Specific microRNAs for Heart Failure: Reference Values in Whole Blood
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Samples
2.3. Determination of Classical Biomarkers
2.4. Isolation of miRNA
2.5. Determination of miRNA
2.6. Statistical Analysis
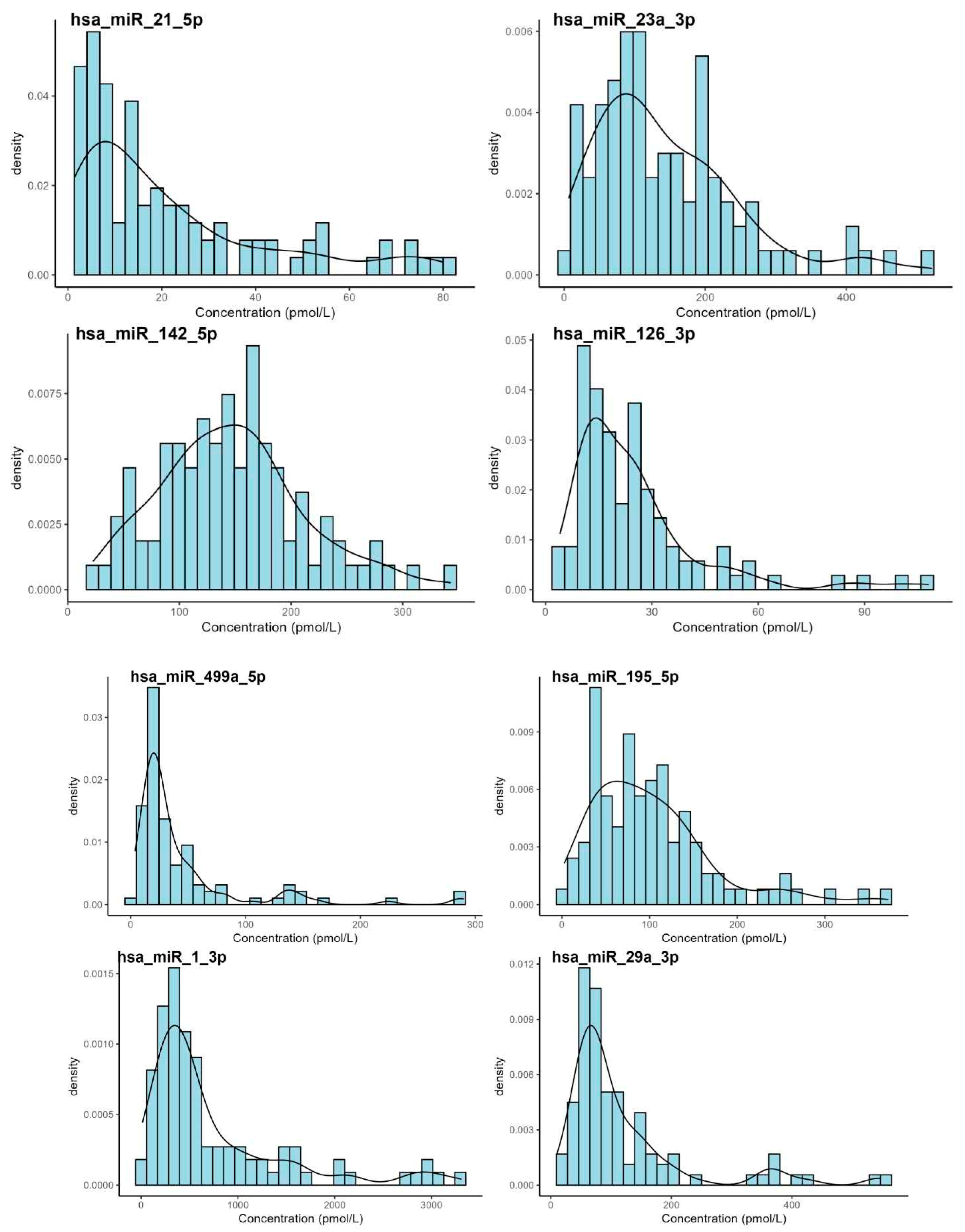
3. Results
4. Discussion
