Ultrasensitive Detection of Rare Mutations via Amplifying–Cleaving–Enriching in Acute Myeloid Leukemia
Abstract
1. Introduction
2. Methods
2.1. Sample Collection and DNA Extraction
2.2. Plasmid DNA Standards Preparation
2.3. Initial and Derived Cleaved Amplified Polymorphic Sequences PCR
2.4. Restriction Enzyme Cleavage
2.5. Magnetic Isolation
2.6. Sanger Sequencing
2.7. Quantitative Real-Time PCR and Allele-Specific PCR
2.8. Droplet Digital PCR Assay
2.9. Statistical Analyses
3. Results
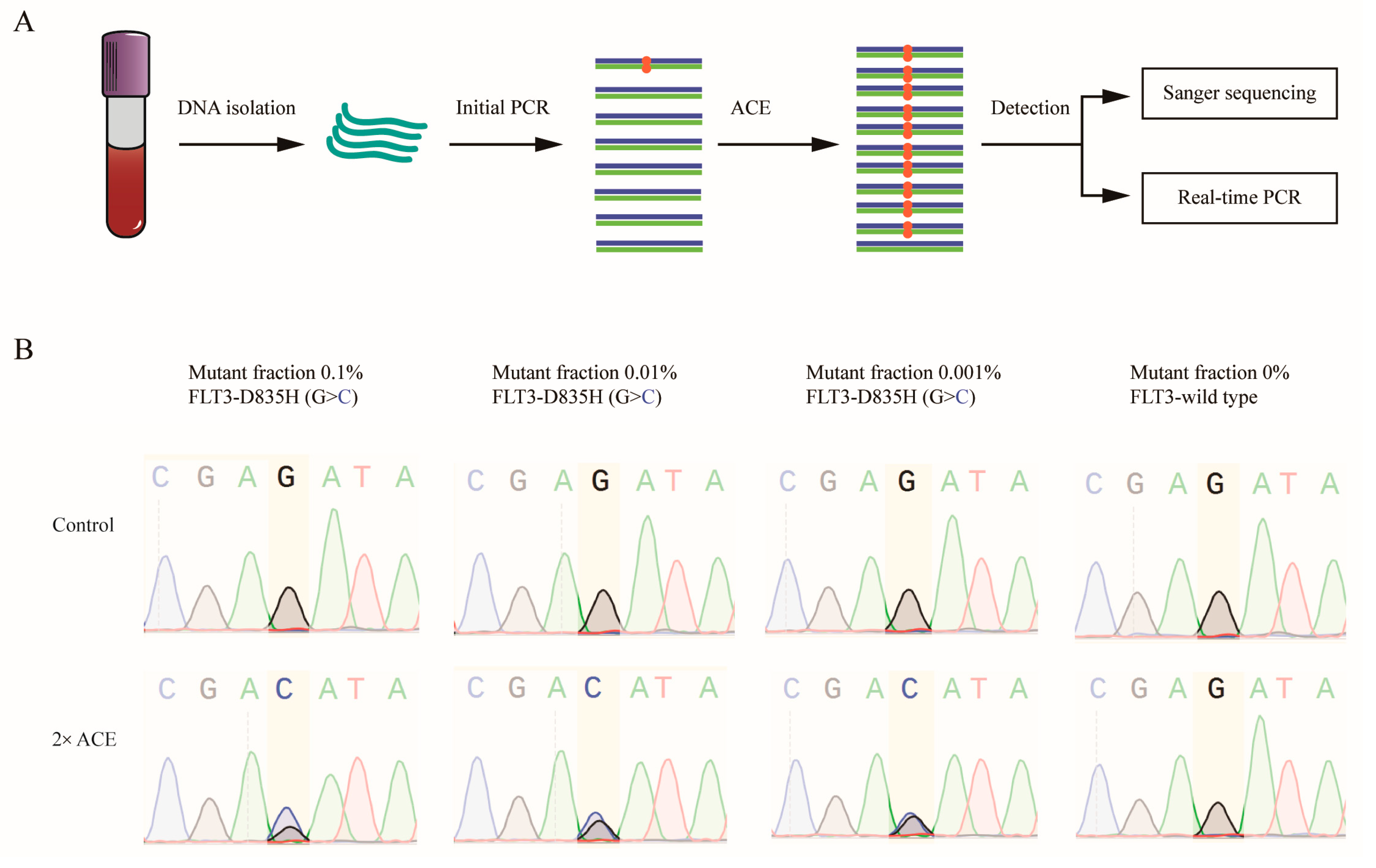
3.1. Design of Amplifying–Cleaving–Enriching (ACE)
3.2. Optimization of Streptavidin-Labeled Dynabeads
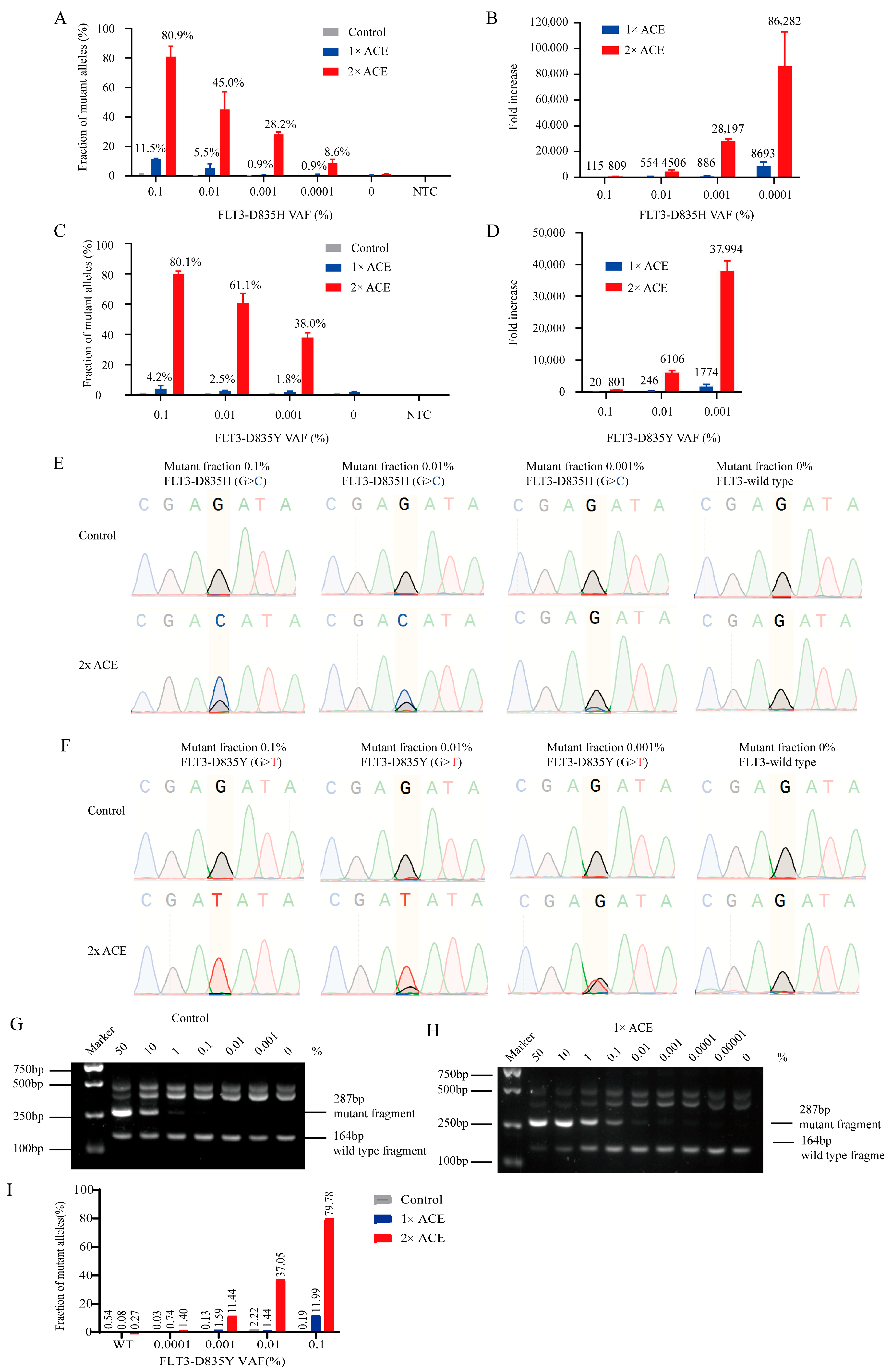
3.3. ACE Specifically Enriched Mutant Sequences of FLT3-TKD with Plasmid Standards
3.4. ACE Specifically Enriched Mutant Sequences of FLT3-TKD in Blood DNAs
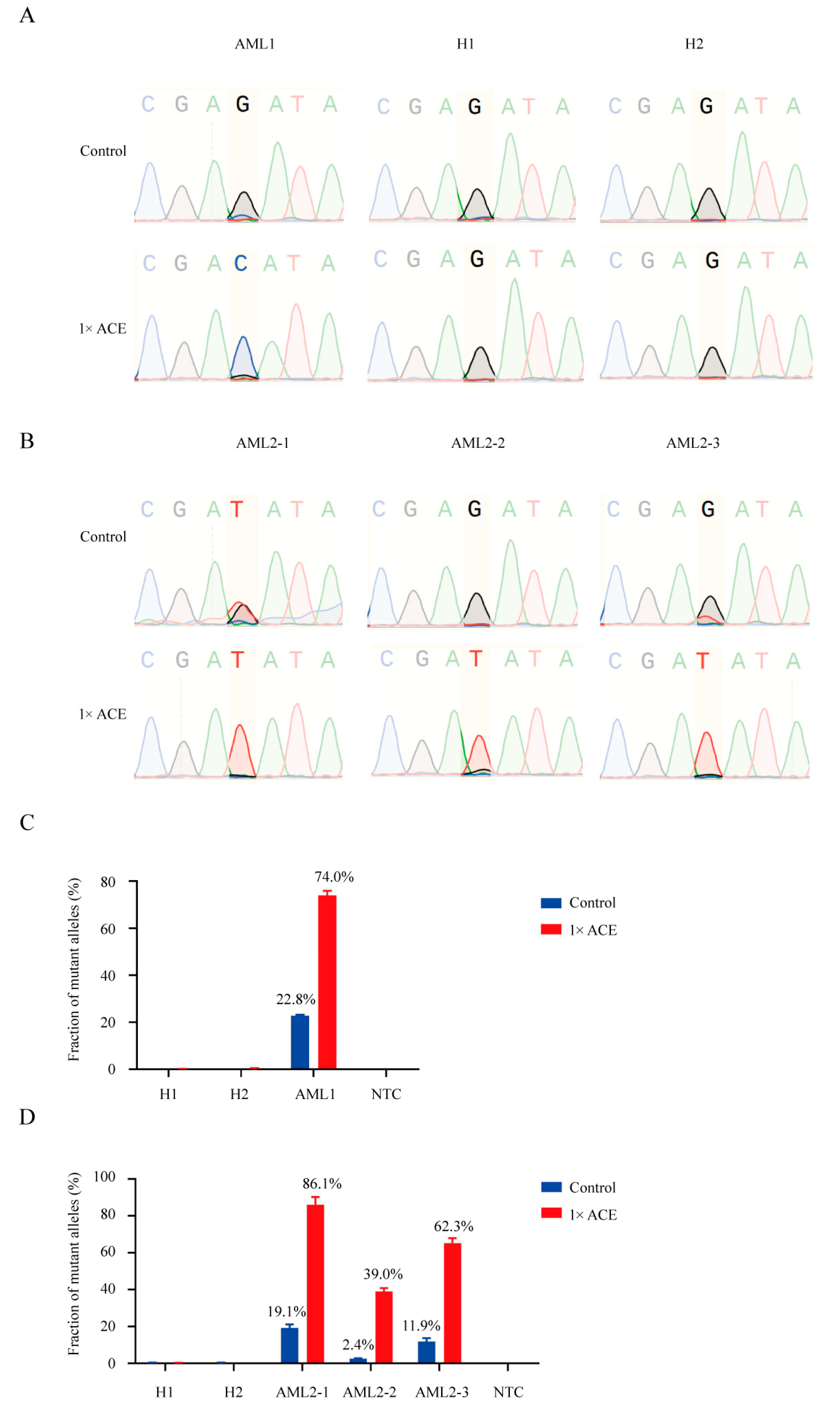
3.5. ACE Detected MRD in an AML Patient During Complete Remission
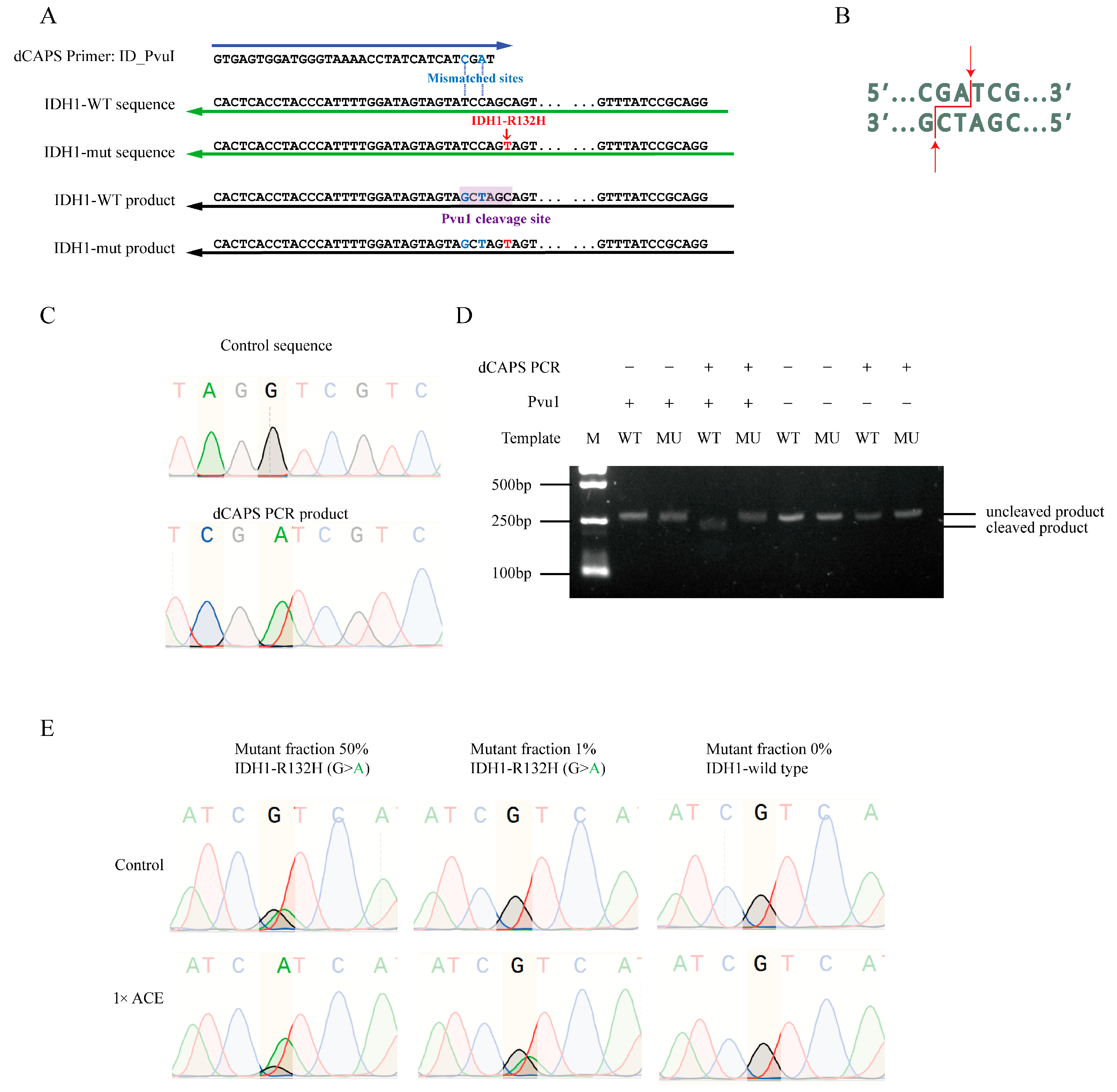
3.6. dCAPS PCR for Creation of Wild-Type-Specific Restriction Cleaving Sites
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Pan, Y.; Guo, Y.; Zhao, W.; Ho, W.T.; Wang, J.; Xu, M.; Yang, F.C.; Zhao, Z.J. Tyrosine kinase inhibitors targeting FLT3 in the treatment of acute myeloid leukemia. Stem Cell Investig. 2017, 4, 48. [Google Scholar] [CrossRef]
- Linet, M.S.; Curtis, R.E.; Schonfeld, S.J.; Vo, J.B.; Morton, L.M.; Dores, G.M. Survival of adult AML patients treated with chemotherapy in the U.S. population by age, race and ethnicity, sex, calendar-year period, and AML subgroup, 2001–2019. eClinicalMedicine 2024, 71, 102549. [Google Scholar] [CrossRef]
