Bulk RNAseq Analysis of Cardiac Myosin-Specific CD4+ and CD8+ T Cells Reveals Distinct Transcriptomic Profiles Between Myocarditis-Resistant and Susceptible Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Isolation of T lymphocytes and Sorting of CD4+ and CD8+ T Cells by Flow Cytometry
2.3. RNA Isolation and Bulk RNA Sequencing
2.4. Transcriptomic Analysis
2.5. Differentially Expressed Genes (DEGs) Analysis
2.6. Enrichment Analysis
2.7. Real-Time qPCR
2.8. TF Network Analysis
2.9. Statistical Analysis
3. Results and Discussion
3.1. Expression of Transcriptomic Profiles Differs between C57BL/6 and A/J Mice in CD4+ and CD8+ T Cells
3.2. DEG Analysis Revealed Overlapping Transcripts between CD4+ and CD8+ T Cells, but Their Expression Differed
3.3. GO and KEGG Enrichment Analyses Revealed Biological and Metabolic Pathways Common to CD4+ and CD8+ T Cells
3.4. GSEA-Enrichment Analyses Revealed Distinct Pathways in CD4+ and CD8+ T Cells
3.5. TF Network Analysis Revealed Regulatory Circuits of Importance to T cell Responses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATD | Autoimmune thyroid disease |
| Bmf | Bcl-2 modifying factor |
| Ct | Cycle threshold |
| Cxcr2 | C-X-C Motif Chemokine Receptor 2 |
| Cyp2r1 | Cytochrome P450 Family 2 Subfamily R Member 1 |
| DEG | Differentially expressed genes |
| EAE | Experimental autoimmune encephalomyelitis |
| EBV | Epstein–Barr virus |
| ES | Enrichment score |
| FDR | False discovery rate |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase |
| GO | Gene Ontology |
| GRN | Gene regulatory networks |
| GSEA | Gene set enrichment analysis |
| GVHD | Graft-versus-host disease |
| GzmK | Granzyme K |
| HSV | Herpes simplex virus |
| IFN | Interferon |
| IL | Interleukin |
| Il12rb2 | Interleukin 12 receptor subunit beta 2 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MHC | Major histocompatibility complex |
| Myhc | Myosin heavy chain |
| NES | Normalized enrichment score |
| Padj | Adjusted P-value |
| PCA | Principal component analysis |
| qPCR | Quantitative polymerase chain reaction |
| T1D | Type 1 diabetes |
| TCR | T cell receptor |
| TF | Transcription factor |
| Tg | Transgenic |
| Treg | T regulatory |
References
- Yates, A.J. Theories and quantification of thymic selection. Front. Immunol. 2014, 5, 13. [Google Scholar] [CrossRef]
- Bains, I.; van Santen, H.M.; Seddon, B.; Yates, A.J. Models of self-peptide sampling by developing T cells identify candidate mechanisms of thymic selection. PLoS Comput. Biol. 2013, 9, e1003102. [Google Scholar] [CrossRef]
- Moran, A.E.; Hogquist, K.A. T-cell receptor affinity in thymic development. Immunology 2012, 135, 261–267. [Google Scholar] [CrossRef]
- Klein, L.; Kyewski, B.; Allen, P.M.; Hogquist, K.A. Positive and negative selection of the T cell repertoire: What thymocytes see (and don’t see). Nat. Rev. Immunol. 2014, 14, 377–391. [Google Scholar] [CrossRef]
- Qi, Y.; Zhang, R.; Lu, Y.; Zou, X.; Yang, W. Aire and Fezf2, two regulators in medullary thymic epithelial cells, control autoimmune diseases by regulating TSAs: Partner or complementer? Front. Immunol. 2022, 13, 948259. [Google Scholar] [CrossRef]
