Drug-Metabolizing Gene Expression Identity: Comparison Across Liver Tissues and Model Cell Lines
Abstract
1. Introduction
2. Materials and Methods
2.1. Citation Analysis of Cell Lines
2.2. Datasets
2.3. Statistics
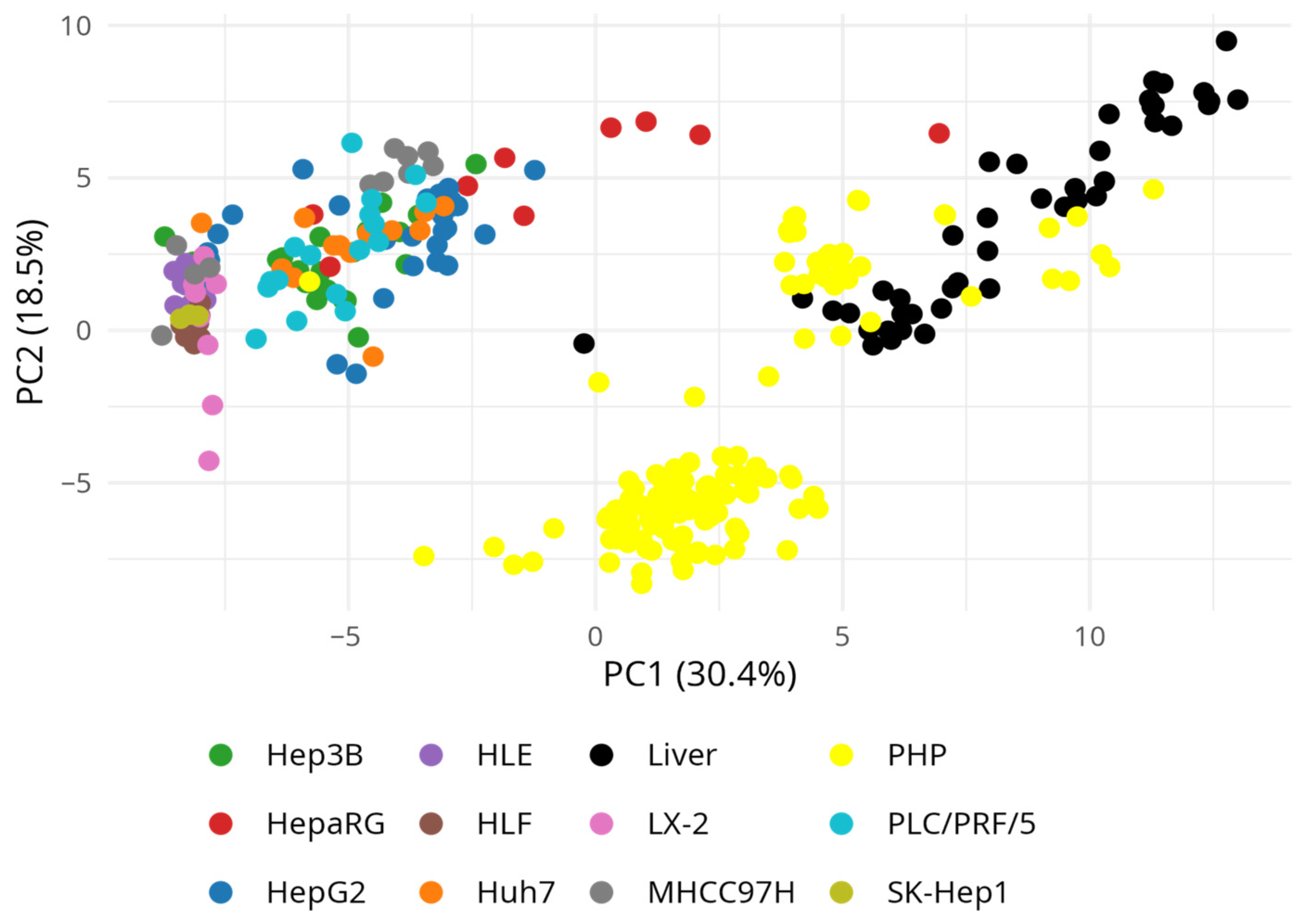
3. Results
3.1. Cell Line Authentication Challenges in Hepatic Research
3.2. Drug Metabolism Genes
| Approved Gene Symbol | Approved Gene Name | Chromosomal Location |
|---|---|---|
| Glutathione S-transferases (GSTs) [38] | ||
| GSTA1 | Glutathione S-transferase (alpha) A1 | 6p12 |
| GSTA2 | Glutathione S-transferase A2 | 6p12.2 |
| GSTA3 | Glutathione S-transferase A3 | 6p12 |
| GSTA4 | Glutathione S-transferase A4 | 6p12 |
| GSTA5 | Glutathione S-transferase A5 | 6p12.1 |
| GSTK1 | Glutathione S-transferase kappa 1 | 7q34 |
| GSTM1 | Glutathione S-transferase M1 | 1p13.3 |
| GSTM2 | Glutathione S-transferase M2 | 1p13 |
| GSTM3 | Glutathione S-transferase M3 | 1p13.3 |
| GSTM4 | Glutathione S-transferase M4 | 1p13.3 |
| GSTM5 | Glutathione S-transferase M5 | 1p13.3 |
| GSTO1 | Glutathione S-transferase omega 1 | 10q25.1 |
| GSTO2 | Glutathione S-transferase omega 2 | 10q25.1 |
| GSTP1 | Glutathione S-transferase (pi) P1 | 11q13.2 |
| GSTT1 | Glutathione S-transferase theta 1 | 22q11.23 |
| GSTT2 | Glutathione S-transferase theta 2 | 22q11.2 |
| GSTZ1 | Glutathione S-transferase (zeta) Z1 | 14q24.3 |
| MGST1 | Microsomal glutathione S-transferase 1 | 12p12.3 |
| MGST2 | Microsomal glutathione S-transferase 2 | 4q31.1 |
| MGST3 | Microsomal glutathione S-transferase 3 | 1q23 |
| PTGES | Prostaglandin E synthase | 9q34.11 |
| Cytochromes P450 (CYP) [39,40] | ||
| CYP1A1 | Cytochrome P450 family 1 subfamily A member 1 | 15q24.1 |
| CYP1A2 | Cytochrome P450 family 1 subfamily A member 2 | 15q24.1 |
| CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | 2p22.2 |
| CYP2A6 | Cytochrome P450 family 2 subfamily A member 6 | 19q13.2 |
| CYP2B6 | Cytochrome P450 family 2 subfamily B member 6 | 19q13.2 |
| CYP2C8 | Cytochrome P450 family 2 subfamily C member 8 | 10q23.33 |
| CYP2C9 | Cytochrome P450 family 2 subfamily C member 9 | 10q24.1 |
| CYP2C19 | Cytochrome P450 family 2 subfamily C member 19 | 10q23.33 |
| CYP2D6 | Cytochrome P450 family 2 subfamily D member 6 | 22q13.2 |
| CYP2J2 | Cytochrome P450 family 2 subfamily J member 2 | 1p32.1 |
| CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 | 7q22.1 |
| CYP3A5 | Cytochrome P450 family 3 subfamily A member 5 | 7q22.1 |
| CYP2E1 | Cytochrome P450 Family 2 Subfamily E Member 1 | 10q26.3 |
| POR | Cytochrome P450 oxidoreductase | 7q11.23 |