- Khwaja, A.; Bjorkholm, M.; Gale, R.E.; Levine, R.L.; Jordan, C.T.; Ehninger, G.; Bloomfield, C.D.; Estey, E.; Burnett, A.; Cornelissen, J.J.; et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers 2016, 2, 16010. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.K.; Chen, Y.B. Maintenance therapy for AML after allogeneic HCT. Front. Oncol. 2022, 12, 895771. [Google Scholar] [CrossRef]
- Newell, L.F.; Cook, R.J. Advances in acute myeloid leukemia. BMJ 2021, 375, n2026. [Google Scholar] [CrossRef]
- Huerga-Dominguez, S.; Villar, S.; Prosper, F.; Alfonso-Pierola, A. Updates on the Management of Acute Myeloid Leukemia. Cancers 2022, 14, 4756. [Google Scholar] [CrossRef]
- Short, N.J.; Daver, N.; Dinardo, C.D.; Kadia, T.; Nasr, L.F.; Macaron, W.; Yilmaz, M.; Borthakur, G.; Montalban-Bravo, G.; Garcia-Manero, G.; et al. Azacitidine, Venetoclax, and Gilteritinib in Newly Diagnosed and Relapsed or Refractory FLT3-Mutated AML. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2024, 42, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Levis, M.J.; Hamadani, M.; Logan, B.; Jones, R.J.; Singh, A.K.; Litzow, M.; Wingard, J.R.; Papadopoulos, E.B.; Perl, A.E.; Soiffer, R.J.; et al. Gilteritinib as Post-Transplant Maintenance for AML with Internal Tandem Duplication Mutation of FLT3. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2024, 42, 1766–1775. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.M.; Yoo, S.J.; Kim, H.J.; Ahn, J.S.; Koh, Y.; Jang, J.H.; Yoon, S.S. IDH1/2 mutations in acute myeloid leukemia. Blood Res. 2022, 57, 13–19. [Google Scholar] [CrossRef]
- Fruchtman, H.; Avigan, Z.M.; Waksal, J.A.; Brennan, N.; Mascarenhas, J.O. Management of isocitrate dehydrogenase 1/2 mutated acute myeloid leukemia. Leukemia 2024, 38, 927–935. [Google Scholar] [CrossRef]
- Zhuang, X.; Pei, H.Z.; Li, T.; Huang, J.; Guo, Y.; Zhao, Y.; Yang, M.; Zhang, D.; Chang, Z.; Zhang, Q.; et al. The Molecular Mechanisms of Resistance to IDH Inhibitors in Acute Myeloid Leukemia. Front. Oncol. 2022, 12, 931462. [Google Scholar] [CrossRef]
