Isolation and Characterization of the Adamantinomatous Craniopharyngioma Primary Cells with Cancer-Associated Fibroblast Features
Abstract
1. Introduction
2. Materials and Methods
2.1. ACP Clinical Samples
2.2. ACP Primary Cell Culture
2.3. Mouse Xenograft Model
2.4. H&E Staining
2.5. Immunofluorescence Staining
2.6. Immunohistochemical Staining
2.7. Whole Exome Sequencing
2.8. Short Tandem Repeat Analysis
2.9. Transcriptome Sequencing
2.10. Bioinformatics Analysis
3. Results
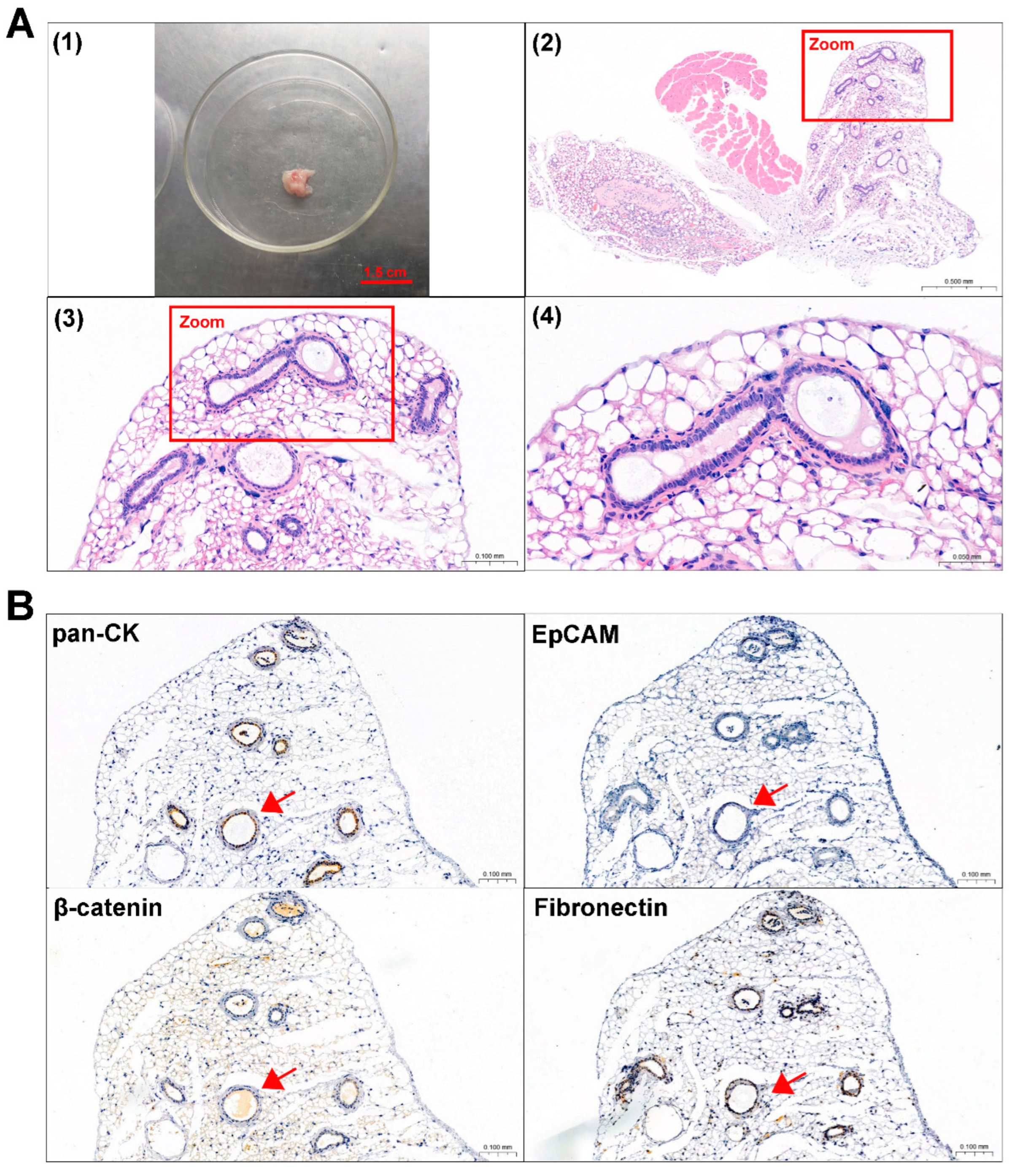
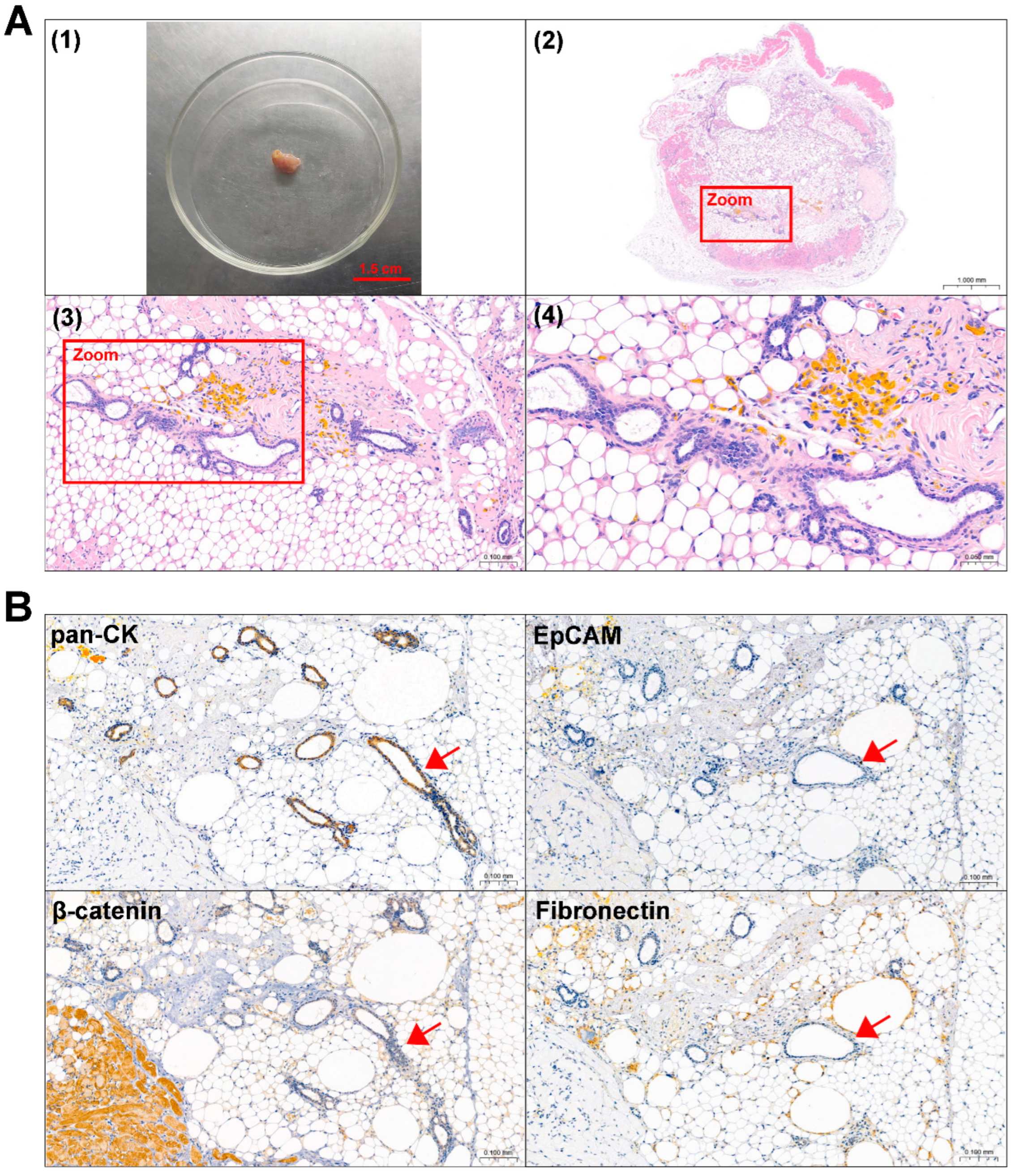
3.1. Isolation and Culture of ACP Primary Cells
| Patients | Age (Years) | Sex | Pathological Diagnosis | TUMOR Morphology | IHC | |||
|---|---|---|---|---|---|---|---|---|
| pan-CK | β-Catenin | BRAFV600E | Ki-67 | |||||
| 0913 | 19 | female | primary ACP | cystic degeneration, calcification | + | + | − | +2% |
| 0824 | 10 | female | recurrent ACP | cystic degeneration, calcification, ossification, cholesterol crystal | + | + | − | +15% |

3.2. Short Tandem Repeat Analysis
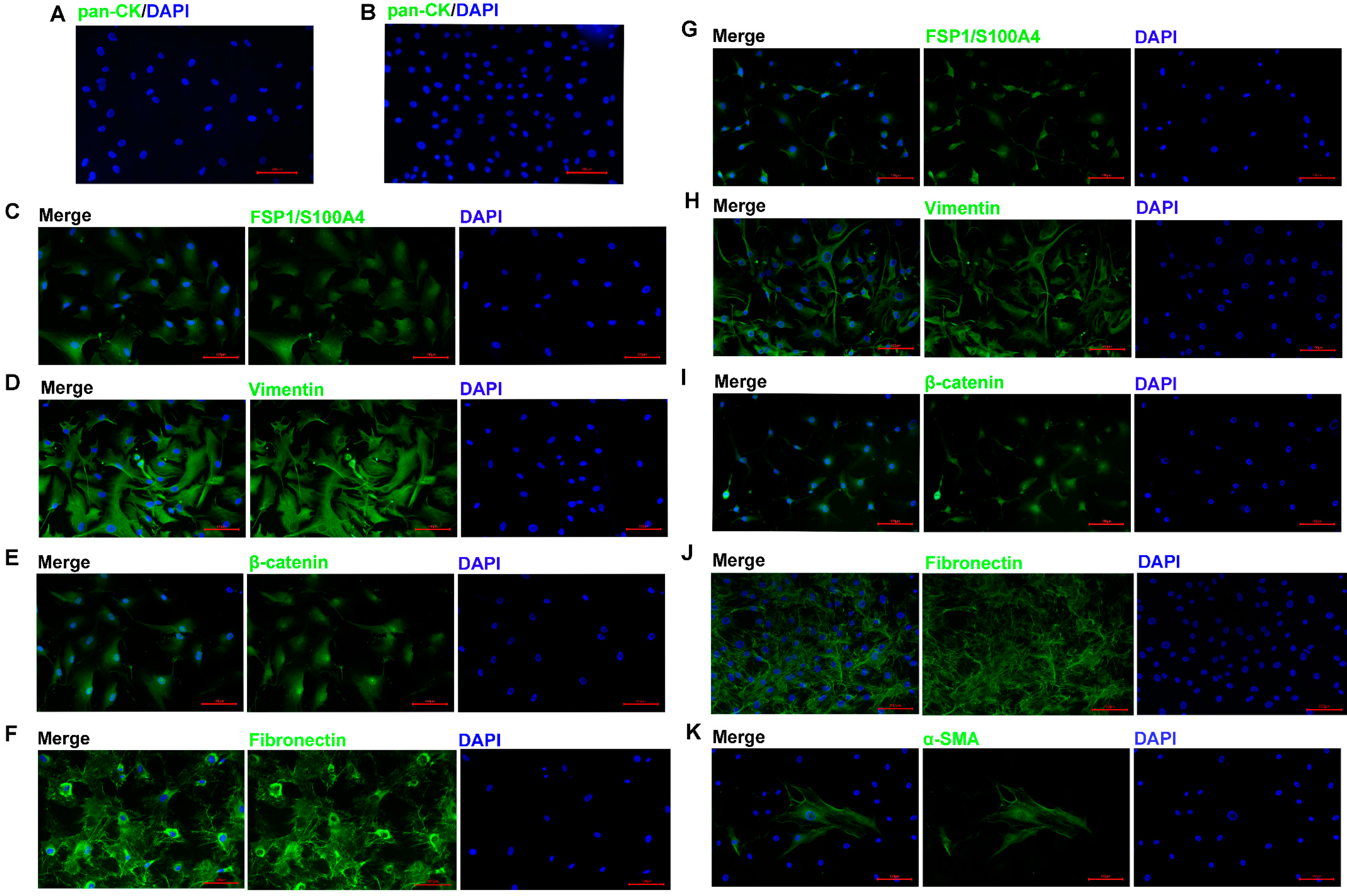
3.3. Identification of ACP Primary Cells with CAF Features
3.4. ACP Primary Cells with CAF Features Derived from ACP Tumor Tissues
3.5. Mouse Subcutaneous Xenograft Formation of ACP Primary Cells
3.6. Transcriptional Characteristics of ACP Primary Cells
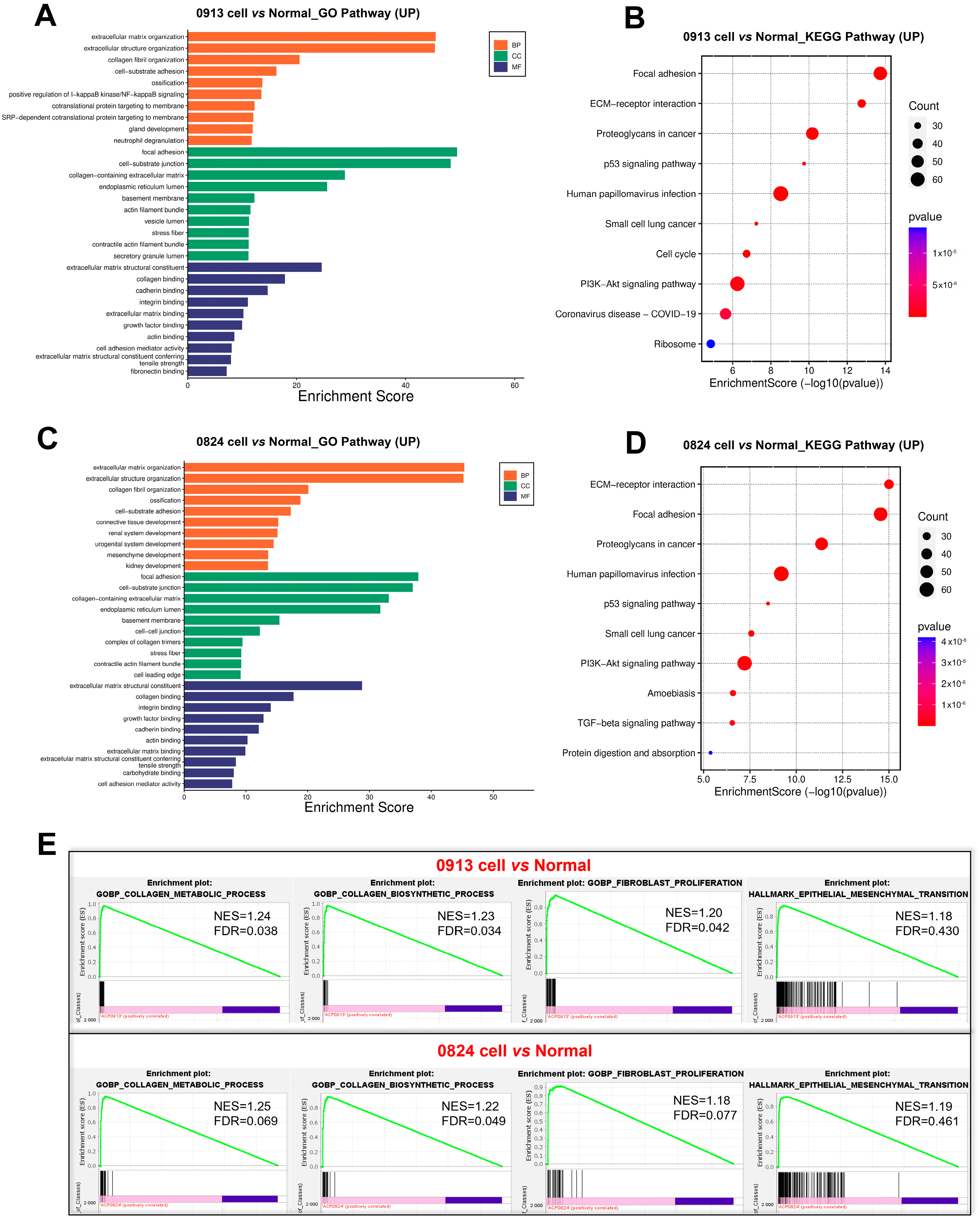
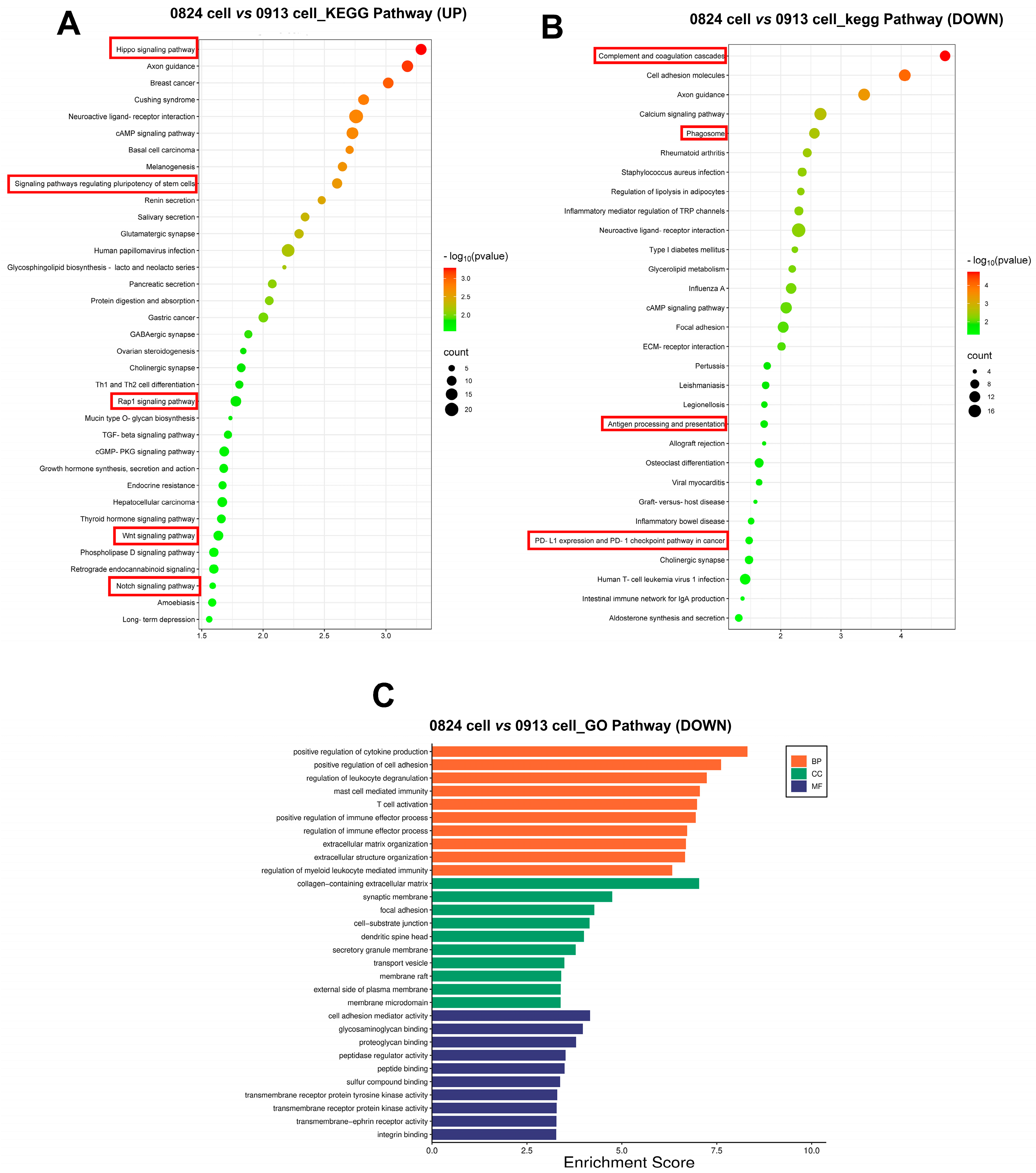
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACP | Adamantinomatous craniopharyngioma |
| STR | Short tandem repeat |
| WES | Whole exome sequencing |
| CAF | Cancer-associated fibroblast |
| GTR | Gross total resection |
| PDC | Patient-derived tumor cell |
| PDO | Patient-derived tumor cell organ |
| PDX | Patient-derived tumor xenograft |
| PBS | Phosphate-buffered saline |
| TCGA | The cancer genome atlas |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| GSEA | Gene set enrichment analysis |
| DEG | Differential expressed gene |
| SNP | Single-nucleotide polymorphism |
| SMG | Significantly mutated gene |
| ECM | Extracellular matrix |
| EMT | Epithelial-mesenchymal transition |
| MET | Mesenchymal-epithelial transition |
References
- Müller, H.L.; Merchant, T.E.; Warmuth-Metz, M.; Martinez-Barbera, J.-P.; Puget, S. Craniopharyngioma. Nat. Rev. Dis. Prim. 2019, 5, 75. [Google Scholar] [CrossRef]
- Müller, H.L. Craniopharyngioma. Endocr. Rev. 2014, 35, 513–543. [Google Scholar] [CrossRef] [PubMed]
- Zacharia, B.E.; Bruce, S.S.; Goldstein, H.; Malone, H.R.; Neugut, A.I.; Bruce, J.N. Incidence, treatment and survival of patients with craniopharyngioma in the surveillance, epidemiology and end results program. Neuro-Oncology 2012, 14, 1070–1078. [Google Scholar] [CrossRef]