- Naito, T.; Tanaka, H.; Naoe, Y.; Taniuchi, I. Transcriptional control of T-cell development. Int. Immunol. 2011, 23, 661–668. [Google Scholar] [CrossRef]
- Vaillant, F.; Blyth, K.; Andrew, L.; Neil, J.C.; Cameron, E.R. Enforced expression of Runx2 perturbs T cell development at a stage coincident with beta-selection. J. Immunol. 2002, 169, 2866–2874. [Google Scholar] [CrossRef]
- Lv, H.; Havari, E.; Pinto, S.; Gottumukkala, R.V.; Cornivelli, L.; Raddassi, K.; Matsui, T.; Rosenzweig, A.; Bronson, R.T.; Smith, R.; et al. Impaired thymic tolerance to alpha-myosin directs autoimmunity to the heart in mice and humans. J. Clin. Investig. 2011, 121, 1561–1573. [Google Scholar] [CrossRef]
- Xing, Y.; Hogquist, K.A. T-cell tolerance: Central and peripheral. Cold Spring Harb. Perspect. Biol. 2012, 4, a006957. [Google Scholar] [CrossRef]
- Bour-Jordan, H.; Esensten, J.H.; Martinez-Llordella, M.; Penaranda, C.; Stumpf, M.; Bluestone, J.A. Intrinsic and extrinsic control of peripheral T-cell tolerance by costimulatory molecules of the CD28/ B7 family. Immunol. Rev. 2011, 241, 180–205. [Google Scholar] [CrossRef]
- Green, D.R.; Droin, N.; Pinkoski, M. Activation-induced cell death in T cells. Immunol. Rev. 2003, 193, 70–81. [Google Scholar] [CrossRef]
- Nurieva, R.I.; Liu, X.; Dong, C. Molecular mechanisms of T-cell tolerance. Immunol. Rev. 2011, 241, 133–144. [Google Scholar] [CrossRef]
- Abbas, A.K.; Lohr, J.; Knoechel, B.; Nagabhushanam, V. T cell tolerance and autoimmunity. Autoimmun. Rev. 2004, 3, 471–475. [Google Scholar] [CrossRef]
- Malhotra, D.; Linehan, J.L.; Dileepan, T.; Lee, Y.J.; Purtha, W.E.; Lu, J.V.; Nelson, R.W.; Fife, B.T.; Orr, H.T.; Anderson, M.S.; et al. Tolerance is established in polyclonal CD4(+) T cells by distinct mechanisms, according to self-peptide expression patterns. Nat. Immunol. 2016, 17, 187–195. [Google Scholar] [CrossRef]
- Lutz, M.B.; Kurts, C. Induction of peripheral CD4+ T-cell tolerance and CD8+ T-cell cross-tolerance by dendritic cells. Eur. J. Immunol. 2009, 39, 2325–2330. [Google Scholar] [CrossRef]
- Sur, M.; Rasquinha, M.T.; Arumugam, R.; Massilamany, C.; Gangaplara, A.; Mone, K.; Lasrado, N.; Yalaka, B.; Doiphode, A.; Gurumurthy, C.; et al. Transgenic Mice Expressing Functional TCRs Specific to Cardiac Myhc-alpha 334-352 on Both CD4 and CD8 T Cells Are Resistant to the Development of Myocarditis on C57BL/6 Genetic Background. Cells 2023, 12, 2346. [Google Scholar] [CrossRef]
- Sur, M.; Rasquinha, M.T.; Mone, K.; Massilamany, C.; Lasrado, N.; Gurumurthy, C.; Sobel, R.A.; Reddy, J. Investigation into Cardiac Myhc-alpha 334-352-Specific TCR Transgenic Mice Reveals a Role for Cytotoxic CD4 T Cells in the Development of Cardiac Autoimmunity. Cells 2024, 13, 234. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Pola-Sanchez, E.; Hernandez-Martinez, K.M.; Perez-Estrada, R.; Selem-Mojica, N.; Simpson, J.; Abraham-Juarez, M.J.; Herrera-Estrella, A.; Villalobos-Escobedo, J.M. RNA-Seq Data Analysis: A Practical Guide for Model and Non-Model Organisms. Curr. Protoc. 2024, 4, e1054. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Oshlack, A.; Robinson, M.D.; Young, M.D. From RNA-seq reads to differential expression results. Genome Biol. 2010, 11, 220. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, M.C.; Konate, M.M.; Chen, L.; Das, B.; Karlovich, C.; Williams, P.M.; Evrard, Y.A.; Doroshow, J.H.; McShane, L.M. TPM, FPKM, or Normalized Counts? A Comparative Study of Quantification Measures for the Analysis of RNA-seq Data from the NCI Patient-Derived Models Repository. J. Transl. Med. 2021, 19, 269. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Lasrado, N.; Gangaplara, A.; Massilamany, C.; Arumugam, R.; Shelbourn, A.; Rasquinha, M.T.; Basavalingappa, R.H.; Delhon, G.; Xiang, S.H.; Pattnaik, A.K.; et al. Attenuated strain of CVB3 with a mutation in the CAR-interacting region protects against both myocarditis and pancreatitis. Sci. Rep. 2021, 11, 12432. [Google Scholar] [CrossRef]
- Mone, K.; Singh, S.; Abdullatif, F.; Sur, M.; Rasquinha, M.T.; Seravalli, J.; Zinniel, D.K.; Mukhopadhyay, I.; Barletta, R.G.; Gebregiworgis, T.; et al. Immunization with Complete Freund’s Adjuvant Reveals Trained Immunity-like Features in A/J Mice. Vaccines 2025, 13, 768. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E.; et al. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Liu, Z.P.; Wu, C.; Miao, H.; Wu, H. RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database 2015, 2015, bav095. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Zhao, B.; Erwin, A.; Xue, B. How many differentially expressed genes: A perspective from the comparison of genotypic and phenotypic distances. Genomics 2018, 110, 67–73. [Google Scholar] [CrossRef]
- Raphael, I.; Joern, R.R.; Forsthuber, T.G. Memory CD4(+) T Cells in Immunity and Autoimmune Diseases. Cells 2020, 9, 531. [Google Scholar] [CrossRef]
- Dittel, B.N. CD4 T cells: Balancing the coming and going of autoimmune-mediated inflammation in the CNS. Brain Behav. Immun. 2008, 22, 421–430. [Google Scholar] [CrossRef]
- Tennakoon, D.K.; Mehta, R.S.; Ortega, S.B.; Bhoj, V.; Racke, M.K.; Karandikar, N.J. Therapeutic induction of regulatory, cytotoxic CD8+ T cells in multiple sclerosis. J. Immunol. 2006, 176, 7119–7129. [Google Scholar] [CrossRef]
- Guo, C.L.; Wang, C.S.; Wang, X.H.; Yu, D.; Liu, Z. GZMK(+)CD8(+) T cells: Multifaceted roles beyond cytotoxicity. Trends Immunol. 2025, 46, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, R.; Kambara, H.; Ma, F.; Luo, H.R. The role of CXCR2 in acute inflammatory responses and its antagonists as anti-inflammatory therapeutics. Curr. Opin. Hematol. 2019, 26, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wu, Q.; Shi, J.; Guo, W.; Jiang, X.; Zhou, B.; Ren, C. LILRB4, from the immune system to the disease target. Am. J. Transl. Res. 2020, 12, 3149–3166. [Google Scholar] [PubMed]
- Renoux, F.; Stellato, M.; Haftmann, C.; Vogetseder, A.; Huang, R.; Subramaniam, A.; Becker, M.O.; Blyszczuk, P.; Becher, B.; Distler, J.H.W.; et al. The AP1 Transcription Factor Fosl2 Promotes Systemic Autoimmunity and Inflammation by Repressing Treg Development. Cell Rep. 2020, 31, 107826. [Google Scholar] [CrossRef]