- The establishment of reference intervals for selected miRNAs (hsa-miR-1-3p, hsa-miR-21-5p, hsa-miR-23a-3p, hsa-miR-29a, hsa-miR-126-3p, hsa-miR-142-3p, hsa-miR-195-5p, hsa-miR-499a-5p) in a healthy population,
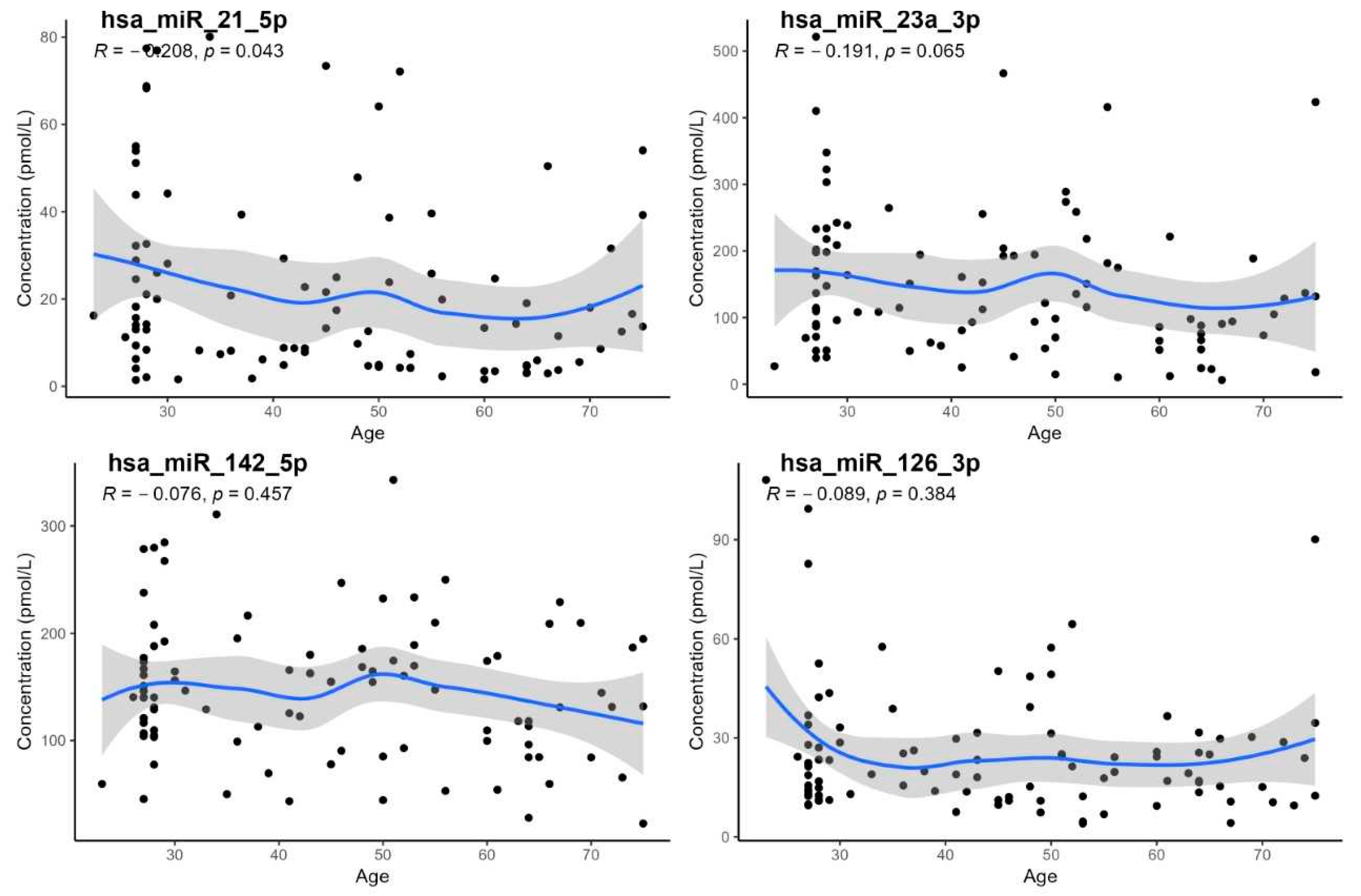
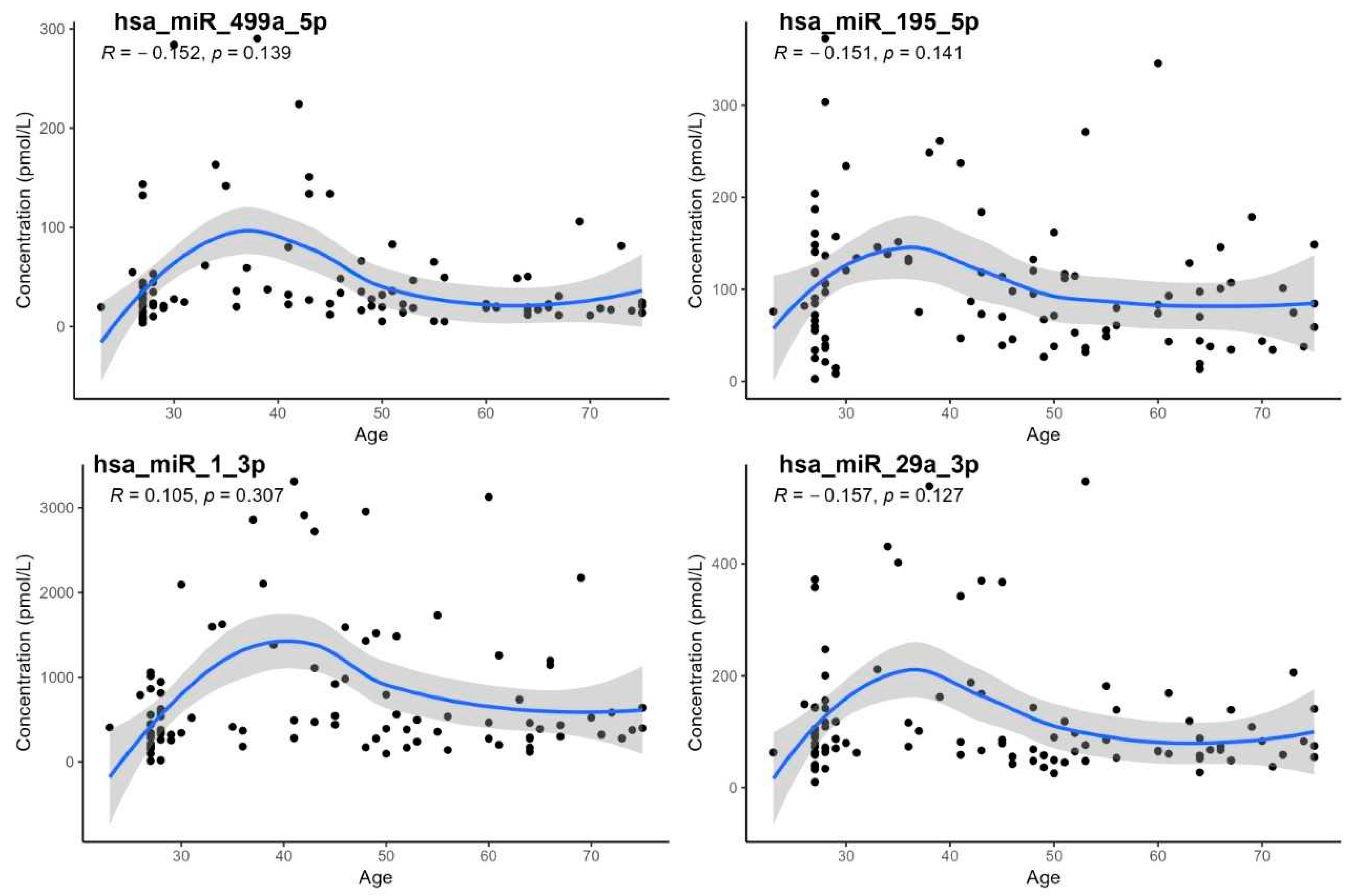
- The demonstration of a statistically significant age dependency only for hsa-miR-21-5p; however, age-specific reference intervals could not be calculated due to the low number of subjects in individual age groups,
- The lack of confirmation of age dependency for hsa-miR-29a, despite some earlier studies having reported such an association [17].