- Nielsen, E.H.; Feldt-Rasmussen, U.; Poulsgaard, L.; Kristensen, L.Ø.; Astrup, J.; Jørgensen, J.O.; Bjerre, P.; Andersen, M.; Andersen, C.; Lindholm, J.; et al. Incidence of craniopharyngioma in Denmark (n = 189) and estimated world incidence of craniopharyngioma in children and adults. J. Neuro-Oncol. 2011, 104, 755–763. [Google Scholar] [CrossRef]
- Massimi, L.; Palombi, D.; Musarra, A.; Bianchi, F.; Frassanito, P.; Tamburrini, G.; Di Rocco, C. Adamantinomatous craniopharyngioma: Evolution in the management. Child’s Nerv. Syst. 2023, 39, 2613–2632. [Google Scholar] [CrossRef] [PubMed]
- Larkin, S.J.; Ansorge, O. Pathology and pathogenesis of craniopharyngiomas. Pituitary 2013, 16, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Burghaus, S.; Hölsken, A.; Buchfelder, M.; Fahlbusch, R.; Riederer, B.M.; Hans, V.; Blümcke, I.; Buslei, R. A tumor-specific cellular environment at the brain invasion border of adamantinomatous craniopharyngiomas. Virchows Arch. 2010, 456, 287–300. [Google Scholar] [CrossRef]
- Apps, J.R.; Hutchinson, J.C.; Arthurs, O.J.; Virasami, A.; Joshi, A.; Zeller-Plumhoff, B.; Moulding, D.; Jacques, T.S.; Sebire, N.J.; Martinez-Barbera, J.P. Imaging Invasion: Micro-CT imaging of adamantinomatous craniopharyngioma highlights cell type specific spatial relationships of tissue invasion. Acta Neuropathol. Commun. 2016, 4, 57. [Google Scholar] [CrossRef]
- Visser, J.; Hukin, J.; Sargent, M.; Steinbok, P.; Goddard, K.; Fryer, C. Late mortality in pediatric patients with craniopharyngioma. J. Neuro-Oncol. 2010, 100, 105–111. [Google Scholar] [CrossRef]
- Wijnen, M.; Olsson, D.S.; Heuvel-Eibrink, M.M.v.D.; Hammarstrand, C.; Janssen, J.A.M.J.L.; van der Lely, A.J.; Johannsson, G.; Neggers, S.J.C.M.M. Excess morbidity and mortality in patients with craniopharyngioma: A hospital-based retrospective cohort study. Eur. J. Endocrinol. 2018, 178, 93–102. [Google Scholar] [CrossRef]
- Wu, J.; Wu, X.; Yang, Y.Q.; Ding, H.; Yang, L.; Bao, Y.Y.; Zhou, L.; Yang, C.X.; Hong, T. Association of histological subtype with risk of recurrence in craniopharyngioma patients: A systematic review and meta-analysis. Neurosurg. Rev. 2022, 45, 139–150. [Google Scholar] [CrossRef]
- Buslei, R.; Nolde, M.; Hofmann, B.; Meissner, S.; Eyupoglu, I.Y.; Siebzehnrübl, F.; Hahnen, E.; Kreutzer, J.; Fahlbusch, R. Common mutations of β-catenin in adamantinomatous craniopharyngiomas but not in other tumours originating from the sellar region. Acta Neuropathol. 2005, 109, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Shibata, T.; Kokubu, A.; Morishita, Y.; Noguchi, M.; Nakanishi, Y.; Sakamoto, M.; Hirohashi, S. Craniopharyngiomas of adamantinomatous type harbor β-catenin gene mutations. Am. J. Pathol. 2002, 161, 1997–2001. [Google Scholar] [CrossRef]