| Alcohol dehydrogenases (ADH) [41,42,43] | ||
| ADH1A | Alcohol Dehydrogenase 1A (Class I), alpha subunit | 4q23 |
| ADH1B | Alcohol Dehydrogenase 1B (Class I), beta subunit | 4q23 |
| ADH1C | Alcohol Dehydrogenase 1C (Class I), gamma subunit | 4q23 |
| ADH4 | Alcohol Dehydrogenase 4 (Class II) | 4q23 |
| ADH5 | Alcohol Dehydrogenase 5 (Class III) | 4q23 |
| ADH6 | Alcohol Dehydrogenase 6 (Class V) | 4q23 |
| ADH7 | Alcohol Dehydrogenase 7 (Class IV) | 4q23 |
| ALDH1A1 | Aldehyde dehydrogenase 1 family member A1 | 9q21 |
| ALDH1A2 | Aldehyde dehydrogenase 1 family member A2 | 15q21.2 |
| ALDH1A3 | Aldehyde dehydrogenase 1 family member A3 | 15q26.3 |
| ALDH1B1 | Aldehyde dehydrogenase 1 family member B1 | 9p13.1 |
| ALDH1L1 | Aldehyde dehydrogenase 1 family member L1 | 3q21.3 |
| ALDH1L2 | Aldehyde dehydrogenase 1 family member L2 | 12q23.3 |
| ALDH2 | Aldehyde dehydrogenase 2 family (mitochondrial) | 12q24.2 |
| ALDH3A1 | Aldehyde dehydrogenase 3 family member A1 | 17p11.2 |
| ALDH3A2 | Aldehyde dehydrogenase 3 family member A2 | 17p11.2 |
| ALDH3B1 | Aldehyde dehydrogenase 3 family member B1 | 11q13.2 |
| ALDH3B2 | Aldehyde dehydrogenase 3 family member B2 | 11q13.2 |
| ALDH4A1 | Aldehyde dehydrogenase 4 family member A1 | 1p36.13 |
| ALDH5A1 | Aldehyde dehydrogenase 5 family member A1 | 6p22.3 |
| ALDH6A1 | Aldehyde dehydrogenase 6 family member A1 | 14q24.3 |
| ALDH7A1 | Aldehyde dehydrogenase 7 family member A1 | 5q31 |
| ALDH8A1 | Aldehyde dehydrogenase 8 family member A1 | 6q23.3 |
| ALDH9A1 | Aldehyde dehydrogenase 9 family member A1 | 1q24.1 |
| ALDH16A1 | Aldehyde dehydrogenase 16 family member A1 | 19q13.33 |
| ALDH18A1 | Aldehyde dehydrogenase 18 family member A1 | 10q24.1 |
| Carboxylesterases (CES) [43] | ||
| CES1 | Carboxylesterase 1 | 16q12.2 |
| CES2 | Carboxylesterase 2 | 16q22.1 |
| CES3 | Carboxylesterase 3 | 16q22.1 |
| CES4A | Carboxylesterase 4A | 16q22.1 |
| CES7 | Carboxylesterase 7 | 16q22.1 |
| Flavin-containing monooxygenases (FMO) [44] | ||
| FMO1 | Flavin containing monooxygenase 1 | 1q24.3 |
| FMO2 | Flavin containing monooxygenase 2 | 1q24.3 |
| FMO3 | Flavin containing monooxygenase 3 | 1q24.3 |
| FMO4 | Flavin containing monooxygenase 4 | 1q24.3 |
| FMO5 | Flavin containing monooxygenase 5 | 1q21.1 |
| N-acetyltransferases (NATs) [45] | ||
| NAT1 | N-acetyltransferase 1 | 8p22 |
| NAT2 | N-acetyltransferase 2, arylamine N-acetyltransferase | 8p22 |
| Methyltransferases (MTs) [34] | ||
| TPMT | Thiopurine s-methyltransferase | 6p22.3 |
| TMT1B | Thiol Methyltransferase 1B | 12q13.2 |
| COMT | Catechol-O-methyltransferase | 22q11.21 |