- Ge, S.S.; Liu, S.B.; Xue, S.L. Developments and challenges of FLT3 inhibitors in acute myeloid leukemia. Front. Oncol. 2022, 12, 996438. [Google Scholar] [CrossRef]
- Woods, A.; Norsworthy, K.J.; Wang, X.; Vallejo, J.; Chiu Yuen Chow, E.; Li, R.J.; Sun, J.; Charlab, R.; Jiang, X.; Pazdur, R.; et al. FDA Approval Summary: Ivosidenib in Combination with Azacitidine for Treatment of Patients with Newly Diagnosed Acute Myeloid Leukemia with an IDH1 Mutation. Clin. Cancer Res. An. Off. J. Am. Assoc. Cancer Res. 2024, 30, 1226–1231. [Google Scholar] [CrossRef] [PubMed]
- Cumbo, C.; Tarantini, F.; Anelli, L.; Zagaria, A.; Specchia, G.; Musto, P.; Albano, F. FLT3 mutational analysis in acute myeloid leukemia: Advantages and pitfalls with different approaches. Blood Rev. 2022, 54, 100928. [Google Scholar] [CrossRef]
- Guo, Y.; Sun, H.; Zhang, D.; Zhao, Y.; Shi, M.; Yang, M.; Xing, S.; Fu, X.; Bin, T.; Lu, B.; et al. Development of a highly sensitive method for detection of FLT3D835Y. Biomark. Res. 2020, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Guo, Y.; Zhao, W.; Tina Ho, W.T.; Fu, X.; Zhao, Z.J. Identification of an orally available compound with potent and broad FLT3 inhibition activity. Oncogene 2016, 35, 2971–2978. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, Y.; Zhao, Y.; Yu, L.; Chang, Z.; Pei, H.; Huang, J.; Chen, C.; Xue, H.; Xu, X.; et al. Expression of a recombinant FLT3 ligand and its emtansine conjugate as a therapeutic candidate against acute myeloid leukemia cells with FLT3 expression. Microb. Cell Fact. 2021, 20, 67. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, Y.; Xu, X.; Fu, X.; Zhao, Z.J. SU11652 Inhibits tyrosine kinase activity of FLT3 and growth of MV-4-11 cells. J. Hematol. Oncol. 2012, 5, 72. [Google Scholar] [CrossRef]
