resonance |
| FLASH | Fast low angle shot |
| TR | Repetition time |
| TE | Echo time |
| FOV | Field-of-view |
| CINE | Cinematic |
| SAX | Short axis |
| EF | Ejection fraction |
| ED | End diastole |
| ES | End systole |
| HCM | Hypertrophic cardiomyopathy |
| LV | Left ventricle/left ventricular |
| LGE | Late gadolinium enhancement |
| Mybpc3 | Cardiac myosin-binding protein c |
| MYH7 | β-myosin heavy chain |
| OMIM | Online Mendelian Inheritance in Man |
| ECG | Electrocardiogram |
| SD | Standard deviation |
References
- Marian, A.J.; Braunwald, E. Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy. Circ. Res. 2017, 121, 749–770. [Google Scholar] [CrossRef] [PubMed]
- Semsarian, C.; Ingles, J.; Maron, M.S.; Maron, B.J. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2015, 65, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Olivotto, I.; Cecchi, F.; Poggesi, C.; Yacoub, M.H. Patterns of disease progression in hypertrophic cardiomyopathy: An individualized approach to clinical staging. Circ. Heart Fail. 2012, 5, 535–546. [Google Scholar] [CrossRef]
- Argiro, A.; Zampieri, M.; Marchi, A.; Cappelli, F.; Del Franco, A.; Mazzoni, C.; Cecchi, F.; Olivotto, I. Stage-specific therapy for hypertrophic cardiomyopathy. Eur. Heart J. Suppl. 2023, 25, C155–C161. [Google Scholar] [CrossRef] [PubMed]
- Gerull, B.; Klaassen, S.; Brodehl, A. The Genetic Landscape of Cardiomyopathies. In Genetic Causes of Cardiac Disease; Erdmann, J., Moretti, A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 45–91. [Google Scholar]
- Motwani, M.; Kidambi, A.; Uddin, A.; Sourbron, S.; Greenwood, J.P.; Plein, S. Quantification of myocardial blood flow with cardiovascular magnetic resonance throughout the cardiac cycle. J. Cardiovasc. Magn. Reson. 2015, 17, 4. [Google Scholar] [CrossRef]
- Radjenovic, A.; Biglands, J.D.; Larghat, A.; Ridgway, J.P.; Ball, S.G.; Greenwood, J.P.; Jerosch-Herold, M.; Plein, S. Estimates of systolic and diastolic myocardial blood flow by dynamic contrast-enhanced MRI. Magn. Reson. Med. 2010, 64, 1696–1703. [Google Scholar] [CrossRef]
- Wassenaar, P.A.; Eleswarpu, C.N.; Schroeder, S.A.; Mo, X.; Raterman, B.D.; White, R.D.; Kolipaka, A. Measuring age-dependent myocardial stiffness across the cardiac cycle using MR elastography: A reproducibility study. Magn. Reson. Med. 2016, 75, 1586–1593. [Google Scholar] [CrossRef]
- Huang, X.; Yue, Y.; Wang, Y.; Deng, Y.; Liu, L.; Di, Y.; Sun, S.; Chen, D.; Fan, L.; Cao, J. Assessment of left ventricular systolic and diastolic abnormalities in patients with hypertrophic cardiomyopathy using real-time three-dimensional echocardiography and two-dimensional speckle tracking imaging. Cardiovasc. Ultrasound 2018, 16, 23. [Google Scholar] [CrossRef]
- Pelliccia, F.; Cecchi, F.; Olivotto, I.; Camici, P.G. Microvascular Dysfunction in Hypertrophic Cardiomyopathy. J. Clin. Med. 2022, 11, 6560. [Google Scholar] [CrossRef]