- Barragan-Galvez, J.C.; Hernandez-Flores, A.; Lopez-Ortega, O.; Rodriguez-Alvarez, A.A.; Maravillas-Montero, J.L.; Ortiz-Navarrete, V. The constant domain of CRTAM is essential for high-affinity interaction with Nectin-like 2. Biochem. Biophys. Rep. 2024, 39, 101813. [Google Scholar] [CrossRef]
- Hermans, D.; van Beers, L.; Broux, B. Nectin Family Ligands Trigger Immune Effector Functions in Health and Autoimmunity. Biology 2023, 12, 452. [Google Scholar] [CrossRef]
- Zhang, Z.; Bahabayi, A.; Liu, D.; Hasimu, A.; Zhang, Y.; Guo, S.; Liu, R.; Zhang, K.; Li, Q.; Xiong, Z.; et al. KLRB1 defines an activated phenotype of CD4+ T cells and shows significant upregulation in patients with primary Sjogren’s syndrome. Int. Immunopharmacol. 2024, 133, 112072. [Google Scholar] [CrossRef]
- Liu, W.; Huber, S.A. Cross-talk between cd1d-restricted nkt cells and gammadelta cells in t regulatory cell response. Virol. J. 2011, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Boivin, W.A.; Cooper, D.M.; Hiebert, P.R.; Granville, D.J. Intracellular versus extracellular granzyme B in immunity and disease: Challenging the dogma. Lab. Investig. 2009, 89, 1195–1220. [Google Scholar] [CrossRef] [PubMed]
- Stark, R.; Wesselink, T.H.; Behr, F.M.; Kragten, N.A.M.; Arens, R.; Koch-Nolte, F.; van Gisbergen, K.; van Lier, R.A.W. T (RM) maintenance is regulated by tissue damage via P2RX7. Sci. Immunol. 2018, 3, eaau1022. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Sun, Y.; Zhao, L.; Shi, X. NLRP3 inflammasome and its role in autoimmune diseases: A promising therapeutic target. Biomed. Pharmacother. 2024, 175, 116679. [Google Scholar] [CrossRef]
- Karampetsou, M.P.; Comte, D.; Suarez-Fueyo, A.; Katsuyama, E.; Yoshida, N.; Kono, M.; Kyttaris, V.C.; Tsokos, G.C. Signaling Lymphocytic Activation Molecule Family Member 1 Engagement Inhibits T Cell-B Cell Interaction and Diminishes Interleukin-6 Production and Plasmablast Differentiation in Systemic Lupus Erythematosus. Arthritis Rheumatol. 2019, 71, 99–108. [Google Scholar] [CrossRef]
- Blum, J.A.; Schmid, C.; Burri, C.; Hatz, C.; Olson, C.; Fungula, B.; Kazumba, L.; Mangoni, P.; Mbo, F.; Deo, K.; et al. Cardiac alterations in human African trypanosomiasis (T.b. gambiense) with respect to the disease stage and antiparasitic treatment. PLoS Negl. Trop. Dis. 2009, 3, e383. [Google Scholar] [CrossRef]
- Sabino, E.C.; Nunes, M.C.P.; Blum, J.; Molina, I.; Ribeiro, A.L.P. Cardiac involvement in Chagas disease and African trypanosomiasis. Nat. Rev. Cardiol. 2024, 21, 865–879. [Google Scholar] [CrossRef]
- Huang, A.; Lin, B.; Ji, X.; Chen, S. A case report of severe myocarditis combined with erythema multiforme caused by herpes simplex virus-1. Front. Cardiovasc. Med. 2025, 12, 1421364. [Google Scholar] [CrossRef]
- Dutta, P.; Saha, D.; Earle, M.; Prasad, C.P.; Singh, M.; Darswal, M.; Aggarwal, V.; Naik, N.; Yadav, R.; Shankar, A.; et al. Unveiling HPV’s hidden link: Cardiovascular diseases and the viral intrigue. Indian. Heart J. 2024, 76, 1–5. [Google Scholar] [CrossRef]