| HNMT | Histamine N-methyltransferase | 2q22.1 |
| Sulfotransferases (SULT) [46,47] | ||
| SULT1A1 | Sulfotransferase family 1A member 1 | 16p11.2 |
| SULT1A2 | Sulfotransferase family 1A member 2 | 16p11.2 |
| SULT1A3 | Sulfotransferase family 1A member 3 | 16p11.2 |
| SULT1A4 | Sulfotransferase family 1A member 4 | 16p11.2 |
| SULT1B1 | Sulfotransferase family 1B member 1 | 4q13.3 |
| SULT1C2 | Sulfotransferase family 1C member 2 | 2q12.3 |
| SULT1C3 | Sulfotransferase family 1C member 3 | 2q12.3 |
| SULT1C4 | Sulfotransferase family 1C member 4 | 2q12.3 |
| SULT1E1 | Sulfotransferase family 1E member 1 | 4q13.3 |
| SULT2A1 | Sulfotransferase family 2A member 1 | 19q13.33 |
| SULT2B1 | Sulfotransferase family 2B member 1 | 19q13.33 |
| SULT4A1 | Sulfotransferase family 4A member 1 | 22q13.31 |
| SULT6B1 | Sulfotransferase family 6B member 1 | 2p22.2 |
| UDP-Glucuronosyltransferases (UGT) [48] | ||
| UGT1A1 | UDP glucuronosyltransferase family 1 member A1 | 2q37.1 |
| UGT1A3 | UDP glucuronosyltransferase family 1 member A3 | 2q37.1 |
| UGT1A4 | UDP glucuronosyltransferase family 1 member A4 | 2q37.1 |
| UGT1A5 | UDP glucuronosyltransferase family 1 member A5 | 2q37.1 |
| UGT1A6 | UDP glucuronosyltransferase family 1 member A6 | 2q37.1 |
| UGT1A7 | UDP glucuronosyltransferase family 1 member A7 | 2q37.1 |
| UGT1A8 | UDP glucuronosyltransferase family 1 member A8 | 2q37.1 |
| UGT1A9 | UDP glucuronosyltransferase family 1 member A9 | 2q37.1 |
| UGT1A10 | UDP glucuronosyltransferase family 1 member A10 | 2q37.1 |
| UGT2A1 | UDP glucuronosyltransferase family 2 member A1 | 4q13.3 |
| UGT2A2 | UDP glucuronosyltransferase family 2 member A2 | 4q13.3 |
| UGT2A3 | UDP glucuronosyltransferase family 2 member A3 | 4q13.3 |
| UGT2B4 | UDP glucuronosyltransferase family 2 member B4 | 4q13.2 |
| UGT2B7 | UDP glucuronosyltransferase family 2 member B7 | 4q13.2 |
| UGT2B10 | UDP glucuronosyltransferase family 2 member B10 | 4q13.2 |
| UGT2B11 | UDP glucuronosyltransferase family 2 member B11 | 4q13.2 |
| UGT2B15 | UDP glucuronosyltransferase family 2 member B15 | 4q13.2 |
| UGT2B17 | UDP glucuronosyltransferase family 2 member B17 | 4q13.2 |
| UGT2B28 | UDP glucuronosyltransferase family 2 member B28 | 4q13.2 |
| UGT3A1 | UDP glycosyltransferase family 3 member A1 | 5p13.2 |
| UGT3A2 | UDP glycosyltransferase family 3 member A2 | 5p13.2 |
| UGT8 (UGT8A1) | UDP glycosyltransferase 8 | 4q26 |
3.3. Core Facts on Drug Metabolism Genes
3.4. Drug-Metabolizing Gene Expression Across Hepatic Models Using GEO Transcriptomic Datasets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CYP | Cytochrome P450 |