- Hengartner, A.C.; Prince, E.; Vijmasi, T.; Hankinson, T.C. Adamantinomatous craniopharyngioma: Moving toward targeted therapies. Neurosurg. Focus 2020, 48, E7. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Barbera, J.P.; Andoniadou, C.L. Biological Behaviour of Craniopharyngiomas. Neuroendocrinology 2020, 110, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Yang, J.; Liang, R.; Zan, X.; Fan, R.; Shan, B.; Liu, H.; Li, L.; Wang, Y.; Wu, M.; et al. Single-cell RNA sequencing highlights intratumor heterogeneity and intercellular network featured in adamantinomatous craniopharyngioma. Sci. Adv. 2023, 9, eadc8933. [Google Scholar] [CrossRef]
- Stribbling, S.M.; Ryan, A.J. The cell-line-derived subcutaneous tumor model in preclinical cancer research. Nat. Protoc. 2022, 17, 2108–2128. [Google Scholar] [CrossRef]
- Bleijs, M.; van de Wetering, M.; Clevers, H.; Drost, J. Xenograft and organoid model systems in cancer research. EMBO J. 2019, 38, e101654. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, C.; Fan, J.; Peng, J.; Pan, J.; Qi, S.; Liu, Y. Feasibility of primary human cell cultures as a model for adamantinomatous craniopharyngioma research: Evidence from RNA-Seq analysis. Oncol. Lett. 2020, 19, 2346–2354. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Guo, R.; Cao, Y.; Pan, J.; Yu, H.; Qiu, X.; Shi, J.; Fan, J.; Qi, S.; et al. Establishment and characterisation of STAM4, a novel human adamantinomatous craniopharyngioma cell line, through human telomerase reverse transcriptase ectopic expression-mediated immortalisation. Neuropathol. Appl. Neurobiol. 2024, 50, e12958. [Google Scholar] [CrossRef]
- Tang, M.; Chen, C.; Wang, G.; Wang, Y.; Zhang, Z.; Li, H.; Lu, Q.; Wang, Z.; Zhao, S.; Yang, C.; et al. Evaluation of B7-H3 targeted immunotherapy in a 3D organoid Model of craniopharyngioma. Biomolecules 2022, 12, 1744. [Google Scholar] [CrossRef]
- Hölsken, A.; Schwarz, M.; Gillmann, C.; Pfister, C.; Uder, M.; Doerfler, A.; Buchfelder, M.; Schlaffer, S.; Fahlbusch, R.; Buslei, R.; et al. Characterization of the murine orthotopic adamantinomatous craniopharyngioma PDX model by MRI in correlation with histology. PLoS ONE 2018, 13, e0197895. [Google Scholar] [CrossRef] [PubMed]
- Andoniadou, C.L.; Matsushima, D.; Gharavy, S.N.M.; Signore, M.; Mackintosh, A.I.; Schaeffer, M.; Gaston-Massuet, C.; Mollard, P.; Jacques, T.S.; Le Tissier, P.; et al. Sox2+ stem/progenitor Cells in the adult mouse pituitary support organ homeostasis and have tumor-inducing potential. Cell Stem Cell 2013, 13, 433–445. [Google Scholar] [CrossRef] [PubMed]