- Watanabe, M.; Panetta, G.L.; Piccirillo, F.; Spoto, S.; Myers, J.; Serino, F.M.; Costantino, S.; Di Sciascio, G. Acute Epstein-Barr related myocarditis: An unusual but life-threatening disease in an immunocompetent patient. J. Cardiol. Cases 2020, 21, 137–140. [Google Scholar] [CrossRef]
- Walenta, K.; Kindermann, I.; Gartner, B.; Kandolph, R.; Link, A.; Bohm, M. Dangerous kisses: Epstein-barr virus myocarditis mimicking myocardial infarction. Am. J. Med. 2006, 119, e3–e6. [Google Scholar] [CrossRef]
- Verbruggen, L.A.; Rebmann, V.; Demanet, C.; De Cock, S.; Grosse-Wilde, H. Soluble HLA-G in rheumatoid arthritis. Hum. Immunol. 2006, 67, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Bortolotti, D.; Bolzani, S.; Fainardi, E. HLA-G Molecules in Autoimmune Diseases and Infections. Front. Immunol. 2014, 5, 592. [Google Scholar] [CrossRef]
- Joosten, S.A.; Sullivan, L.C.; Ottenhoff, T.H. Characteristics of HLA-E Restricted T-Cell Responses and Their Role in Infectious Diseases. J. Immunol. Res. 2016, 2016, 2695396. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- House, S.L.; Wang, J.; Castro, A.M.; Weinheimer, C.; Kovacs, A.; Ornitz, D.M. Fibroblast growth factor 2 is an essential cardioprotective factor in a closed-chest model of cardiac ischemia-reperfusion injury. Physiol. Rep. 2015, 3, e12278. [Google Scholar] [CrossRef]
- Salomon, B.L.; Leclerc, M.; Tosello, J.; Ronin, E.; Piaggio, E.; Cohen, J.L. Tumor Necrosis Factor alpha and Regulatory T Cells in Oncoimmunology. Front. Immunol. 2018, 9, 444. [Google Scholar] [CrossRef]
- Kass, D.A.; Champion, H.C.; Beavo, J.A. Phosphodiesterase type 5: Expanding roles in cardiovascular regulation. Circ. Res. 2007, 101, 1084–1095. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Li, G.; Li, L.; Da, Y. Pin1 maintains the effector program of pathogenic Th17 cells in autoimmune neuroinflammation. J. Autoimmun. 2024, 147, 103262. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, E.S.; Russell, F.A.; Spina, D.; McDougall, J.J.; Graepel, R.; Gentry, C.; Staniland, A.A.; Mountford, D.M.; Keeble, J.E.; Malcangio, M.; et al. A distinct role for transient receptor potential ankyrin 1, in addition to transient receptor potential vanilloid 1, in tumor necrosis factor alpha-induced inflammatory hyperalgesia and Freund’s complete adjuvant-induced monarthritis. Arthritis Rheum. 2011, 63, 819–829. [Google Scholar] [CrossRef]
- Barzaghi, F.; Passerini, L.; Bacchetta, R. Immune dysregulation, polyendocrinopathy, enteropathy, x-linked syndrome: A paradigm of immunodeficiency with autoimmunity. Front. Immunol. 2012, 3, 211. [Google Scholar] [CrossRef] [PubMed]
- Hou, C.H.; Tang, C.H.; Hsu, C.J.; Hou, S.M.; Liu, J.F. CCN4 induces IL-6 production through alphavbeta5 receptor, PI3K, Akt, and NF-kappaB singling pathway in human synovial fibroblasts. Arthritis Res. Ther. 2013, 15, R19. [Google Scholar] [CrossRef] [PubMed]
- Larrick, J.W.; Wright, S.C. Cytotoxic mechanism of tumor necrosis factor-alpha. FASEB J. 1990, 4, 3215–3223. [Google Scholar] [CrossRef] [PubMed]
- Wensveen, F.M.; Jelencic, V.; Polic, B. NKG2D: A Master Regulator of Immune Cell Responsiveness. Front. Immunol. 2018, 9, 441. [Google Scholar] [CrossRef]