- Raphael, C.E.; Mitchell, F.; Kanaganayagam, G.S.; Liew, A.C.; Di Pietro, E.; Vieira, M.S.; Kanapeckaite, L.; Newsome, S.; Gregson, J.; Owen, R.; et al. Cardiovascular magnetic resonance predictors of heart failure in hypertrophic cardiomyopathy: The role of myocardial replacement fibrosis and the microcirculation. J. Cardiovasc. Magn. Reson. 2021, 23, 26. [Google Scholar] [CrossRef]
- Kellman, P.; Xue, H.; Spottiswoode, B.S.; Sandino, C.M.; Hansen, M.S.; Abdel-Gadir, A.; Treibel, T.A.; Rosmini, S.; Mancini, C.; Bandettini, W.P.; et al. Free-breathing T2* mapping using respiratory motion corrected averaging. J. Cardiovasc. Magn. Reson. 2015, 17, 3. [Google Scholar] [CrossRef]
- Juan, Y.H.; Huang, P.C.; Lin, G.; Liu, M.H.; Lin, Y.C.; Wang, J.J.; Ng, K.K.; Cheung, Y.C.; Wang, C.H.; Ng, S.H. Oxygen-sensitive T2* magnetic resonance imaging to correlate heart function and ischemic etiology of post-hospitalized chronic heart failure patients. Eur. J. Radiol. 2020, 128, 109036. [Google Scholar] [CrossRef] [PubMed]
- Ando, K.; Nagao, M.; Watanabe, E.; Sakai, A.; Suzuki, A.; Nakao, R.; Ishizaki, U.; Sakai, S.; Hagiwara, N. Association between myocardial hypoxia and fibrosis in hypertrophic cardiomyopathy: Analysis by T2* BOLD and T1 mapping MRI. Eur. Radiol. 2020, 30, 4327–4336. [Google Scholar] [CrossRef] [PubMed]
- van Nierop, B.J.; Bax, N.A.; Nelissen, J.L.; Arslan, F.; Motaal, A.G.; de Graaf, L.; Zwanenburg, J.J.; Luijten, P.R.; Nicolay, K.; Strijkers, G.J. Assessment of Myocardial Fibrosis in Mice Using a T2*-Weighted 3D Radial Magnetic Resonance Imaging Sequence. PLoS ONE 2015, 10, e0129899. [Google Scholar] [CrossRef] [PubMed]
- Robbers, L.; Nijveldt, R.; Beek, A.M.; Teunissen, P.F.A.; Hollander, M.R.; Biesbroek, P.S.; Everaars, H.; van de Ven, P.M.; Hofman, M.B.M.; van Royen, N.; et al. The influence of microvascular injury on native T1 and T2* relaxation values after acute myocardial infarction: Implications for non-contrast-enhanced infarct assessment. Eur. Radiol. 2018, 28, 824–832. [Google Scholar] [CrossRef]
- Zhang, M.K.; Zhang, Z.; Xue, H.; Fan, L.; Wen, Y. Microvascular Rarefaction and Myocardial Fibrosis in Hypertrophic Obstructive Cardiomyopathy: A Histopathological Comparison of Pediatric and Adult Patients. Heart Surg. Forum 2022, 25, E042–E047. [Google Scholar] [CrossRef]
- Snel, G.J.H.; van den Boomen, M.; Hernandez, L.M.; Nguyen, C.T.; Sosnovik, D.E.; Velthuis, B.K.; Slart, R.; Borra, R.J.H.; Prakken, N.H.J. Cardiovascular magnetic resonance native T2 and T2(*) quantitative values for cardiomyopathies and heart transplantations: A systematic review and meta-analysis. J. Cardiovasc. Magn. Reson. 2020, 22, 34. [Google Scholar] [CrossRef]
- Huelnhagen, T.; Ku, M.C.; Reimann, H.M.; Serradas Duarte, T.; Pohlmann, A.; Flemming, B.; Seeliger, E.; Eichhorn, C.; Ferrari, V.A.; Prothmann, M.; et al. Myocardial Effective Transverse Relaxation Time T 2(*) is Elevated in Hypertrophic Cardiomyopathy: A 7.0 T Magnetic Resonance Imaging Study. Sci. Rep. 2018, 8, 3974. [Google Scholar] [CrossRef]