Limitation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALT | alanine aminotransferase |
| AST | aspartate aminotransferase |
| AUC | area under the receiver operating characteristic curve |
| BMI | body mass index |
| CRP | C-reactive protein |
| DCM | dilated cardiomyopathy |
| ERK-MAP kinase | extracellular signal-regulated kinase–mitogen-activated protein kinase |
| FPG | fasting plasma glucose |
| Hb | hemoglobin |
| HbA1c | glycated hemoglobin A1c |
| HCM | hypertrophic cardiomyopathy |
| Hct | hematocrit |
| HRP | horseradish peroxidase |
| HFrEF | heart failure with reduced ejection fraction |
| IL-6 | interleukin 6 |
| Leu | leukocyte |
| LVEF | left ventricular ejection fraction |
| miRNA | microRNA |
| miREIA | microRNA enzymatic immunoassay |
| MYH7 | gene encoding β-myosin heavy chain |
| NT-proBNP | N-terminal pro-B-type natriuretic peptide |
| PLT | platelet |
| RS | Spearman’s rank correlation coefficient |
| ROC | receiver operating characteristic |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
| TMB | tetramethylbenzidine (chromogenic substrate) |
| WBC | white blood cell |
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kamińska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 865–874. [Google Scholar] [CrossRef]
- Çakmak, H.A.; Demir, M. MicroRNA and Cardiovascular Diseases. Balk. Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef]
- Gargiulo, P.; Marzano, F.; Salvatore, M.; Esposito, G.; Bossone, E.; Perrone-Filardi, P. MicroRNAs: Diagnostic, prognostic and therapeutic role in heart failure—A review. ESC Heart Fail. 2023, 10, 753–761. [Google Scholar] [CrossRef]
- Ponikowski, P.; Voors, A.A.; Anker, S.D.; Bueno, H.; Cleland, J.G.F.; Coats, A.J.S.; Falk, V.; González-Juanatey, J.R.; Harjola, V.-P.; Jankowska, E.A.; et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur. Heart J. 2016, 37, 2129–2200. [Google Scholar] [CrossRef]
- Sigutova, R.; Evin, L.; Stejskal, D.; Ploticova, V.; Svagera, Z. Specific microRNAs and heart failure: Time for the next step toward application? Biomed. Pap. 2022, 166, 359–368. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Solberg, H.E. International Federation of Clinical Chemistry (IFCC), Scientific Committee, Clinical Section, Expert Panel on Theory of Reference Values, and International Committee for Standardization in Haematology (ICSH), Standing Committee on Reference Values. Approved Recommendation (1986) on the theory of reference values. Part 1. The concept of reference values. J. Clin. Chem. Clin. Biochem. 1987, 25, 337–342. [Google Scholar] [PubMed]
- Horowitz, G.L. Defining, Establishing, and Verifying Reference Intervals in the Clinical Laboratory, 3rd ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2010. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing [Computer Software], version 4.1.2; R Foundation for Statistical Computing: Vienna, Austria, 2021. Available online: https://www.r-project.org/ (accessed on 12 May 2025).