- Arakaki, R.; Yamada, A.; Kudo, Y.; Hayashi, Y.; Ishimaru, N. Mechanism of activation-induced cell death of T cells and regulation of FasL expression. Crit. Rev. Immunol. 2014, 34, 301–314. [Google Scholar] [CrossRef]
- Lasrado, N.; Borcherding, N.; Arumugam, R.; Starr, T.K.; Reddy, J. Dissecting the cellular landscape and transcriptome network in viral myocarditis by single-cell RNA sequencing. iScience 2022, 25, 103865. [Google Scholar] [CrossRef]
- Kim, H.P.; Leonard, W.J. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: A role for DNA methylation. J. Exp. Med. 2007, 204, 1543–1551. [Google Scholar] [CrossRef]
- Cui, X.; Li, C.G.; Gao, H.; Cheng, M.; Jiang, F. Boosting regulatory T cell-dependent immune tolerance by activation of p53. Int. Immunopharmacol. 2023, 125, 111167. [Google Scholar] [CrossRef]
- Martinez, G.J.; Zhang, Z.; Chung, Y.; Reynolds, J.M.; Lin, X.; Jetten, A.M.; Feng, X.H.; Dong, C. Smad3 differentially regulates the induction of regulatory and inflammatory T cell differentiation. J. Biol. Chem. 2009, 284, 35283–35286. [Google Scholar] [CrossRef]
- Wan, Y.Y. GATA3: A master of many trades in immune regulation. Trends Immunol. 2014, 35, 233–242. [Google Scholar] [CrossRef]
- Durant, L.; Watford, W.T.; Ramos, H.L.; Laurence, A.; Vahedi, G.; Wei, L.; Takahashi, H.; Sun, H.W.; Kanno, Y.; Powrie, F.; et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity 2010, 32, 605–615. [Google Scholar] [CrossRef]
- Noack, M.; Miossec, P. Th17 and regulatory T cell balance in autoimmune and inflammatory diseases. Autoimmun. Rev. 2014, 13, 668–677. [Google Scholar] [CrossRef] [PubMed]
- Messina, N.; Fulford, T.; O’Reilly, L.; Loh, W.X.; Motyer, J.M.; Ellis, D.; McLean, C.; Naeem, H.; Lin, A.; Gugasyan, R.; et al. The NF-kappaB transcription factor RelA is required for the tolerogenic function of Foxp3(+) regulatory T cells. J. Autoimmun. 2016, 70, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Barnabei, L.; Laplantine, E.; Mbongo, W.; Rieux-Laucat, F.; Weil, R. NF-kappaB: At the Borders of Autoimmunity and Inflammation. Front. Immunol. 2021, 12, 716469. [Google Scholar] [CrossRef] [PubMed]
- Arbelaez, C.A.; Palle, P.; Charaix, J.; Bettelli, E. STAT1 signaling protects self-reactive T cells from control by innate cells during neuroinflammation. JCI Insight 2022, 7, e148222. [Google Scholar] [CrossRef]
- Van Der Byl, W.; Nussing, S.; Peters, T.J.; Ahn, A.; Li, H.; Ledergor, G.; David, E.; Koh, A.S.; Wagle, M.V.; Deguit, C.D.T.; et al. The CD8(+) T cell tolerance checkpoint triggers a distinct differentiation state defined by protein translation defects. Immunity 2024, 57, 1324–1344.e1328. [Google Scholar] [CrossRef]
- Moon, Y.M.; Lee, S.Y.; Kwok, S.K.; Lee, S.H.; Kim, D.; Kim, W.K.; Her, Y.M.; Son, H.J.; Kim, E.K.; Ryu, J.G.; et al. The Fos-Related Antigen 1-JUNB/Activator Protein 1 Transcription Complex, a Downstream Target of Signal Transducer and Activator of Transcription 3, Induces T Helper 17 Differentiation and Promotes Experimental Autoimmune Arthritis. Front. Immunol. 2017, 8, 1793. [Google Scholar] [CrossRef]