- Huelnhagen, T.; Hezel, F.; Serradas Duarte, T.; Pohlmann, A.; Oezerdem, C.; Flemming, B.; Seeliger, E.; Prothmann, M.; Schulz-Menger, J.; Niendorf, T. Myocardial effective transverse relaxation time T2* Correlates with left ventricular wall thickness: A 7.0 T MRI study. Magn. Reson. Med. 2017, 77, 2381–2389. [Google Scholar] [CrossRef]
- Hezel, F.; Thalhammer, C.; Waiczies, S.; Schulz-Menger, J.; Niendorf, T. High spatial resolution and temporally resolved T2* mapping of normal human myocardium at 7.0 Tesla: An ultrahigh field magnetic resonance feasibility study. PLoS ONE 2012, 7, e52324. [Google Scholar] [CrossRef]
- Niendorf, T.; Schulz-Menger, J.; Paul, K.; Huelnhagen, T.; Ferrari, V.A.; Hodge, R. High Field Cardiac Magnetic Resonance Imaging. Circ. Cardiovasc. Imaging 2017, 10, e005460. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Frauenrath, T.; Hezel, F.; Krombach, G.A.; Kremer, U.; Koppers, B.; Butenweg, C.; Goemmel, A.; Utting, J.F.; Schulz-Menger, J.; et al. Comparison of left ventricular function assessment using phonocardiogram- and electrocardiogram-triggered 2D SSFP CINE MR imaging at 1.5 T and 3.0 T. Eur. Radiol. 2010, 20, 1344–1355. [Google Scholar] [CrossRef] [PubMed]
- Lanzer, P.; Barta, C.; Botvinick, E.H.; Wiesendanger, H.U.; Modin, G.; Higgins, C.B. ECG-synchronized cardiac MR imaging: Method and evaluation. Radiology 1985, 155, 681–686. [Google Scholar] [CrossRef]
- Stab, D.; Roessler, J.; O’Brien, K.; Hamilton-Craig, C.; Barth, M. ECG Triggering in Ultra-High Field Cardiovascular MRI. Tomography 2016, 2, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Frauenrath, T.; Hezel, F.; Heinrichs, U.; Kozerke, S.; Utting, J.F.; Kob, M.; Butenweg, C.; Boesiger, P.; Niendorf, T. Feasibility of cardiac gating free of interference with electro-magnetic fields at 1.5 Tesla, 3.0 Tesla and 7.0 Tesla using an MR-stethoscope. Investig. Radiol. 2009, 44, 539–547. [Google Scholar] [CrossRef]
- Oumaima, L. Full cardiac cycle coverage T2* mapping detects early myocardial changes in hypertrophic cardiomyopathy. In Proceedings of the International Society for Magnetic Resonance in Medicine Annual Meeting 31, Toronto, ON, Canada, 3–8 June 2023. [Google Scholar]
- Shalikar, S. Improving Cardiac Phase-Resolved T2*-Mapping of the Murine Heart: Artifact Reduction and Enhanced Accuracy. In Proceedings of the International Society for Magnetic Resonance in Medicine Annual Meeting 32, Singapore, 4–9 May 2024. [Google Scholar]
- Vignier, N.; Schlossarek, S.; Fraysse, B.; Mearini, G.; Kramer, E.; Pointu, H.; Mougenot, N.; Guiard, J.; Reimer, R.; Hohenberg, H.; et al. Nonsense-mediated mRNA decay and ubiquitin-proteasome system regulate cardiac myosin-binding protein C mutant levels in cardiomyopathic mice. Circ. Res. 2009, 105, 239–248. [Google Scholar] [CrossRef]
- National Electrical Manufacturers Association. Determination of Image uniformity in Diagnostic Magnetic Resonance Images; NEMA: Rosslyn, VA, USA, 2008. [Google Scholar]
