PRRs-Dependent and Independent Mechanisms of STING Signaling in Inflammatory and Autoimmune Diseases
Abstract
1. Introduction
2. PRRs Dependent STING Signaling
2.1. DNA-cGAS-STING Signaling
2.2. DNA-IFI16-STING Signaling
2.3. DNA-(DNA-PK)-STING Signaling
2.4. DNA-DDX41-STING Signaling
3. PRRs-Independent STING Signaling
3.1. STING Mutations
3.2. ER Homeostasis
3.3. STING Trafficking
3.4. STING Stabilization
3.5. STING Functions as a More Central Node than PRRs
| Activation Mode | Key Regulators | Mechanism | Disease Systems & Models | Reference |
|---|---|---|---|---|
| STING mutations | V155M, N154S, V147L, C206Y, G207E, R281Q, R284S, H72N, G166E site of STING | Mutation altering amino acids at the dimerization interface or ligand-binding domain. | SAVI associated disease | [61,62] |
| STING mutations | C88Y, R96L, V193I, C88Y, R96L, V193I | Mutations in the transmembrane domain alter STING topology and cause constitutive activation. | SAVI associated disease | [107] |
| ER stress | TOLLIP (positive regulator) | Transmembrane domain mutations disrupt STING topology, leading to constitutive activation. | Autoinflammatory disease | [58] |
| ER stress | HRD1 (negative regulator) | ERAD directly interacts with, and ubiquitinates, ER-resident resting STING, leading to its proteasomal degradation. | Viral infection and tumor models | [55] |
| ER stress | UNC13D (negative regulator) | UNC13D colocalizes with STING on the ER, inhibiting STING oligomerization and activation. | Cell level | [108] |
| STING trafficking | STIM1, NSP2, ISD017 (negative regulator) | STIM1 binds resting STING, retaining it on the ER and suppressing spontaneous activation; NSP2 and ISD017 further potentiate this STIM1-mediated inhibition. | Autoinflammatory disease | [82,83,109] |
| STING trafficking | STEEP (positive regulator) | Inducing COPII-mediated ER to Golgi trafficking of STING. | SAVI associated disease | [84] |
| STING trafficking | iRhom2, ADAM17 (positive regulator) | ADAM17 selectively stabilizes iRhom2 as a positive regulator of STING. | Viral infection and tumor models | [56,85] |
| STING trafficking | SCAP (positive regulator) | SCAP facilitates STING translocation to the Golgi. | nonalcoholic fatty liver disease | [87] |
| STING trafficking | COP II (positive regulator) | STING phosphorylation cues COPII transport; post-fusion coat loss at the ERGIC/cis-Golgi enables IRF3 recruitment for TBK1-mediated phosphorylation. | SAVI associated disease | [110] |
| STING trafficking | COP I (negative regulator) | Mutations in COP components result in aberrant ER localization of STING. | COPA syndrome | [81,91] |
| STING trafficking | ACBD3 (positive regulator) | ACBD3 facilitates COP II-independent ER exit of STING and its trafficking to the Golgi. | Cell level | [88] |
| STING trafficking | YIPF5 (positive regulator) | YIPF5 promotes STING signaling by recruiting it to COPII vesicles and driving ER-to-Golgi trafficking. | Viral infection | [111] |
| STING stabilization | CYLD, USP18, USP20, eIF3f (negative regulator) | STING stability is regulated by ubiquitination, and lysosomal degradation mechanisms. | Viral infection | [56,92,93] |
| STING stabilization | TOLLIP (positive regulator) | TOLLIP inhibits the lysosomal degradation of STING. | Huntington’s disease & viral infection | [58,112] |
| IRE1α (negative regulator) | IRE1α positively regulates the lysosomal degradation of STING. | Huntington’s disease | [58,113] | |
| STING stabilization | NPC1 | NPC1 interacts with STING and recruits it to the lysosome for degradation in both human and mouse cells. | Niemann-Pick disease type C | [86] |
| STING stabilization | ESCRT | The ESCRT complex facilitates ubiquitin-mediated degradation of STING. | Cell level | [95,96] |
4. STING Activation in Inflammatory and Autoimmune Diseases
4.1. Digestive System Diseases
4.2. Cardiovascular Diseases
4.3. Kidney Diseases
4.4. Pulmonary Diseases
4.5. Neurodegenerative Diseases