- Ge, S.S.; Qiu, Q.C.; Dai, H.P.; Shen, X.D.; Wu, T.M.; Du, J.H.; Wan, C.L.; Shen, H.J.; Wu, D.P.; Xue, S.L.; et al. Mutation spectrum of FLT3 and significance of non-canonical FLT3 mutations in haematological malignancy. Br. J. Haematol. 2023, 202, 539–549. [Google Scholar] [CrossRef]
- Smith, C.C.; Wang, Q.; Chin, C.S.; Salerno, S.; Damon, L.E.; Levis, M.J.; Perl, A.E.; Travers, K.J.; Wang, S.; Hunt, J.P.; et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature 2012, 485, 260–263. [Google Scholar] [CrossRef]
- Study Offers Insights on Prognostic Significance of IDH Mutations Across Age Groups in AML. Oncologist 2021, 26 (Suppl. S1), S13–S14. [CrossRef]
- Marcucci, G.; Maharry, K.; Wu, Y.Z.; Radmacher, M.D.; Mrozek, K.; Margeson, D.; Holland, K.B.; Whitman, S.P.; Becker, H.; Schwind, S.; et al. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: A Cancer and Leukemia Group B study. J. Clin. Oncol. 2010, 28, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.P.; Gonen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- Green, C.L.; Evans, C.M.; Zhao, L.; Hills, R.K.; Burnett, A.K.; Linch, D.C.; Gale, R.E. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood 2011, 118, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Heuser, M.; Freeman, S.D.; Ossenkoppele, G.J.; Buccisano, F.; Hourigan, C.S.; Ngai, L.L.; Tettero, J.M.; Bachas, C.; Baer, C.; Béné, M.C.; et al. 2021 Update on MRD in acute myeloid leukemia: A consensus document from the European LeukemiaNet MRD Working Party. Blood 2021, 138, 2753–2767. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Wei, A.H.; Iland, H.J.; DiNardo, C.D.; Reynolds, J. MRD intervention in AML: A new therapeutic horizon. Blood, 2025; in press. [Google Scholar] [CrossRef]