- Tanaka, S.; Tanaka, K.; Magnusson, F.; Chung, Y.; Martinez, G.J.; Wang, Y.H.; Nurieva, R.I.; Kurosaki, T.; Dong, C. CCAAT/enhancer-binding protein alpha negatively regulates IFN-gamma expression in T cells. J. Immunol. 2014, 193, 6152–6160. [Google Scholar] [CrossRef]
- Tan, T.G.; Mathis, D.; Benoist, C. Singular role for T-BET+CXCR3+ regulatory T cells in protection from autoimmune diabetes. Proc. Natl. Acad. Sci. USA 2016, 113, 14103–14108. [Google Scholar] [CrossRef]
- Corcoran, S.E.; O’Neill, L.A. HIF1alpha and metabolic reprogramming in inflammation. J. Clin. Investig. 2016, 126, 3699–3707. [Google Scholar] [CrossRef]
- Sharman, S.J.; Coomber, K.; Mayshak, R.; Curtis, A.; Hyder, S.; Walker, A.; Liknaitzky, P.; Miller, P. Situational Characteristics Uniquely Associated with Children’s Exposure to Intimate Partner Violence. J. Interpers. Violence 2021, 36, NP11087–NP11105. [Google Scholar] [CrossRef]
- Singh, P.; Ravanan, P.; Talwar, P. Death Associated Protein Kinase 1 (DAPK1): A Regulator of Apoptosis and Autophagy. Front. Mol. Neurosci. 2016, 9, 46. [Google Scholar] [CrossRef]
- Shohat, G.; Spivak-Kroizman, T.; Cohen, O.; Bialik, S.; Shani, G.; Berrisi, H.; Eisenstein, M.; Kimchi, A. The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J. Biol. Chem. 2001, 276, 47460–47467. [Google Scholar] [CrossRef]
- Zhou, Y.; Chen, X.; Zu, X. ZBTB7A as a therapeutic target for cancer. Biochem. Biophys. Res. Commun. 2024, 736, 150888. [Google Scholar] [CrossRef]
- Cui, X.; Song, Y.; Han, J.; Yuan, Z. The multifaceted role of SMAD4 in immune cell function. Biochem. Biophys. Rep. 2025, 41, 101902. [Google Scholar] [CrossRef]
- Andersen, L.; Gulich, A.F.; Alteneder, M.; Preglej, T.; Orola, M.J.; Dhele, N.; Stolz, V.; Schebesta, A.; Hamminger, P.; Hladik, A.; et al. The Transcription Factor MAZR/PATZ1 Regulates the Development of FOXP3(+) Regulatory T Cells. Cell Rep. 2019, 29, 4447–4459.e4446. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, M.; Yang, P.; Chen, D.; Zhou, D.; Ren, Y.; Zhang, Z.; Ruan, C.; Da, Y.; Zhang, R. Sp3 ameliorated experimental autoimmune encephalomyelitis by triggering Socs3 in Th17 cells. J. Adv. Res. 2025, in press. [Google Scholar] [CrossRef]
- Manoharan, I.; Swafford, D.; Shanmugam, A.; Patel, N.; Prasad, P.D.; Thangaraju, M.; Manicassamy, S. Activation of Transcription Factor 4 in Dendritic Cells Controls Th1/Th17 Responses and Autoimmune Neuroinflammation. J. Immunol. 2021, 207, 1428–1436. [Google Scholar] [CrossRef]
- Cook, M.E.; Jarjour, N.N.; Lin, C.C.; Edelson, B.T. Transcription Factor Bhlhe40 in Immunity and Autoimmunity. Trends Immunol. 2020, 41, 1023–1036. [Google Scholar] [CrossRef]
- Yasuda, K.; Shimodan, S.; Maehara, N.; Hirota, A.; Iijima, R.; Nishijima, A.; Mori, H.; Toyama, R.; Ito, A.; Yoshikawa, Y.; et al. AIM/CD5L ameliorates autoimmune arthritis by promoting removal of inflammatory DAMPs at the lesions. J. Autoimmun. 2024, 142, 103149. [Google Scholar] [CrossRef]