| PHP | Primary Hepatocytes |
| HCC | Hepatocellular Carcinoma |
| HB | Hepatoblastoma |
| AFP | Alpha-fetoprotein |
| HBV | Hepatitis B Virus |
| HBsAg | Hepatitis B Surface Antigen |
| HSC | Hepatic Stellate Cell |
| DME | Drug-metabolizing enzymes |
| ADH/ALDH | Alcohol and Aldehyde Dehydrogenase |
| FMO | Flavin-containing Monooxygenase |
| CES | Carboxylesterase |
| UGT | UDP-glucuronosyltransferase |
| SULT | Sulfotransferase |
| NAT | N-acetyltransferase |
| MT | Methyltransferase |
| GST | Glutathione-S-transferases |
| PCA | Principal Component Analysis |
| OOC | Organ-On-A-Chip |
References
- Watari, R.; Kakiki, M.; Oshikata, A.; Takezawa, T.; Yamasaki, C.; Ishida, Y.; Tateno, C.; Kuroda, Y.; Ishida, S.; Kusano, K. A Long-Term Culture System Based on a Collagen Vitrigel Membrane Chamber That Supports Liver-Specific Functions of Hepatocytes Isolated from Mice with Humanized Livers. J. Toxicol. Sci. 2018, 43, 521–529. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Wang, Y.; Jin, W.; Zhang, Z.; Jin, L.; Qian, J.; Zheng, L. CYP3A4 and CYP3A5: The Crucial Roles in Clinical Drug Metabolism and the Significant Implications of Genetic Polymorphisms. PeerJ 2024, 12, e18636. [Google Scholar] [CrossRef] [PubMed]
- Kiamehr, M.; Heiskanen, L.; Laufer, T.; Düsterloh, A.; Kahraman, M.; Käkelä, R.; Laaksonen, R.; Aalto-Setälä, K. Dedifferentiation of Primary Hepatocytes Is Accompanied with Reorganization of Lipid Metabolism Indicated by Altered Molecular Lipid and miRNA Profiles. Int. J. Mol. Sci. 2019, 20, 2910. [Google Scholar] [CrossRef]
- Gerets, H.H.J.; Tilmant, K.; Gerin, B.; Chanteux, H.; Depelchin, B.O.; Dhalluin, S.; Atienzar, F.A. Characterization of Primary Human Hepatocytes, HepG2 Cells, and HepaRG Cells at the mRNA Level and CYP Activity in Response to Inducers and Their Predictivity for the Detection of Human Hepatotoxins. Cell Biol. Toxicol. 2012, 28, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Zhou, J.; He, X.-P.; Zhang, Y.-F.; Gao, N.; Tian, X.; Fang, Y.; Wen, Q.; Jia, L.-J.; Jin, H.; et al. Changes in Cytochrome P450s-Mediated Drug Clearance in Patients with Hepatocellular Carcinoma in Vitro and in Vivo: A Bottom-up Approach. Oncotarget 2016, 7, 28612–28623. [Google Scholar] [CrossRef] [PubMed]
- Heintze, T.; Klein, K.; Hofmann, U.; Zanger, U.M. Differential Effects on Human Cytochromes P450 by CRISPR/Cas9-Induced Genetic Knockout of Cytochrome P450 Reductase and Cytochrome B5 in HepaRG Cells. Sci. Rep. 2021, 11, 1000. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 1 September 2025).