- Stejskal, D.; Hlozankova, M.; Sigutova, R.; Andelova, K.; Svagera, Z.; Svestak, M. Comparison of a new immunoassay and PCR-based method for quantification of microRNAs in whole blood. A pilot methodical study. Biomed. Pap. 2019, 163, 39–44. [Google Scholar] [CrossRef]
- Krepelkova, I.; Mrackova, T.; Izakova, J.; Dvorakova, B.; Chalupova, L.; Mikulik, R.; Slaby, O.; Bartos, M.; Ruzicka, V. Evaluation of miRNA detection methods for the analytical characteristic necessary for clinical utilization. Biotechniques 2019, 66, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Townsend, N.; Kazakiewicz, D.; Wright, F.L.; Timmis, A.; Huculeci, R.; Torbica, A.; Gale, C.P.; Achenbach, S.; Weidinger, F.; Vardas, P. Epidemiology of cardiovascular disease in Europe. Nat. Rev. Cardiol. 2022, 19, 133–143. [Google Scholar] [CrossRef]
- Kuai, Z.; Ma, Y.; Gao, W.; Zhang, X.; Wang, X.; Ye, Y.; Zhang, X.; Yuan, J. Potential diagnostic value of circulating miRNAs in HFrEF and bioinformatics analysis. Heliyon 2024, 10, e37929. [Google Scholar] [CrossRef]
- Severino, P.; Mancone, M.; D’Amato, A.; Mariani, M.V.; Prosperi, S.; Alunni Fegatelli, D.; Birtolo, L.I.; Angotti, D.; Milanese, A.; Cerrato, E.; et al. Heart failure ‘the cancer of the heart’: The prognostic role of the HLM score. ESC Heart Fail. 2024, 11, 390–399. [Google Scholar] [CrossRef]
- Florio, M.C.; Magenta, A.; Beji, S.; Lakatta, E.G.; Capogrossi, M.C. Aging, MicroRNAs, and Heart Failure. Curr. Probl. Cardiol. 2020, 45, 100406. [Google Scholar] [CrossRef]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146. [Google Scholar] [CrossRef] [PubMed]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, R.; Viviani Anselmi, C.; Losi, M.A.; Papa, L.; Cavarretta, E.; Da Costa Martins, P.; Contaldi, C.; Saccani Jotti, G.; Franzone, A.; Galastri, L.; et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 2014, 63, 920–927. [Google Scholar] [CrossRef]
- Silverman, M.G.; Yeri, A.; Moorthy, M.V.; Camacho Garcia, F.; Chatterjee, N.A.; Glinge, C.S.A.; Tfelt-Hansen, J.; Salvador, A.M.; Pico, A.R.; Shah, R.; et al. Circulating miRNAs and Risk of Sudden Death in Patients With Coronary Heart Disease. JACC Clin. Electrophysiol. 2020, 6, 70–79. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; Chen, L.; Chen, K.; Zhou, J.; Song, J. MiR-1-3p that correlates with left ventricular function of HCM can serve as a potential target and differentiate HCM from DCM. J. Transl. Med. 2018, 16, 161. [Google Scholar] [CrossRef]
- Rincón, L.M.; Rodríguez-Serrano, M.; Conde, E.; Lanza, V.F.; Sanmartín, M.; González-Portilla, P.; Paz-García, M.; Del Rey, J.M.; Menacho, M.; García Bermejo, M.L.; et al. Serum microRNAs are key predictors of long-term heart failure and cardiovascular death after myocardial infarction. ESC Heart Fail. 2022, 9, 3367–3379. [Google Scholar] [CrossRef] [PubMed]