- Bradford, B.M.; Sester, D.P.; Hume, D.A.; Mabbott, N.A. Defining the anatomical localisation of subsets of the murine mononuclear phagocyte system using integrin alpha X (Itgax, CD11c) and colony stimulating factor 1 receptor (Csf1r, CD115) expression fails to discriminate dendritic cells from macrophages. Immunobiology 2011, 216, 1228–1237. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Hu, G.; Hu, Q.; Chang, Y.; Hu, Y.; Gao, L.; Chen, X.; Yang, P.C.; Zhang, Y.; Li, M.; et al. Single-Cell RNA Sequencing to Dissect the Immunological Network of Autoimmune Myocarditis. Circulation 2020, 142, 384–400. [Google Scholar] [CrossRef]
- Ye, L.; Huang, X.; Tang, Q.; Fang, G.; Bi, Y.; Pang, Y.; Huang, A. Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveal macrophage subpopulation characteristics and the role of STAT1 in rheumatoid arthritis. J. Transl. Med. 2025, 23, 1161. [Google Scholar] [CrossRef]



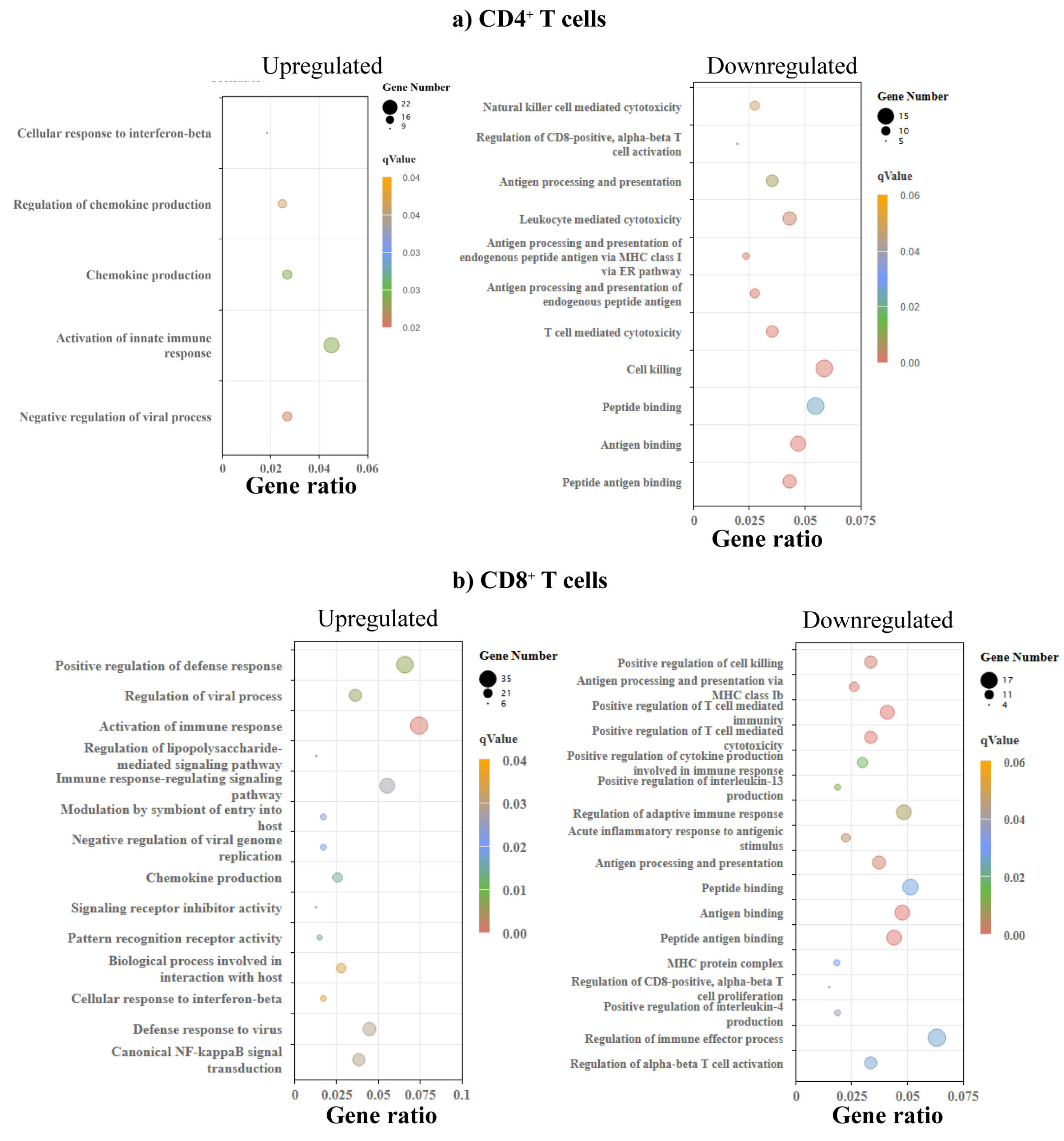


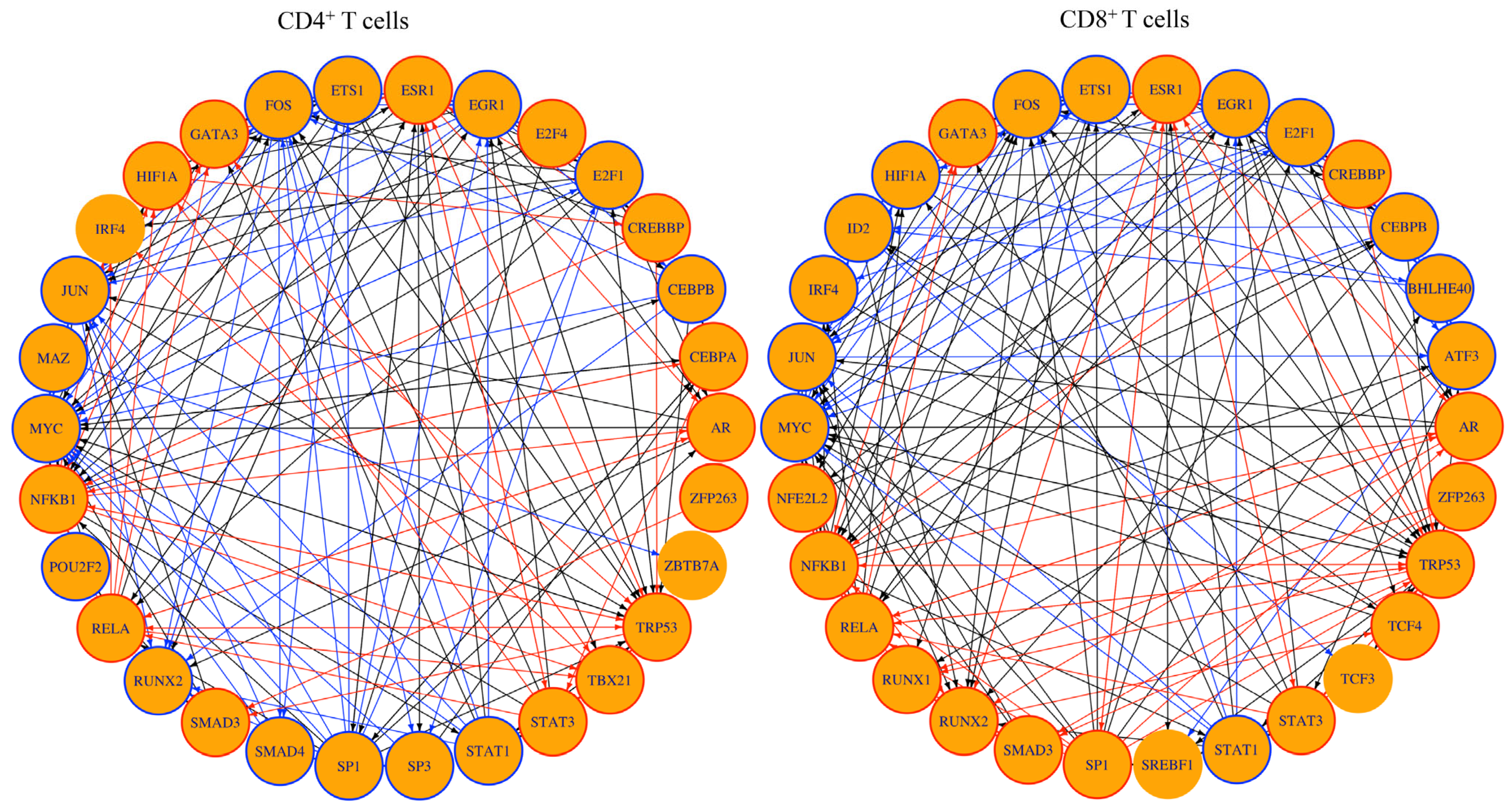
| Pathways | NES | FDR |
|---|---|---|
| GSEA_GO analysis | ||
| Positively enriched in CD4+ T cells | ||
| Negative regulation by host of viral processes | 2.273 | 0.04177 |
| Transcription coactivator activity | 2.121 | 0.05327 |
| Negatively enriched in CD4+ T cells | ||
| Negative regulation of heart contraction | −1.832 | 0.2275 |
| Positive regulation of cardiac muscle hypertrophy | −1.882 | 0.1980 |
| Positively enriched in CD8+ T cells | ||
| Natural killer cell mediated cytotoxicity | 1.979 | 0.08659 |
| Negative regulation of extrinsic apoptotic signaling pathway in absence of ligand | 1.893 | 0.12661 |
| Positive regulation of I kappab kinase nf kappab signaling | 1.927 | 0.09712 |
| Positive regulation of interleukin 2 production | 1.940 | 0.09835 |
| Response to interferon beta | 2.086 | 0.05416 |
| GSEA_KEGG analysis | ||
| Positively enriched in CD4+ T cells | ||
| Natural killer cell mediated cytotoxicity | 1.597 | 0.1258 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, S.; Sur, M.; Mone, K.; Ayad, C.W.; Massilamany, C.; Gangaplara, A.; Reddy, J. Bulk RNAseq Analysis of Cardiac Myosin-Specific CD4+ and CD8+ T Cells Reveals Distinct Transcriptomic Profiles Between Myocarditis-Resistant and Susceptible Mice. Biomedicines 2025, 13, 2725. https://doi.org/10.3390/biomedicines13112725
Singh S, Sur M, Mone K, Ayad CW, Massilamany C, Gangaplara A, Reddy J. Bulk RNAseq Analysis of Cardiac Myosin-Specific CD4+ and CD8+ T Cells Reveals Distinct Transcriptomic Profiles Between Myocarditis-Resistant and Susceptible Mice. Biomedicines. 2025; 13(11):2725. https://doi.org/10.3390/biomedicines13112725
Chicago/Turabian StyleSingh, Shraddha, Meghna Sur, Kiruthiga Mone, Celia Wafa Ayad, Chandirasegran Massilamany, Arunakumar Gangaplara, and Jay Reddy. 2025. "Bulk RNAseq Analysis of Cardiac Myosin-Specific CD4+ and CD8+ T Cells Reveals Distinct Transcriptomic Profiles Between Myocarditis-Resistant and Susceptible Mice" Biomedicines 13, no. 11: 2725. https://doi.org/10.3390/biomedicines13112725
APA StyleSingh, S., Sur, M., Mone, K., Ayad, C. W., Massilamany, C., Gangaplara, A., & Reddy, J. (2025). Bulk RNAseq Analysis of Cardiac Myosin-Specific CD4+ and CD8+ T Cells Reveals Distinct Transcriptomic Profiles Between Myocarditis-Resistant and Susceptible Mice. Biomedicines, 13(11), 2725. https://doi.org/10.3390/biomedicines13112725