- Arzumanian, V.A.; Kiseleva, O.I.; Poverennaya, E.V. The Curious Case of the HepG2 Cell Line: 40 Years of Expertise. Int. J. Mol. Sci. 2021, 22, 13135. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- López-Terrada, D.; Cheung, S.W.; Finegold, M.J.; Knowles, B.B. Hep G2 Is a Hepatoblastoma-Derived Cell Line. Hum. Pathol. 2009, 40, 1512–1515. [Google Scholar] [CrossRef]
- Arzumanian, V.A.; Pyatnitsky, M.A.; Vakhrushev, I.V.; Ptitsyn, K.G.; Radko, S.P.; Zgoda, V.G.; Kiseleva, O.I.; Poveryennaya, E.V. Molecular Profile of the HepG2 Tumor Cell Line. Biomed. Chem. Res. Methods 2024, 7, e00239. [Google Scholar] [CrossRef]
- Tai, Y.; Gao, J.-H.; Zhao, C.; Tong, H.; Zheng, S.-P.; Huang, Z.-Y.; Liu, R.; Tang, C.-W.; Li, J. SK-Hep1: Not Hepatocellular Carcinoma Cells but a Cell Model for Liver Sinusoidal Endothelial Cells. Int. J. Clin. Exp. Pathol. 2018, 11, 2931–2938. [Google Scholar] [PubMed]
- Dor, I.; Namba, M.; Sato, J. Establishment and Some Biological Characteristics of Human Hepatoma Cell Lines. Gann 1975, 66, 385–392. [Google Scholar] [PubMed]
- Wakizaka, K.; Kamiyama, T.; Kakisaka, T.; Orimo, T.; Nagatsu, A.; Aiyama, T.; Shichi, S.; Taketomi, A. Expression of Wnt5a and ROR2, Components of the Noncanonical Wnt-Signaling Pathway, Is Associated with Tumor Differentiation in Hepatocellular Carcinoma. Ann. Surg. Oncol. 2024, 31, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Ohashi, R.; Naito, K.; Kanki, K. Hedgehog Signal Inhibitor GANT61 Inhibits the Malignant Behavior of Undifferentiated Hepatocellular Carcinoma Cells by Targeting Non-Canonical GLI Signaling. Int. J. Mol. Sci. 2020, 21, 3126. [Google Scholar] [CrossRef]
- Sigafoos, A.N.; Paradise, B.D.; Fernandez-Zapico, M.E. Hedgehog/GLI Signaling Pathway: Transduction, Regulation, and Implications for Disease. Cancers 2021, 13, 3410. [Google Scholar] [CrossRef]
- Daemer, R.J.; Feinstone, S.M.; Alexander, J.J.; Tully, J.G.; London, W.T.; Wong, D.C.; Purcell, R.H. PLC/PRF/5 (Alexander) Hepatoma Cell Line: Further Characterization and Studies of Infectivity. Infect. Immun. 1980, 30, 607–611. [Google Scholar] [CrossRef]
- Alexander, J.J.; Bey, E.M.; Geddes, E.W.; Lecatsas, G. Establishment of a Continuously Growing Cell Line from Primary Carcinoma of the Liver. S. Afr. Med. J. Suid-Afr. Tydskr. Vir Geneeskd. 1976, 50, 2124–2128. [Google Scholar] [PubMed]
- Miao, J.; Chen, G.G.; Chun, S.; Yun, J.; Chak, E.C.W.; Ho, R.L.K.; Lai, P.B.S. Adenovirus-mediated tBid Overexpression Results in Therapeutic Effects on P53-resistant Hepatocellular Carcinoma. Int. J. Cancer 2006, 119, 1985–1993. [Google Scholar] [CrossRef]
- He, M.; Zhao, M.; Shen, B.; Prise, K.M.; Shao, C. Radiation-Induced Intercellular Signaling Mediated by Cytochrome-c via a P53-Dependent Pathway in Hepatoma Cells. Oncogene 2011, 30, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Yang, X.; Qi, S.; Liu, H.; Jiang, H.; Hoppmann, S.; Cao, Q.; Chua, M.-S.; So, S.K.; Cheng, Z. Molecular Imaging of Hepatocellular Carcinoma Xenografts with Epidermal Growth Factor Receptor Targeted Affibody Probes. BioMed Res. Int. 2013, 2013, 759057. [Google Scholar] [CrossRef]
- Kawaguchi, K.; Honda, M.; Yamashita, T.; Shirota, Y.; Kaneko, S. Differential Gene Alteration among Hepatoma Cell Lines Demonstrated by cDNA Microarray-Based Comparative Genomic Hybridization. Biochem. Biophys. Res. Commun. 2005, 329, 370–380. [Google Scholar] [CrossRef]
- Kasai, F.; Hirayama, N.; Ozawa, M.; Satoh, M.; Kohara, A. HuH-7 Reference Genome Profile: Complex Karyotype Composed of Massive Loss of Heterozygosity. Hum. Cell 2018, 31, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Dial, S.; Shi, L.; Branham, W.; Liu, J.; Fang, J.-L.; Green, B.; Deng, H.; Kaput, J.; Ning, B. Similarities and Differences in the Expression of Drug-Metabolizing Enzymes between Human Hepatic Cell Lines and Primary Human Hepatocytes. Drug Metab. Dispos. Biol. Fate Chem. 2011, 39, 528–538. [Google Scholar] [CrossRef]