- Gaston-Massuet, C.; Andoniadou, C.L.; Signore, M.; Jayakody, S.A.; Charolidi, N.; Kyeyune, R.; Vernay, B.; Jacques, T.S.; Taketo, M.M.; Le Tissier, P.; et al. Increased Wingless (Wnt) signaling in pituitary progenitor/stem cells gives rise to pituitary tumors in mice and humans. Proc. Natl. Acad. Sci. USA 2011, 108, 11482–11487. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, X.; Ainiwan, Y.; Li, M.; Pan, J.; Chen, Y.; Xiao, Z.; Wang, Z.; Xiao, X.; Tang, J.; et al. Axl as a potential therapeutic target for adamantinomatous craniopharyngiomas: Based on single nucleus RNA-seq and spatial transcriptome profiling. Cancer Lett. 2024, 592, 216905. [Google Scholar] [CrossRef]
- Kehrberg, R.J.; Bhyravbhatla, N.; Batra, S.K.; Kumar, S. Epigenetic regulation of cancer-associated fibroblast heterogeneity. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2023, 1878, 188901. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K. FAK-Dependent Cell Motility and Cell Elongation. Cells 2020, 9, 192. [Google Scholar] [CrossRef]
- Kuburich, N.A.; Hollander, P.D.; Pietz, J.T.; Mani, S.A. Vimentin and cytokeratin: Good alone, bad together. Semin. Cancer Biol. 2022, 86, 816–826. [Google Scholar] [CrossRef]
- Coelho-Rato, L.S.; Parvanian, S.; Modi, M.K.; Eriksson, J.E. Vimentin at the core of wound healing. Trends Cell Biol. 2024, 34, 239–254. [Google Scholar] [CrossRef]
- Qi, S.; Zhou, J.; Pan, J.; Zhang, C.; Silky, C.; Yan, X. Epithelial–mesenchymal transition and clinicopathological correlation in craniopharyngioma. Histopathology 2012, 61, 711–725. [Google Scholar] [CrossRef]
- Deng, H.; Lei, T.; Liu, S.; Hao, W.; Hu, M.; Xiang, X.; Ye, L.; Chen, D.; Li, Y.; Liu, F. Proteomics study of primary and recurrent adamantinomatous craniopharyngiomas. Clin. Proteom. 2024, 21, 29. [Google Scholar] [CrossRef]
- Chen, M.; Zheng, S.-H.; Liu, Y.; Shi, J.; Qi, S.-T. Periostin activates pathways involved in epithelial–mesenchymal transition in adamantinomatous craniopharyngioma. J. Neurol. Sci. 2016, 360, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, J.; Liu, H.; Zhao, C.; Tu, Z.; Xu, D.; Zhang, E.; Zhou, Z.; Qi, X.; Wang, X.; et al. Single-cell and spatial sequencing identifies senescent and germinal tumor cells in adamantinomatous craniopharyngiomas. Cell Biosci. 2024, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Bakir, B.; Chiarella, A.M.; Pitarresi, J.R.; Rustgi, A.K. EMT, MET, Plasticity, and Tumor Metastasis. Trends Cell Biol. 2015, 30, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.W.; Weinberg, R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat. Rev. Cancer 2021, 21, 325–338. [Google Scholar] [CrossRef]