- Orvain, C.; Ali, N.; Othus, M.; Rodríguez-Arbolí, E.; Milano, F.; Le, C.M.; Sandmaier, B.M.; Scott, B.L.; Appelbaum, F.R.; Walter, R.B. Relative prognostic value of flow cytometric measurable residual disease before allogeneic hematopoietic cell transplantation for adults with MDS/AML or AML. Am. J. Hematol. 2024, 99, 862–870. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Janne, P.A.; Makrigiorgos, G.M. Coamplification at lower denaturation temperature-PCR increases mutation-detection selectivity of TaqMan-based real-time PCR. Clin. Chem. 2009, 55, 748–756. [Google Scholar] [CrossRef]
- Wu, L.R.; Chen, S.X.; Wu, Y.; Patel, A.A.; Zhang, D.Y. Multiplexed enrichment of rare DNA variants via sequence-selective and temperature-robust amplification. Nat. Biomed. Eng. 2017, 1, 714–723, Erratum in Nat. Biomed. Eng. 2017, 1, 1005. https://doi.org/10.1038/s41551-017-0156-z. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef]
- He, R.; Wang, L.; Wang, F.; Li, W.; Liu, Y.; Li, A.; Wang, Y.; Mao, W.; Zhai, C.; Ma, L. Pyrococcus furiosus Argonaute-mediated nucleic acid detection. Chem. Commun. 2019, 55, 13219–13222. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Yu, J.; Hwang, G.H.; Kim, S.; Kim, H.S.; Ye, S.; Kim, K.; Park, J.; Park, D.Y.; Cho, Y.K.; et al. CUT-PCR: CRISPR-mediated, ultrasensitive detection of target DNA using PCR. Oncogene 2017, 36, 6823–6829. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Hegge, J.W.; Mauk, M.G.; Chen, J.; Till, J.E.; Bhagwat, N.; Azink, L.T.; Peng, J.; Sen, M.; Mays, J.; et al. Highly specific enrichment of rare nucleic acid fractions using Thermus thermophilus argonaute with applications in cancer diagnostics. Nucleic Acids Res. 2020, 48, e19. [Google Scholar] [CrossRef]
- Liu, Q.; Guo, X.; Xun, G.; Li, Z.; Chong, Y.; Yang, L.; Wang, H.; Zhang, F.; Luo, S.; Cui, L.; et al. Argonaute integrated single-tube PCR system enables supersensitive detection of rare mutations. Nucleic Acids Res. 2021, 49, e75. [Google Scholar] [CrossRef] [PubMed]