- Jiao, M.; You, H.-Z.; Yang, X.-Y.; Yuan, H.; Li, Y.-L.; Liu, W.-X.; Jin, M.; Du, J. Circulating microRNA signature for the diagnosis of childhood dilated cardiomyopathy. Sci. Rep. 2018, 8, 724. [Google Scholar] [CrossRef]
- Grau-Perez, M.; Martinez-Arroyo, O.; Rubia-Martinez, M.; Flores-Chova, A.; Rodriguez-Hernandez, Z.; Fernández-Navarro, P.; Gonzalez-Neira, A.; Alonso, M.R.; Pita, G.; Pineda, S.; et al. Association of miR-126-3p, miR-1260b and miR-374a-5p with the incidence of heart failure in a population-based cohort: The Hortega Follow-Up Study. Eur. J. Intern. Med. 2025, 135, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Agnese, V.; Sciacca, S.; Scardulla, C.; Cipriani, M.; Pilato, M.; Oh, J.K.; Pasta, S.; Maalouf, J.; Conaldi, P.G.; et al. MicroRNA-30d and -483-3p for bi-ventricular remodelling and miR-126-3p for pulmonary hypertension in advanced heart failure. ESC Heart Fail. 2024, 11, 155–166. [Google Scholar] [CrossRef]
- Fan, X.; Yang, G.; Wang, Y.; Shi, H.; Nitschke, K.; Sattler, K.; Abumayyaleh, M.; Cyganek, L.; Nuhn, P.; Worst, T.; et al. Exosomal mir-126-3p derived from endothelial cells induces ion channel dysfunction by targeting RGS3 signaling in cardiomyocytes: A novel mechanism in Takotsubo cardiomyopathy. Stem. Cell Res. Ther. 2025, 16, 36. [Google Scholar] [CrossRef]
- Baulina, N.; Pisklova, M.; Kiselev, I.; Chumakova, O.; Zateyshchikov, D.; Favorova, O. Circulating miR-499a-5p Is a Potential Biomarker of MYH7-Associated Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2022, 23, 3791. [Google Scholar] [CrossRef]
- Wang, L.; Qin, D.; Shi, H.; Zhang, Y.; Li, H.; Han, Q. MiR-195-5p Promotes Cardiomyocyte Hypertrophy by Targeting MFN2 and FBXW7. BioMed Res. Int. 2019, 2019, 1580982. [Google Scholar] [CrossRef]
- Xuan, L.; Zhu, Y.; Liu, Y.; Yang, H.; Wang, S.; Li, Q.; Yang, C.; Jiao, L.; Zhang, Y.; Yang, B.; et al. Up-regulation of miR-195 contributes to cardiac hypertrophy-induced arrhythmia by targeting calcium and potassium channels. J. Cell. Mol. Med. 2021, 25, 1801. [Google Scholar] [CrossRef]
- Wan, J.; Ling, X.; Peng, B.; Ding, G. miR-142-5p regulates CD4+ T cells in human non-small cell lung cancer through PD-L1 expression via the PTEN pathway. Oncol. Rep. 2018, 40, 272–282. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zampetaki, A.; Kiechl, S.; Drozdov, I.; Willeit, P.; Mayr, U.; Prokopi, M.; Mayr, A.; Weger, S.; Oberhollenzer, F.; Bonora, E.; et al. Plasma MicroRNA Profiling Reveals Loss of Endothelial MiR-126 and Other MicroRNAs in Type 2 Diabetes. Circ. Res. 2010, 107, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Dimmeler, S.; Zeiher, A.M. Circulating microRNAs: Novel biomarkers for cardiovascular diseases? Eur. Heart J. 2010, 31, 2705–2707. [Google Scholar] [CrossRef] [PubMed]