- Akhmetkaliyev, A.; Alibrahim, N.; Shafiee, D.; Tulchinsky, E. EMT/MET plasticity in cancer and Go-or-Grow decisions in quiescence: The two sides of the same coin? Mol. Cancer 2023, 22, 90. [Google Scholar] [CrossRef]
- Sun, Y.; Jia, X.; Gao, Q.; Liu, X.; Hou, L. The ubiquitin ligase UBE4A inhibits prostate cancer progression by targeting interleukin-like EMT inducer (ILEI). IUBMB Life 2017, 69, 16–21. [Google Scholar] [CrossRef]
- Wang, Z.; Mo, Y.; Tan, Y.; Wen, Z.; Dai, Z.; Zhang, H.; Zhang, X.; Feng, S.; Liang, X.; Song, T.; et al. The ALDH Family Contributes to Immunocyte Infiltration, Proliferation and Epithelial-Mesenchymal Transformation in Glioma. Front. Immunol. 2021, 12, 756606. [Google Scholar] [CrossRef]
- Yin, W.; Zhu, H.; Tan, J.; Xin, Z.; Zhou, Q.; Cao, Y.; Wu, Z.; Wang, L.; Zhao, M.; Jiang, X.; et al. Identification of collagen genes related to immune infiltration and epithelial-mesenchymal transition in glioma. Cancer Cell Int. 2021, 21, 276. [Google Scholar] [CrossRef]
- Quan, J.; Bode, A.M.; Luo, X. ACSL family: The regulatory mechanisms and therapeutic implications in cancer. Eur. J. Pharmacol. 2021, 909, 174397. [Google Scholar] [CrossRef]
- Quan, J.; Cheng, C.; Tan, Y.; Jiang, N.; Liao, C.; Liao, W.; Cao, Y.; Luo, X. Acyl-CoA synthetase long-chain 3-mediated fatty acid oxidation is required for TGFβ1-induced epithelial-mesenchymal transition and metastasis of colorectal carcinoma. Int. J. Biol. Sci. 2022, 18, 2484–2496. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-L.; Li, Z.-F.; Cao, Q.-W.; Wang, W.-Z. SLC12A8 plays a key role in bladder cancer progression and EMT. Open Med. 2021, 16, 058–067. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, H.; Wang, S. SLC12A8 promotes proliferation, invasiveness, migration and epithelial-mesenchymal transition of bladder cancer cells by activating JAK/STAT signaling. J. South. Med. Univ. 2023, 43, 1613–1621. [Google Scholar] [CrossRef]
- Dey, A.; Varelas, X.; Guan, K.-L. Targeting the Hippo pathway in cancer, fibrosis, wound healing and regenerative medicine. Nat. Rev. Drug Discov. 2020, 19, 480–494. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Shah, S.; Brock, E.J.; Ji, K.; Mattingly, R.R. Ras and Rap1: A tale of two GTPases. Semin. Cancer Biol. 2019, 54, 29–39. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef] [PubMed]
- The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7–20. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2012, 28, 423–425. [Google Scholar] [CrossRef]