- Jouan, E.; Le Vée, M.; Denizot, C.; Parmentier, Y.; Fardel, O. Drug Transporter Expression and Activity in Human Hepatoma HuH-7 Cells. Pharmaceutics 2016, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.N.; Li, Y.; Nakamoto, K.; Subileau, E.; Steen, D.; Zhong, X. A Comparison of Whole Genome Gene Expression Profiles of HepaRG Cells and HepG2 Cells to Primary Human Hepatocytes and Human Liver Tissues. Drug Metab. Dispos. Biol. Fate Chem. 2010, 38, 988–994. [Google Scholar] [CrossRef]
- Dubois-Pot-Schneider, H.; Fekir, K.; Coulouarn, C.; Glaise, D.; Aninat, C.; Jarnouen, K.; Le Guével, R.; Kubo, T.; Ishida, S.; Morel, F.; et al. Inflammatory Cytokines Promote the Retrodifferentiation of Tumor-derived Hepatocyte-like Cells to Progenitor Cells. Hepatology 2014, 60, 2077–2090. [Google Scholar] [CrossRef]
- Li, Y.; Tang, Z.Y.; Ye, S.L.; Liu, Y.K.; Chen, J.; Xue, Q.; Chen, J.; Gao, D.M.; Bao, W.H. Establishment of Cell Clones with Different Metastatic Potential from the Metastatic Hepatocellular Carcinoma Cell Line MHCC97. World J. Gastroenterol. 2001, 7, 630–636. [Google Scholar] [CrossRef]
- Taimr, P.; Higuchi, H.; Kocova, E.; Rippe, R.A.; Friedman, S.; Gores, G.J. Activated Stellate Cells Express the TRAIL Receptor-2/Death Receptor-5 and Undergo TRAIL-Mediated Apoptosis. Hepatology 2003, 37, 87–95. [Google Scholar] [CrossRef]
- Weiskirchen, R.; Weimer, J.; Meurer, S.K.; Kron, A.; Seipel, B.; Vater, I.; Arnold, N.; Siebert, R.; Xu, L.; Friedman, S.L.; et al. Genetic Characteristics of the Human Hepatic Stellate Cell Line LX-2. PLoS ONE 2013, 8, e75692. [Google Scholar] [CrossRef]
- Arzumanian, V.; Pyatnitskiy, M.; Poverennaya, E. Comparative Transcriptomic Analysis of Three Common Liver Cell Lines. Int. J. Mol. Sci. 2023, 24, 8791. [Google Scholar] [CrossRef] [PubMed]
- Sissung, T.M.; English, B.C.; Venzon, D.; Figg, W.D.; Deeken, J.F. Clinical Pharmacology and Pharmacogenetics in a Genomics Era: The DMET Platform. Pharmacogenomics 2010, 11, 89–103. [Google Scholar] [CrossRef]
- Liu, X.; Yang, J.; Zhang, Y.; Fang, Y.; Wang, F.; Wang, J.; Zheng, X.; Yang, J. A Systematic Study on Drug-Response Associated Genes Using Baseline Gene Expressions of the Cancer Cell Line Encyclopedia. Sci. Rep. 2016, 6, 22811. [Google Scholar] [CrossRef]
- Pinto, N.; Dolan, M.E. Clinically Relevant Genetic Variations in Drug Metabolizing Enzymes. Curr. Drug Metab. 2011, 12, 487–497. [Google Scholar] [CrossRef]
- Ul Amin Mohsin, N.; Farrukh, M.; Shahzadi, S.; Irfan, M. Drug Metabolism: Phase I and Phase II Metabolic Pathways. In Pharmaceutical Science; Rudrapal, M., Ed.; IntechOpen: London, UK, 2024; Volume 2, ISBN 978-0-85014-147-4. [Google Scholar]
- Iacopetta, D.; Ceramella, J.; Catalano, A.; Scali, E.; Scumaci, D.; Pellegrino, M.; Aquaro, S.; Saturnino, C.; Sinicropi, M.S. Impact of Cytochrome P450 Enzymes on the Phase I Metabolism of Drugs. Appl. Sci. 2023, 13, 6045. [Google Scholar] [CrossRef]
- Her, L.; Zhu, H.-J. Carboxylesterase 1 and Precision Pharmacotherapy: Pharmacogenetics and Nongenetic Regulators. Drug Metab. Dispos. Biol. Fate Chem. 2020, 48, 230–244. [Google Scholar] [CrossRef]
- Nebert, D.W.; Vasiliou, V. Analysis of the Glutathione S-Transferase (GST) Gene Family. Hum. Genom. 2004, 1, 460. [Google Scholar] [CrossRef]
- Zhou, Y.; Lauschke, V.M. The Genetic Landscape of Major Drug Metabolizing Cytochrome P450 Genes—An Updated Analysis of Population-Scale Sequencing Data. Pharmacogenom. J. 2022, 22, 284–293. [Google Scholar] [CrossRef]
- Zanger, U.M.; Schwab, M. Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 2013, 138, 103–141. [Google Scholar] [CrossRef] [PubMed]