- Di Sopra, L.; Piccini, D.; Coppo, S.; Stuber, M.; Yerly, J. An automated approach to fully self-gated free-running cardiac and respiratory motion-resolved 5D whole-heart MRI. Magn. Reson. Med. 2019, 82, 2118–2132. [Google Scholar] [CrossRef]
- Heiberg, E.; Sjogren, J.; Ugander, M.; Carlsson, M.; Engblom, H.; Arheden, H. Design and validation of Segment—Freely available software for cardiovascular image analysis. BMC Med. Imaging 2010, 10, 1. [Google Scholar] [CrossRef]
- Nagao, M.; Matsuo, Y.; Kamitani, T.; Yonezawa, M.; Yamasaki, Y.; Kawanami, S.; Abe, K.; Mukai, Y.; Higo, T.; Yabuuchi, H.; et al. Quantification of myocardial iron deficiency in nonischemic heart failure by cardiac T2* magnetic resonance imaging. Am. J. Cardiol. 2014, 113, 1024–1030. [Google Scholar] [CrossRef]
- Chen, B.H.; Wu, R.; An, D.A.; Shi, R.Y.; Yao, Q.Y.; Lu, Q.; Hu, J.; Jiang, M.; Deen, J.; Chandra, A.; et al. Oxygenation-sensitive cardiovascular magnetic resonance in hypertensive heart disease with left ventricular myocardial hypertrophy and non-left ventricular myocardial hypertrophy: Insight from altered mechanics and cardiac BOLD imaging. J. Magn. Reson. Imaging 2018, 48, 1297–1306. [Google Scholar] [CrossRef]
- Weise Valdes, E.; Barth, P.; Piran, M.; Laser, K.T.; Burchert, W.; Korperich, H. Left-Ventricular Reference Myocardial Strain Assessed by Cardiovascular Magnetic Resonance Feature Tracking and fSENC-Impact of Temporal Resolution and Cardiac Muscle Mass. Front. Cardiovasc. Med. 2021, 8, 764496. [Google Scholar] [CrossRef] [PubMed]
- Dendy, J.M.; Hughes, S.G.; Soslow, J.H.; Clark, D.E.; Paschal, C.B.; Gore, J.C. Myocardial Tissue Oxygenation and Microvascular Blood Volume Measurement Using a Contrast Blood Oxygenation Level-Dependent Imaging Model. Invest. Radiol. 2022, 57, 561–566. [Google Scholar] [CrossRef]
- Ellims, A.H.; Iles, L.M.; Ling, L.H.; Chong, B.; Macciocca, I.; Slavin, G.S.; Hare, J.L.; Kaye, D.M.; Marasco, S.F.; McLean, C.A.; et al. A comprehensive evaluation of myocardial fibrosis in hypertrophic cardiomyopathy with cardiac magnetic resonance imaging: Linking genotype with fibrotic phenotype. Eur. Heart J. Cardiovasc. Imaging 2014, 15, 1108–1116. [Google Scholar] [CrossRef]
- Morrone, D.; Gentile, F.; Aimo, A.; Cameli, M.; Barison, A.; Picoi, M.E.; Guglielmo, M.; Villano, A.; DeVita, A.; Mandoli, G.E.; et al. Perspectives in noninvasive imaging for chronic coronary syndromes. Int. J. Cardiol. 2022, 365, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Gastl, M.; Gotschy, A.; von Spiczak, J.; Polacin, M.; Bonner, F.; Gruner, C.; Kelm, M.; Ruschitzka, F.; Alkadhi, H.; Kozerke, S.; et al. Cardiovascular magnetic resonance T2* mapping for structural alterations in hypertrophic cardiomyopathy. Eur. J. Radiol. Open 2019, 6, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Gastl, M.; Gruner, C.; Labucay, K.; Gotschy, A.; Von Spiczak, J.; Polacin, M.; Boenner, F.; Kelm, M.; Ruschitzka, F.; Alkadhi, H.; et al. Cardiovascular magnetic resonance T2* mapping for the assessment of cardiovascular events in hypertrophic cardiomyopathy. Open Heart 2020, 7, e001152. [Google Scholar] [CrossRef]