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef]
- Chun, S.; Fay, J.C. Identification of deleterious mutations within three human genomes. Genome Res. 2009, 19, 1553–1561. [Google Scholar] [CrossRef] [PubMed]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Faust, G.G.; Hall, I.M. SAMBLASTER: Fast duplicate marking and structural variant read extraction. Bioinformatics 2014, 30, 2503–2505. [Google Scholar] [CrossRef]
- Frankish, A.; Diekhans, M.; Jungreis, I.; Lagarde, J.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Armstrong, J.; Barnes, I.; et al. GENCODE 2021. Nucleic Acids Res. 2021, 49, D916–D923. [Google Scholar] [CrossRef]
- Garber, M.; Guttman, M.; Clamp, M.; Zody, M.C.; Friedman, N.; Xie, X. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 2009, 25, i54–i62. [Google Scholar] [CrossRef] [PubMed]
- Hamosh, A.; Scott, A.F.; Amberger, J.S.; Bocchini, C.A.; McKusick, V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005, 33, D514–D517. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32, D258–D261. [Google Scholar] [CrossRef]
- Huber, C.D.; Kim, B.Y.; Lohmueller, K.E. Population genetic models of GERP scores suggest pervasive turnover of constrained sites across mammalian evolution. PLoS Genet. 2020, 16, e1008827. [Google Scholar] [CrossRef]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Yang, S.; Nykamp, K.; Garcia, J.; Lincoln, S.E.; Topper, S.E. Pathogenic variant burden in the ExAC database: An empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Wright, M.W.; Brister, J.R.; Ciufo, S.; Haddad, D.; McVeigh, R.; Rajput, B.; Robbertse, B.; Smith-White, B.; Ako-Adjei, D.; et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016, 44, D733–D745. [Google Scholar] [CrossRef]
- Pio, M.G.; Siffo, S.; Scheps, K.G.; Molina, M.F.; Adrover, E.; Abelleyro, M.M.; Rivolta, C.M.; Targovnik, H.M. Curating the gnomAD database: Report of novel variants in the thyrogobulin gene using in silico bioinformatics algorithms. Mol. Cell Endocrinol. 2021, 534, 111359. [Google Scholar] [CrossRef]
- Pollard, K.S.; Hubisz, M.J.; Rosenbloom, K.R.; Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010, 20, 110–121. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Saunders, C.T.; Wong, W.S.; Swamy, S.; Becq, J.; Murray, L.J.; Cheetham, R.K. Strelka: Accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012, 28, 1811–1817. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, C.F.; Anthony, K.; Krupa, S.; Buchoff, J.; Day, M.; Hannay, T.; Buetow, K.H. PID: The Pathway Interaction Database. Nucleic Acids Res. 2009, 37, D674–D679. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.; Edwards, K.J.; Day, I.N.; Gaunt, T.R. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Steinhaus, R.; Proft, S.; Schuelke, M.; Cooper, D.N.; Schwarz, J.M.; Seelow, D. MutationTaster2021. Nucleic Acids Res. 2021, 49, W446–W451. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Heywood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD®): Optimizing its use in a clinical diagnostic or research setting. Hum Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef]
- Tarasov, A.; Vilella, A.J.; Cuppen, E.; Nijman, I.J.; Prins, P. Sambamba: Fast processing of NGS alignment formats. Bioinformatics. 2015, 31, 2032–2034. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]

| STR Loci | ACP0913 | Database: Lu-138 (Matching Rate: 73.33%) | ACP0824 | Database: Yub2503c (Matching Rate: 78.57%) |
|---|---|---|---|---|
| Amelogenin | X | X | X | X |
| CSF1PO | 11, 13 | 11, 12 | 12 | 10, 12 |
| D2S1338 | 19, 24 | 16, 19 | ||
| D3S1358 | 16, 17 | 15, 18 | ||
| D5S818 | 10 | 10, 12 | 10, 11 | 10, 11 |
| D7S820 | 8, 11 | 10, 11 | 11, 12 | 11 |
| D8S1179 | 10, 11 | 11, 12 | ||
| D13S317 | 8, 11 | 8, 11 | 8, 11 | 11, 12 |
| D16S539 | 10, 11 | 11 | 10, 12 | 10, 12 |
| D18S51 | 14 | 13, 14 | ||
| D19S433 | 13, 15.2 | 13, 15 | ||
| D21S11 | 30, 31 | 30, 31 | ||
| FGA | 22, 24 | 22 | ||
| PentaD | 11 | 8, 12 | ||
| PentaE | 12, 17 | 5, 16 | ||
| TH01 | 7, 9 | 7, 9 | 7, 9 | 7, 9 |
| TPOX | 8, 11 | 8, 11 | 9, 11 | 9, 11 |
| vWA | 18, 19 | 17, 18 | 14 | 17 |
| D6S1043 | 12, 14 | 19, 21 | ||
| D12S391 | 19, 21 | 18, 19 | ||
| D2S441 | 11, 12 | 12, 14 |
| (a) | |||||||||||
| 0913 Tumor Tissue | 0913 Primary Cells | ||||||||||
| Gene | Position | ID | HGVS_C | HGVS_P | Effect-Priority | Gene | Position | ID | HGVS_C | HGVS_P | Effect-Impact |
| CTNNB1 | chr3 | rs121913413 | exon3: c.122C > T exon4: c.101C > T | p.Thr41Ile p.Thr34Ile | Missense-High | CTNNB1 | / | / | / | / | / |
| ACSL3 | chr2 | rs145194965 | exon5: c.725C > T exon6: c.725C > T exon7: c.725C > T | p.Pro242Leu | Missense-High | ACSL3 | chr2 | rs145194965 | c.725C > T c.269C > T | p.Pro242Leu p.Pro90Leu | Missense- Moderate |
| ALDH16A1 | chr19 | rs759657072 | exon13: c.1642C > T exon14: c.1795C > T | p.Leu548Leu p.Leu599Leu | Synonymous- Low | ALDH16A1 | chr19 | rs759657072 | c.1642C > T c.1795C > T c.1306C > T | p.Leu548Leu p.Leu599Leu p.Leu436Leu | Synonymous- Low |
| COL3A1 | chr2 | rs1800255 | exon30: c.2092G > A | p.Ala698Thr | Missense-Low | COL3A1 | chr2 | rs1800255 | c.2092G > A | p.Ala698Thr | Missense- Moderate |
| chr2 | rs1516446 | exon50: c.4059T > G | p.His1353Gln | Missense-Low | chr2 | rs1516446 | c.4059T > G c.3150T > G | p.His1353Gln p.His1050Gln | Missense- Moderate | ||