- Blombery, P.; Anderson, M.A.; Gong, J.N.; Thijssen, R.; Birkinshaw, R.W.; Thompson, E.R.; Teh, C.E.; Nguyen, T.; Xu, Z.; Flensburg, C.; et al. Acquisition of the Recurrent Gly101Val Mutation in BCL2 Confers Resistance to Venetoclax in Patients with Progressive Chronic Lymphocytic Leukemia. Cancer Discov. 2019, 9, 342–353. [Google Scholar] [CrossRef]
- Winters, A.C.; Minhajuddin, M.; Stevens, B.M.; Major, A.; Bosma, G.; Abbott, D.; Miltgen, N.; Yuan, J.; Treece, A.L.; Siegele, B.J.; et al. Multi-gene measurable residual disease assessed by digital polymerase chain reaction has clinical and biological utility in acute myeloid leukemia patients receiving venetoclax/azacitidine. Haematologica 2024, 109, 1766–1778. [Google Scholar] [CrossRef]
- Chen, W.; Huang, J.; Zhao, Y.; Huang, L.; Yuan, Z.; Gu, M.; Xu, X.; Shi, J.; Luo, Y.; Yu, J.; et al. Measurable residual disease monitoring by ddPCR in the early posttransplant period complements the traditional MFC method to predict relapse after HSCT in AML/MDS: A multicenter retrospective study. J. Transl. Med. 2024, 22, 410. [Google Scholar] [CrossRef]
- Li, E.W.; Tran, N.Y.K.; McCulloch, D.; Krigstein, M.; Catalano, A.; Othman, J.; Abadir, E.; Smith, C.; Iland, H. FLT3-TKD Measurable Residual Disease Detection Using Droplet Digital PCR and Clinical Applications in Acute Myeloid Leukemia. Int. J. Mol. Sci. 2024, 25, 5771. [Google Scholar] [CrossRef]
- Kim, D.; Kim, S.; Song, H.; Gwak, D.; Min, S.; Byun, J.M.; Koh, Y.; Hong, J.; Yoon, S.S.; Yun, H.; et al. Pursuing dynamics of minimal residual leukemic subclones in relapsed and refractory acute myeloid leukemia during conventional therapy. Cancer Med. 2024, 13, e7182. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.C.; Shimony, S.; Luskin, M.R.; Stone, R.M. Biology and Management of Acute Myeloid Leukemia with Mutated NPM1. Am. J. Hematol. 2025, 100, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Maier, J.; Lange, T.; Cross, M.; Wildenberger, K.; Niederwieser, D.; Franke, G.N. Optimized Digital Droplet PCR for BCR-ABL. J. Mol. Diagn. 2019, 21, 27–37. [Google Scholar] [CrossRef] [PubMed]
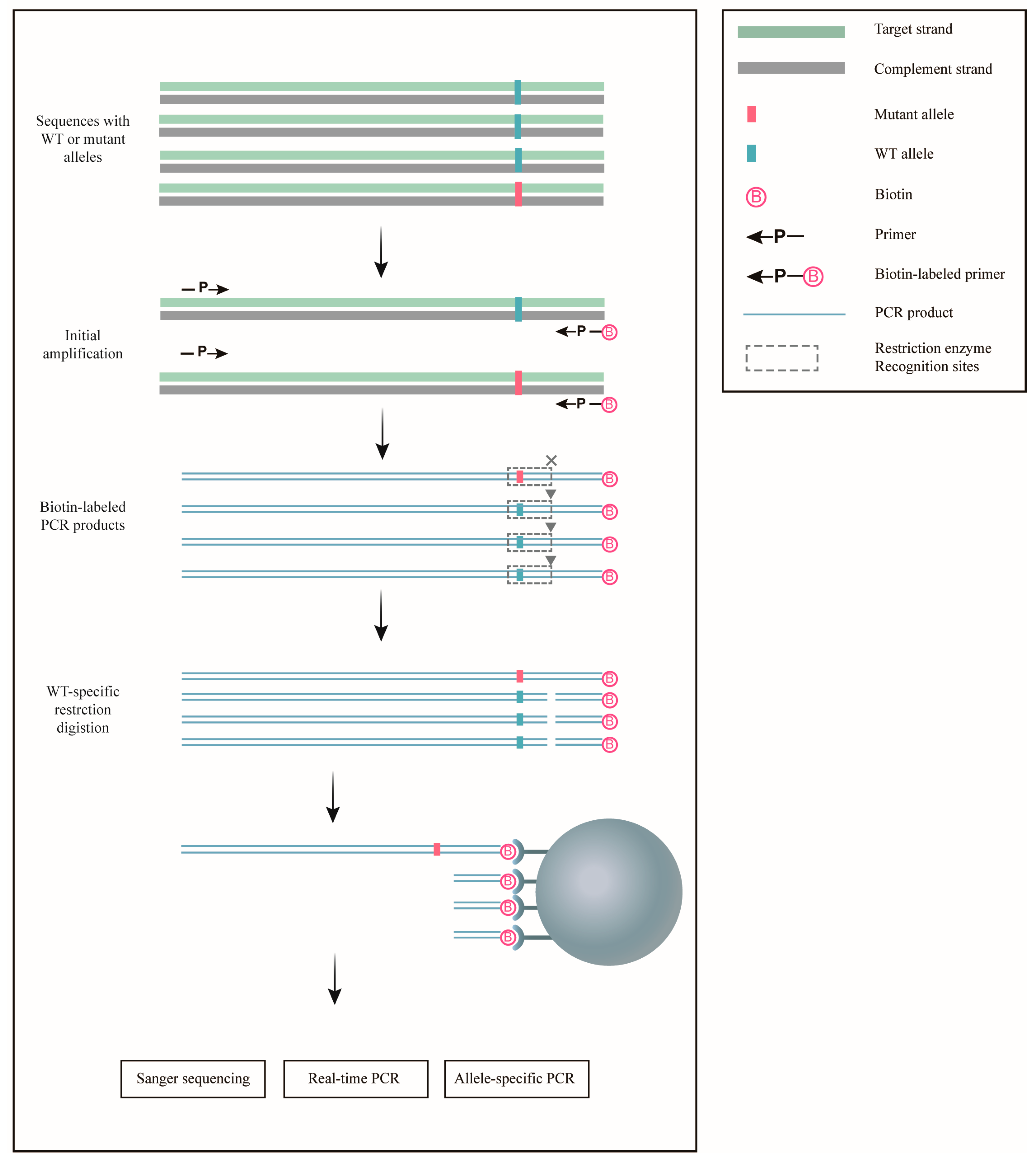
| Genes | Amino Acid Changes | DNA Changes | Original Sequences | Derived Sequences | Enzymes | dCAPS Primer (Red Labels Indicate Modifications from the Original Sequences) |
|---|---|---|---|---|---|---|
| BRAF | V600E | chr7: g.140753336A>T | ACAGT | ACTAGT | SpeI-HF | AAAAATAGGTGATTTTGGTCTAGCTACTAG |
| KRAS | G12D | chr12: g.25245350C>T | GCTGGT | ACCGGT | AgeI-HF | CTGAATATAAACTTGTGGTAGTTGGAACCG |
| NRAS | Q61R | chr1: g.114713908T>C | TGGACA | TGTACA | BsrGI-HF | GTTTGTTGGACATACTGGATACAGCTGTAC |
| EGFR | L858R | chr7: g.55191822T>G | TGGCCA | - | MscI | - |
| EGFR | T790M | chr7: g.55181378C>T | GCTCATCAC | GTGCATCAC | DraIII-HF | TCTGCCTCACCTCCACCGTGCAGTGCATCA |
| JAK2 | V617F | chr9: g.5073770G>T | GGAGTATGTGTCTGT | - | BsaXI | - |
| KIT | D816V | chr4: g.54733155A>T | GAGAC | - | BsmAI | - |
| KIT | N822K | chr4: g.54733174T>G | TTATGT | TTATAA | SspI-HF | ATGGGTACTCACGTTTCCTTTAACCTTATA (reversed) |
| PTPN11 | E76K | chr12: g.112450406G>A | TGGCTG | CAGCTG | PvuII-HF | TATGGAGGGGAGAAATTTGCCACTTCAGCT |
| PTPN11 | D61Y | chr12: g.112450361G>T | CACTGGTG | CACTGTGTG | DraIII-HF | TCACCCACATCAAGATTCAGAACACTGTGT |
| Method | Nuclease | Sensitivity | Specificity | Enrichment Efficiency |
|---|---|---|---|---|
| COLD-PCR [29] | - | 0.01% with NGS | Medium | 500-fold |
| BDA [30] | - | 0.01% with qPCR | Medium | 10,000-fold |