- Mearini, G.; Stimpel, D.; Geertz, B.; Weinberger, F.; Kramer, E.; Schlossarek, S.; Mourot-Filiatre, J.; Stoehr, A.; Dutsch, A.; Wijnker, P.J.; et al. Mybpc3 gene therapy for neonatal cardiomyopathy enables long-term disease prevention in mice. Nat. Commun. 2014, 5, 5515. [Google Scholar] [CrossRef]
- Mearini, G.; Stimpel, D.; Kramer, E.; Geertz, B.; Braren, I.; Gedicke-Hornung, C.; Precigout, G.; Muller, O.J.; Katus, H.A.; Eschenhagen, T.; et al. Repair of Mybpc3 mRNA by 5′-trans-splicing in a Mouse Model of Hypertrophic Cardiomyopathy. Mol. Ther. Nucleic Acids 2013, 2, e102. [Google Scholar] [CrossRef]
- Carrier, L.; Mearini, G.; Stathopoulou, K.; Cuello, F. Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology. Gene 2015, 573, 188–197. [Google Scholar] [CrossRef]
- Farrell, E.T.; Grimes, A.C.; de Lange, W.J.; Armstrong, A.E.; Ralphe, J.C. Increased Postnatal Cardiac Hyperplasia Precedes Cardiomyocyte Hypertrophy in a Model of Hypertrophic Cardiomyopathy. Front. Physiol. 2017, 8, 414. [Google Scholar] [CrossRef]
- Pereda, D.; García-Lunar, I.; Sierra, F.; Sánchez-Quintana, D.; Santiago, E.; Ballesteros, C.; Encalada, J.F.; Sánchez-González, J.; Fuster, V.; Ibáñez, B.; et al. Magnetic Resonance Characterization of Cardiac Adaptation and Myocardial Fibrosis in Pulmonary Hypertension Secondary to Systemic-To-Pulmonary Shunt. Circ. Cardiovasc. Imaging 2016, 9, e004566. [Google Scholar] [CrossRef] [PubMed]
- Faragli, A.; Tanacli, R.; Kolp, C.; Lapinskas, T.; Stehning, C.; Schnackenburg, B.; Lo Muzio, F.P.; Perna, S.; Pieske, B.; Nagel, E.; et al. Cardiovascular magnetic resonance feature tracking in pigs: A reproducibility and sample size calculation study. Int. J. Cardiovasc. Imaging 2020, 36, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Lapinskas, T.; Grune, J.; Zamani, S.M.; Jeuthe, S.; Messroghli, D.; Gebker, R.; Meyborg, H.; Kintscher, U.; Zaliunas, R.; Pieske, B.; et al. Cardiovascular magnetic resonance feature tracking in small animals—A preliminary study on reproducibility and sample size calculation. BMC Med. Imaging 2017, 17, 51. [Google Scholar] [CrossRef] [PubMed]


 ), while no significant correlation was found in the Mybpc3-WT mice (blue
), while no significant correlation was found in the Mybpc3-WT mice (blue  ). (B) No significant correlation was detected between the T2* and the LV ejection fraction in either group. Associations were assessed using Spearman’s rank correlation coefficient (ρ), and were considered significant at p < 0.05. Abbreviations: LV: left ventricle; KI: Mybpc3-KI; WT: wild type.
). (B) No significant correlation was detected between the T2* and the LV ejection fraction in either group. Associations were assessed using Spearman’s rank correlation coefficient (ρ), and were considered significant at p < 0.05. Abbreviations: LV: left ventricle; KI: Mybpc3-KI; WT: wild type.