| (b) | |||||||||||
| 0824 Tumor Tissue | 0824 Primary Cells | ||||||||||
| Gene | Position | ID | HGVS_C | HGVS_P | Effect-Priority | Gene | Position | ID | HGVS_C | HGVS_P | Effect-Impact |
| CTNNB1 | chr3 | rs121913399 | exon3: c.100G > A exon4: c.79G > A | p.Gly34Arg p.Gly27Arg | Missense-High | CTNNB1 | chr3 | rs2293303 | c.2340C > T | p.Asp780Asp | Synonymous- Low |
| chr3 | rs2293303 | exon15: c.2340C > T | p.Asp780Asp | Synonymous- Low | |||||||
| UBE4A | chr11 | rs782399567 | exon16: c.2552A > G exon16: c.2573A > G | p.Asn851Ser p.Asn858Ser | Missense-High | UBE4A | chr11 | rs782399567 | c.2552A > G c.2573A > G c.968A > G | p.Asn851Ser p.Asn858Ser p.Asn323Ser | Missense- Moderate |
| ALDH16A1 | chr19 | rs766834756 | exon1: c.76C > A | p.His26Asn | Missense-High | ALDH16A1 | chr19 | rs766834756 | c.76C > A | p.His26Asn | Missense- Moderate |
| chr19 | rs1320303 | exon6: c.679C > G | p.Leu227Val | Missense-Low | chr19 | rs1320303 | c.679C > G c.190C > G | p.Leu227Val p.Leu64Val | Missense- Moderate | ||
| SLC12A8 | chr3 | rs761900953 | exon9: c.1294C > T exon10: c.1294C > T | p.His432Tyr p.His432Tyr | Missense-High | SLC12A8 | chr3 | rs2981482 | c.1991G > A c.1394G > A | p.Arg664Gln p.Arg465Gln | Missense- Moderate |
| chr3 | rs2981482 | exon13: c.1991G > A exon14: c.1991G > A | p.Arg664Gln | Missense-Low | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.; Lei, T.; Wang, Y.; Yu, Z.; Liu, S.; Ye, L.; Li, W.; Yang, Q.; Jin, H.; Liu, F.; et al. Isolation and Characterization of the Adamantinomatous Craniopharyngioma Primary Cells with Cancer-Associated Fibroblast Features. Biomedicines 2025, 13, 912. https://doi.org/10.3390/biomedicines13040912
Chen D, Lei T, Wang Y, Yu Z, Liu S, Ye L, Li W, Yang Q, Jin H, Liu F, et al. Isolation and Characterization of the Adamantinomatous Craniopharyngioma Primary Cells with Cancer-Associated Fibroblast Features. Biomedicines. 2025; 13(4):912. https://doi.org/10.3390/biomedicines13040912
Chicago/Turabian StyleChen, Dongting, Ting Lei, Yulin Wang, Zaitao Yu, Siqi Liu, Ling Ye, Wanfang Li, Qin Yang, Hongtao Jin, Fangjun Liu, and et al. 2025. "Isolation and Characterization of the Adamantinomatous Craniopharyngioma Primary Cells with Cancer-Associated Fibroblast Features" Biomedicines 13, no. 4: 912. https://doi.org/10.3390/biomedicines13040912
APA StyleChen, D., Lei, T., Wang, Y., Yu, Z., Liu, S., Ye, L., Li, W., Yang, Q., Jin, H., Liu, F., & Li, Y. (2025). Isolation and Characterization of the Adamantinomatous Craniopharyngioma Primary Cells with Cancer-Associated Fibroblast Features. Biomedicines, 13(4), 912. https://doi.org/10.3390/biomedicines13040912