- Edenberg, H.J. The Genetics of Alcohol Metabolism: Role of Alcohol Dehydrogenase and Aldehyde Dehydrogenase Variants. Alcohol Res. Health 2007, 30, 5–13. [Google Scholar] [PubMed]
- Chen, C.-H.; Kraemer, B.R.; Lee, L.; Mochly-Rosen, D. Annotation of 1350 Common Genetic Variants of the 19 ALDH Multigene Family from Global Human Genome Aggregation Database (gnomAD). Biomolecules 2021, 11, 1423. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zou, L.; Jin, Q.; Hou, J.; Ge, G.; Yang, L. Human Carboxylesterases: A Comprehensive Review. Acta Pharm. Sin. B 2018, 8, 699–712. [Google Scholar] [CrossRef] [PubMed]
- McCombie, R.R.; Dolphin, C.T.; Povey, S.; Phillips, I.R.; Shephard, E.A. Localization of Human Flavin-Containing Monooxygenase Genes FMO2 and FMO5 to Chromosome 1q. Genomics 1996, 34, 426–429. [Google Scholar] [CrossRef]
- Manca, A.; Calcagno, A.; D’Avolio, A.; Cusato, J. Pharmacogenetics of First-Line Antitubercular Drugs: An Update. Ther. Drug Monit. 2025. [Google Scholar] [CrossRef] [PubMed]
- Hebbring, S.J.; Moyer, A.M.; Weinshilboum, R.M. Sulfotransferase Gene Copy Number Variation: Pharmacogenetics and Function. Cytogenet. Genome Res. 2008, 123, 205–210. [Google Scholar] [CrossRef]
- Kurogi, K.; Suiko, M.; Sakakibara, Y. Evolution and Multiple Functions of Sulfonation and Cytosolic Sulfotransferases across Species. Biosci. Biotechnol. Biochem. 2024, 88, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Jarrar, Y.; Lee, S.-J. The Functionality of UDP-Glucuronosyltransferase Genetic Variants and Their Association with Drug Responses and Human Diseases. J. Pers. Med. 2021, 11, 554. [Google Scholar] [CrossRef]
- Ramírez, B.; Niño-Orrego, M.J.; Cárdenas, D.; Ariza, K.E.; Quintero, K.; Contreras Bravo, N.C.; Tamayo-Agudelo, C.; González, M.A.; Laissue, P.; Fonseca Mendoza, D.J. Copy Number Variation Profiling in Pharmacogenetics CYP-450 and GST Genes in Colombian Population. BMC Med. Genom. 2019, 12, 110. [Google Scholar] [CrossRef]
- Ahmed, S.; Zhou, Z.; Zhou, J.; Chen, S.-Q. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genom. Proteom. Bioinform. 2016, 14, 298–313. [Google Scholar] [CrossRef]
- Štancl, P.; Gršković, P.; Držaić, S.; Vičić, A.; Karlić, R.; Korać, P. RNA-Sequencing Identification of Genes Supporting HepG2 as a Model Cell Line for Hepatocellular Carcinoma or Hepatocytes. Genes 2024, 15, 1460. [Google Scholar] [CrossRef]
- Zou, J.; Li, H.; Huang, Q.; Liu, X.; Qi, X.; Wang, Y.; Lu, L.; Liu, Z. Dopamine-Induced SULT1A3/4 Promotes EMT and Cancer Stemness in Hepatocellular Carcinoma. Tumor Biol. 2017, 39, 101042831771927. [Google Scholar] [CrossRef]
- Jennen, D.G.J.; Magkoufopoulou, C.; Ketelslegers, H.B.; van Herwijnen, M.H.M.; Kleinjans, J.C.S.; van Delft, J.H.M. Comparison of HepG2 and HepaRG by Whole-Genome Gene Expression Analysis for the Purpose of Chemical Hazard Identification. Toxicol. Sci. 2010, 115, 66–79. [Google Scholar] [CrossRef]
- Aninat, C.; Piton, A.; Glaise, D.; Le Charpentier, T.; Langouët, S.; Morel, F.; Guguen-Guillouzo, C.; Guillouzo, A. Expression of cytochromes p450, conjugating enzymes and nuclear receptors in human hepatoma HepaRG cells. Drug Metab. Dispos. 2006, 34, 75–83. [Google Scholar] [CrossRef]
- Turpeinen, M.; Tolonen, A.; Chesne, C.; Guillouzo, A.; Uusitalo, J.; Pelkonen, O. Functional Expression, Inhibition and Induction of CYP Enzymes in HepaRG Cells. Toxicol. Vitro Int. J. Publ. Assoc. BIBRA 2009, 23, 748–753. [Google Scholar] [CrossRef]
- Duivenvoorde, L.P.M.; Louisse, J.; Pinckaers, N.E.T.; Nguyen, T.; Van Der Zande, M. Comparison of Gene Expression and Biotransformation Activity of HepaRG Cells under Static and Dynamic Culture Conditions. Sci. Rep. 2021, 11, 10327. [Google Scholar] [CrossRef]