 ), while no significant correlation was found in the Mybpc3-WT mice (blue
), while no significant correlation was found in the Mybpc3-WT mice (blue  ). (B) No significant correlation was detected between the T2* and the LV ejection fraction in either group. Associations were assessed using Spearman’s rank correlation coefficient (ρ), and were considered significant at p < 0.05. Abbreviations: LV: left ventricle; KI: Mybpc3-KI; WT: wild type.
). (B) No significant correlation was detected between the T2* and the LV ejection fraction in either group. Associations were assessed using Spearman’s rank correlation coefficient (ρ), and were considered significant at p < 0.05. Abbreviations: LV: left ventricle; KI: Mybpc3-KI; WT: wild type.
| Study Type | In Vivo | Phantom and In Vivo |
|---|---|---|
| Parameters | CINE Imaging | T2* mapping |
| TR (ms) | 8.5 | 14 |
| First TE (ms) | 1.58 | 1.5 |
| Echo Spacing (ms) | --- | 1.6 |
| Number of Echo Images | --- | 7 |
| FOV (mm × mm) Slice Thickness (mm) Number of Slices | 11 × 22 0.8 16 | 30 × 30 0.8 10 |
| Matrix Size | 192 × 384 | 128 × 128 |
| Excitation Flip Angle (°) | 20° | 10° |
| Receiver Bandwidth (kHz) | 98 | 133 |
| Number of Measurements | --- | 300 |
| Scan Time | 10 m6 s0 ms | |
| Sequence | IntraGate FLASH | MGE–monopolar |
| Nominal Average 1 | 27 | |
| Number of Cardiac Phases | 16 | 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laghzali, O.; Shalikar, S.; Liu, S.; Lehmann, S.; Periquito, J.d.S.; Pohlmann, A.; Waiczies, S.; Carrier, L.; Yang, H.-J.; Niendorf, T.; et al. Cardiac Phase-Resolved T2* Magnetic Resonance Imaging Reveals Differences Between Normal Hearts and a Humanized Mouse Model of Hypertrophic Cardiomyopathy. Biomedicines 2025, 13, 1193. https://doi.org/10.3390/biomedicines13051193
Laghzali O, Shalikar S, Liu S, Lehmann S, Periquito JdS, Pohlmann A, Waiczies S, Carrier L, Yang H-J, Niendorf T, et al. Cardiac Phase-Resolved T2* Magnetic Resonance Imaging Reveals Differences Between Normal Hearts and a Humanized Mouse Model of Hypertrophic Cardiomyopathy. Biomedicines. 2025; 13(5):1193. https://doi.org/10.3390/biomedicines13051193
Chicago/Turabian StyleLaghzali, Oumaima, Shahriar Shalikar, Siqin Liu, Sandra Lehmann, Joao dos Santos Periquito, Andreas Pohlmann, Sonia Waiczies, Lucie Carrier, Hsin-Jung Yang, Thoralf Niendorf, and et al. 2025. "Cardiac Phase-Resolved T2* Magnetic Resonance Imaging Reveals Differences Between Normal Hearts and a Humanized Mouse Model of Hypertrophic Cardiomyopathy" Biomedicines 13, no. 5: 1193. https://doi.org/10.3390/biomedicines13051193
APA StyleLaghzali, O., Shalikar, S., Liu, S., Lehmann, S., Periquito, J. d. S., Pohlmann, A., Waiczies, S., Carrier, L., Yang, H.-J., Niendorf, T., & Ku, M.-C. (2025). Cardiac Phase-Resolved T2* Magnetic Resonance Imaging Reveals Differences Between Normal Hearts and a Humanized Mouse Model of Hypertrophic Cardiomyopathy. Biomedicines, 13(5), 1193. https://doi.org/10.3390/biomedicines13051193