| HOLMESv2 [31] | Cas12 | 0.1% with fluorescence detection | High | - |
| SHERLOCKv2 [32] | Cas12, Cas13 and Csm6 | 0.6% with fluorescence detection | High | - |
| PAND [33] | PfAgo | 0.1% with fluorescence detection | High | - |
| Cut-PCR [34] | Cas9 | 0.01% with NGS | High | 18-fold |
| NAVIGATER [35] | TtAgo | 0.01% with XNA-PCR | High | 60-fold (0.5%VAF) |
| A-Star [36] | PfAgo | 0.01% with TaqMan qRT-PCR | High | 5500-fold |
| ACE | Restriction Endonucleases | 0.0001% with Real-Time PCR/Sanger sequence/AS-PCR/ddPCR | High | 80,000-fold |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhuang, X.; Ma, L.; Yu, L.; Zhao, Y.; Zhang, D.; Li, C.; Liu, C.; Xiao, Y.; Chang, Z.; Li, S.; et al. Ultrasensitive Detection of Rare Mutations via Amplifying–Cleaving–Enriching in Acute Myeloid Leukemia. Biomedicines 2025, 13, 3026. https://doi.org/10.3390/biomedicines13123026
Zhuang X, Ma L, Yu L, Zhao Y, Zhang D, Li C, Liu C, Xiao Y, Chang Z, Li S, et al. Ultrasensitive Detection of Rare Mutations via Amplifying–Cleaving–Enriching in Acute Myeloid Leukemia. Biomedicines. 2025; 13(12):3026. https://doi.org/10.3390/biomedicines13123026
Chicago/Turabian StyleZhuang, Xiaomei, Lingling Ma, Liuting Yu, Yuming Zhao, Dengyang Zhang, Chunmou Li, Chaoxing Liu, Yan Xiao, Zhiguang Chang, Shuping Li, and et al. 2025. "Ultrasensitive Detection of Rare Mutations via Amplifying–Cleaving–Enriching in Acute Myeloid Leukemia" Biomedicines 13, no. 12: 3026. https://doi.org/10.3390/biomedicines13123026
APA StyleZhuang, X., Ma, L., Yu, L., Zhao, Y., Zhang, D., Li, C., Liu, C., Xiao, Y., Chang, Z., Li, S., Chen, C., Chen, Y., Zhou, G., Zhao, Z. J., & Guo, Y. (2025). Ultrasensitive Detection of Rare Mutations via Amplifying–Cleaving–Enriching in Acute Myeloid Leukemia. Biomedicines, 13(12), 3026. https://doi.org/10.3390/biomedicines13123026