- Sajadian, S.O.; Tripura, C.; Samani, F.S.; Ruoss, M.; Dooley, S.; Baharvand, H.; Nussler, A.K. Vitamin C Enhances Epigenetic Modifications Induced by 5-Azacytidine and Cell Cycle Arrest in the Hepatocellular Carcinoma Cell Lines HLE and Huh7. Clin. Epigenet. 2016, 8, 46. [Google Scholar] [CrossRef]
- Zhang, B.; Ting, W.-J.; Gao, J.; Kang, Z.-F.; Huang, C.-Y.; Weng, Y.-J. Erk Phosphorylation Reduces the Thymoquinone Toxicity in Human Hepatocarcinoma. Environ. Toxicol. 2021, 36, 1990–1998. [Google Scholar] [CrossRef] [PubMed]
- Salman, E.D.; Kadlubar, S.A.; Falany, C.N. Expression and Localization of Cytosolic Sulfotransferase (SULT) 1A1 and SULT1A3 in Normal Human Brain. Drug Metab. Dispos. Biol. Fate Chem. 2009, 37, 706–709. [Google Scholar] [CrossRef]
- Voulgaridou, G.-P.; Theologidis, V.; Xanthis, V.; Papagiannaki, E.; Tsochantaridis, I.; Fadouloglou, V.E.; Pappa, A. Identification of a Peptide Ligand for Human ALDH3A1 through Peptide Phage Display: Prediction and Characterization of Protein Interaction Sites and Inhibition of ALDH3A1 Enzymatic Activity. Front. Mol. Biosci. 2023, 10, 1161111. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Cai, J.; Yang, Z.; He, X.; Xing, Z.; Zu, J.; Xie, E.; Henry, L.; Chong, C.R.; John, E.M.; et al. Trends in Hepatocellular Carcinoma Mortality Rates in the US and Projections Through 2040. JAMA Netw. Open 2024, 7, e2445525. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Huo, Y.; Zhang, Z.; Luo, Y.; Liu, X.; Chen, Q.; Wu, P.; Shi, W.; Wu, T.; Tang, C.; et al. Generation of αGal-Enhanced Bifunctional Tumor Vaccine. Acta Pharm. Sin. B 2022, 12, 3177–3186. [Google Scholar] [CrossRef]
- Okuyama, S.; Mine, A.; Nakamura, T.; Ohasi, Y.; Seto, M.; Tada, M. Transgenic HepaRG Cells Expressing CYP2D6 as an Improved Model of Primary Human Hepatocytes. Pharmacol. Res. Perspect. 2022, 10, e00939. [Google Scholar] [CrossRef]
- Zhuge, J.; Luo, Y.; Yu, Y.-N. Heterologous Expression of Human Cytochrome P450 2E1 in HepG2 Cell Line. World J. Gastroenterol. 2003, 9, 2732–2736. [Google Scholar] [CrossRef] [PubMed]
- Satoh, D.; Iwado, S.; Abe, S.; Kazuki, K.; Wakuri, S.; Oshimura, M.; Kazuki, Y. Establishment of a Novel Hepatocyte Model That Expresses Four Cytochrome P450 Genes Stably via Mammalian-Derived Artificial Chromosome for Pharmacokinetics and Toxicity Studies. PLoS ONE 2017, 12, e0187072. [Google Scholar] [CrossRef] [PubMed]
- Dorr, C.R.; Remmel, R.P.; Muthusamy, A.; Fisher, J.; Moriarity, B.S.; Yasuda, K.; Wu, B.; Guan, W.; Schuetz, E.G.; Oetting, W.S.; et al. CRISPR/Cas9 Genetic Modification of CYP3A5 *3 in HuH-7 Human Hepatocyte Cell Line Leads to Cell Lines with Increased Midazolam and Tacrolimus Metabolism. Drug Metab. Dispos. Biol. Fate Chem. 2017, 45, 957–965. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arzumanian, V.A.; Timofeeva, E.V.; Kiseleva, O.I.; Poverennaya, E.V. Drug-Metabolizing Gene Expression Identity: Comparison Across Liver Tissues and Model Cell Lines. Biomedicines 2025, 13, 2722. https://doi.org/10.3390/biomedicines13112722
Arzumanian VA, Timofeeva EV, Kiseleva OI, Poverennaya EV. Drug-Metabolizing Gene Expression Identity: Comparison Across Liver Tissues and Model Cell Lines. Biomedicines. 2025; 13(11):2722. https://doi.org/10.3390/biomedicines13112722
Chicago/Turabian StyleArzumanian, Viktoriia A., Ekaterina V. Timofeeva, Olga I. Kiseleva, and Ekaterina V. Poverennaya. 2025. "Drug-Metabolizing Gene Expression Identity: Comparison Across Liver Tissues and Model Cell Lines" Biomedicines 13, no. 11: 2722. https://doi.org/10.3390/biomedicines13112722
APA StyleArzumanian, V. A., Timofeeva, E. V., Kiseleva, O. I., & Poverennaya, E. V. (2025). Drug-Metabolizing Gene Expression Identity: Comparison Across Liver Tissues and Model Cell Lines. Biomedicines, 13(11), 2722. https://doi.org/10.3390/biomedicines13112722