- Schober, A.; Blay, R.M.; Maleki, S.S.; Zahedi, F.; Winklmaier, A.E.; Kakar, M.Y.; Baatsch, I.M.; Zhu, M.; Geißler, C.; Fusco, A.E.; et al. MicroRNA-21 controls circadian regulation of apoptosis in atherosclerotic lesions. Circulation 2021, 144, 1059–1073. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Zhang, P.; Zhou, L.; Yin, B.; Pan, H.; Peng, X. Clock-controlled miR-142-3p can target its activator, Bmal1. BMC Mol. Biol. 2012, 13, 27. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhao, X.; Zhu, X.; Cheng, S.; Xie, Y.; Wang, Z.; Liu, Y.; Jiang, Z.; Xiao, J.; Guo, H.; Wang, Y.; et al. MiR-29a/b/c regulate human circadian gene hPER1 expression by targeting its 3′UTR. Acta Biochim. Biophys. Sin. 2014, 46, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.I.d.R.; Silvello, D.; Martinelli, N.C.; Garbin, A.; Biolo, A.; Clausell, N.; Andrades, M.; dos Santos, K.G.; Rohde, L.E. Plasma levels of microRNA-21, -126 and -423-5p alter during clinical improvement and are associated with the prognosis of acute heart failure. Mol. Med. Rep. 2018, 17, 4736–4746. [Google Scholar] [CrossRef] [PubMed]
- Hromádka, M.; Černá, V.; Pešta, M.; Kučerová, A.; Jarkovský, J.; Rajdl, D.; Rokyta, R.; Moťovská, Z. Prognostic value of microRNAs in patients after myocardial infarction: A substudy of PRAGUE-18. Dis. Markers 2019, 2019, 2925019. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]

| Mean | Median | SD | Minimum | Maximum | Normality Test (p-Value) | |
|---|---|---|---|---|---|---|
| Age | 44.9 | 44.0 | 15.8 | 23.0 | 75.0 | <0.001 |
| BMI | 25.6 | 25.0 | 4.80 | 16.7 | 40.9 | <0.001 |
| Leu [109/L] | 6.36 | 6.06 | 1.66 | 4.10 | 8.91 | <0.001 |
| Hb [g/L] | 139 | 139 | 12 | 120 | 169 | 0.594 |
| Hct [%] | 46.1 | 40.8 | 4.19 | 35.1 | 41.7 | <0.001 |
| Urea [mmol/L] | 4.78 | 4.80 | 1.26 | 1.90 | 8.00 | 0.017 |
| Cr [µmol/L] | 74.5 | 72.0 | 17.0 | 54.0 | 105 | 0.022 |
| ALT [µkat/L] | 0.436 | 0.330 | 0.298 | 0.130 | 0.710 | <0.001 |
| AST [µkat/L] | 0.437 | 0.410 | 0.158 | 0.230 | 0.740 | <0.001 |
| NT-proBNP [ng/L] | 68.2 | 46.0 | 45.9 | 35.0 | 252 | <0.001 |
| FPG [mmol/L] | 5.03 | 5.09 | 0.382 | 4.00 | 5.60 | <0.001 |
| HbA1c [mmol/mol] | 35.3 | 35.5 | 4.37 | 29.0 | 41.0 | <0.001 |
| CRP [mg/L] | 2.20 | 1.30 | 2.50 | 0.200 | 7.90 | <0.001 |
| miRNA | Mean | Median | SD | Min. | Max. | Shapiro-Wilk (p-Value) |
|---|---|---|---|---|---|---|
| hsa-miR-21-5p | 25.2 | 14.3 | 26.9 | 1.46 | 128 | <0.001 |
| hsa-miR-23a-3p | 156 | 119 | 132 | 6.31 | 920 | <0.001 |
| hsa-miR-142-5p | 155 | 145 | 114 | 21.1 | 1080 | <0.001 |
| hsa-miR-126-3p | 28.0 | 21.3 | 26.2 | 4.05 | 156 | <0.001 |
| hsa-miR-499a-5p | 54.8 | 24.7 | 82.3 | 3.79 | 511 | <0.001 |
| hsa-miR-195-5p | 106 | 88.6 | 79.4 | 2.68 | 454 | <0.001 |
| hsa-miR-1-3p | 862 | 468 | 1090 | 13.4 | 8740 | <0.001 |
| hsa-miR-29a-3p | 136 | 82.3 | 163 | 9.81 | 1250 | <0.001 |
| Wilcoxon Test, Sex | Correlation on Age | Correlation on BMI | |||
|---|---|---|---|---|---|
| miRNA | p-Value | RS | p-Value | RS | p-Value |
| hsa-miR-21-5p | 0.360 | −0.208 | 0.043 | −0.057 | 0.590 |
| hsa-miR-23a-3p | 0.570 | −0.191 | 0.065 | 0.005 | 0.960 |
| hsa-miR-126-3p | 0.230 | −0.089 | 0.384 | −0.086 | 0.400 |
| hsa-miR-142-5p | 0.650 | −0.076 | 0.457 | 0.016 | 0.880 |
| hsa-miR-195-5p | 0.083 | −0.151 | 0.141 | −0.140 | 0.160 |
| hsa-miR-1-3p | 0.700 | 0.105 | 0.307 | 0.100 | 0.310 |
| hsa-miR-499a-5p | 0.620 | −0.152 | 0.139 | −0.180 | 0.160 |
| hsa-miR-29a-3p | 0.390 | −0.157 | 0.127 | −0.140 | 0.180 |
| miRNA | n | Outliers | 2.5th | 90% Confidence Interval | 97.5th | 90% Confidence Interval |
|---|---|---|---|---|---|---|
| hsa-miR-21-5p | 95 | 4 | 1.45 | 1.04–2.04 | 96.3 | 78.9–120 |
| hsa-miR-23a-3p | 94 | 5 | 13.0 | 7.78–21.2 | 432 | 370–496 |
| hsa-miR-126-3p | 95 | 4 | 5.67 | 4.70–6.84 | 66.5 | 55.2–72.7 |
| hsa-miR-142-5p | 97 | 2 | 37.4 | 26.9–49.9 | 293 | 269–318 |
| hsa-miR-195-5p | 95 | 4 | 11.5 | 7.06–17.5 | 254 | 222–288 |
| hsa-miR-1-3p | 88 | 11 | 50.6 | 34.4–75.5 | 1800 | 1410–2250 |
| hsa-miR-499a-5p | 86 | 13 | 8.90 | 4.71–7.35 | 82.5 | 67.1–98.8 |
| hsa-miR-29a-3p | 87 | 12 | 22.9 | 18.5–28.4 | 210 | 177–243 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sigutova, R.; Evin, L.; Kusnierova, P.; Stejskal, D.; Vsiansky, F.; Bace, E.; Kufova, E.; Kubikova, G.; Svagera, Z.; Branny, M.; et al. Specific microRNAs for Heart Failure: Reference Values in Whole Blood. Biomedicines 2025, 13, 2559. https://doi.org/10.3390/biomedicines13102559
Sigutova R, Evin L, Kusnierova P, Stejskal D, Vsiansky F, Bace E, Kufova E, Kubikova G, Svagera Z, Branny M, et al. Specific microRNAs for Heart Failure: Reference Values in Whole Blood. Biomedicines. 2025; 13(10):2559. https://doi.org/10.3390/biomedicines13102559
Chicago/Turabian StyleSigutova, Radka, Lukas Evin, Pavlina Kusnierova, David Stejskal, Frantisek Vsiansky, Eva Bace, Eliska Kufova, Gabriela Kubikova, Zdenek Svagera, Marian Branny, and et al. 2025. "Specific microRNAs for Heart Failure: Reference Values in Whole Blood" Biomedicines 13, no. 10: 2559. https://doi.org/10.3390/biomedicines13102559
APA StyleSigutova, R., Evin, L., Kusnierova, P., Stejskal, D., Vsiansky, F., Bace, E., Kufova, E., Kubikova, G., Svagera, Z., Branny, M., & Vaclavik, J. (2025). Specific microRNAs for Heart Failure: Reference Values in Whole Blood. Biomedicines, 13(10), 2559. https://doi.org/10.3390/biomedicines13102559