4.6. Autoimmune Diseases
4.7. PRRs-Independent STING Activation Has Been Severely Underestimated
| System Diseases | Activated Cells/Tissue | Trigger | Mechanism of STING Activation | PRRs/Non-PRRs |
|---|---|---|---|---|
| Digestive system diseases | ||||
| Severe acute pancreatitis | Macrophages | mtDNA | mtDNA―cGAS―STING―IRF3/IRF7―NLRP3―SAP [126]. | cGAS |
| Acute pancreatitis | Macrophages | Cytosolic DNA of acinar cells | Cytosolic DNA/cell―free DNA―cGAS―STING―NF kB/IRF3―TNFα/IFNβ―AP [127]. | cGAS |
| Acute pancreatitis | Pancreatic acinar cells | mtDNA of acinar cells | cGAS―STING―NF-κB―AP [208]. | cGAS |
| Acute pancreatitis | Pancreatic β cells | Inflammatory mitochondria | Inflammatory mitochondria from macrophages―cGAS―STING activation [209]. | cGAS |
| Acute liver injury | Macrophages | mtDNA | mtDNA―cGAS―STING―NLRP3―hepatocyte pyroptosis [131]. | cGAS |
| Hepatic inflammation and fibrosis | Macrophages | cGAMP | cGAMP―STING―JNK1/NFκB―hepatocyte and HSC inflammatory and fibrosis [132]. | cGAMP |
| Inflammatory colitis | Monocytes | cGAS/CDNs | cGAS/microbiota―STING―MyD88―IFNβ/IL1β/IL18―inflammatory colitis [135]. | cGAS |
| Insulin resistance and inflammatory colitis | Hepatocytes, adipocytes and macrophages | DNA from mEVs | mEVs―cGAS-STING―insulin resistance and inflammatory colitis [136,210]. | cGAS |
| Colitis | Dendritic cells | Protein-mtDNA complex from damaged colonic epithelial cells | protein-mtDNA complex―STING―IRF3/NF-κB―IL-12 (T helper cell)―chronic colitis [137]. | protein-mtDNA complex (DAMPs) |
| Liver fibrosis | Hepatocytes | TNF-α | TNF-α―STING―IRF3/WDR5/DOT1L―NLRP3―GSDMD [130]. | cGAS |
| Alcoholic liver disease | Hepatocytes | ER stress | ER stress―STING―IRF3―hepatocyte apoptosis [138]. | non-PRRs |
| Liver injury and fibrosis | Hepatocytes | ER stress | ER stress―STING―IRF3―hepatocyte apoptosis [139] | non-PRRs |
| Liver metaflammation | Macrophages | STING trafficking | SCAP―STING―TBK1―NF-κB―metaflammation [87] | non-PRRs |
| Liver injury | Macrophages | Malonyl-CoA/FASN | Malonyl-CoA inhibits STING palmitoylation to alleviate sepsis-induced liver injury [211]. | non-PRRs |
| Cardiovascular diseases | ||||
| Vascular diseases induced by obesity | Aortic endothelial cells | mtDNA | mtDNA―cGAS―STING―IRF3―ICAM-1―endothelial activation/inflammation [142]. | cGAS |
| Atherogenesis | Macrophages | mtDNA | mtDNA―cGAS―STING―TBK1―NF-κB―atherogenesis [146]. | cGAS |
| Cardiac dysfunction | Ventricular myocytes & Macrophages | mtDNA | iNOS―mtDNA―cGAS―STING―IRF3―cardiac dysfunction [145]. | cGAS |
| Vascular endothelial dysfunction | Artery endothelial cells | Self-mtDNA | cGAS―STING―PERK―IRF3/NF-κB―vascular endothelial dysfunction and atherosclerosis [212]. | cGAS |
| SAVI associated myocarditis | Monocytes & endothelial cells | Mutant STING | Mutant STING―NF-κB/IFN-β―inflammation [15,148]. | non-PRRs |
| Pathological cardiac hypertrophy | Cardiac fibroblasts | ER stress | AC or Ang II―ER stress―STING-TBK1-RIF3/ NF-κB heart inflammation and fibrosis [74]. | non-PRRs |
| Myocardial ischemia/reperfusion | Cardiomyocyte | ER stress | ER stress―STING-IRF3―Rubicon―autophagic flux dysfunction [72] | non-PRRs |
| Diabetic retinopathy | Retinal endothelial | ER stress | Hyperlipidemia―IRE1α―XBP1―ER stress―STING―TBK1―NF-κB―pro-inflammatory [78]. | non-PRRs |
| Kidney diseases | ||||
| Acute kidney injury | Tubular cells | mtDNA | mtDNA―cGAS―STING―TBK1―p65―neutrophil infiltration and tissue inflammation [149]. | cGAS |
| Glomerular diseases | Podocyte cells | mtDNA | cGAS―STING―TBK1―IRF3/IFN α―glomerular diseases [213]. | cGAS |
| Kidney inflammation and fibrosis | Macrophages | dsDNA | dsDNA―cGAS―STING―TBK1/IRF3―p65 axis suppresses kidney inflammation and fibrosis [152]. | cGAS |
| Acute kidney injury | Tubular cells | ER stress crosstalk | STING―ER stress―mtROS―NLRP3 inflammasome―acute kidney injury [155]. | non-PRRs |
| SAVI-associated glomerulosclerosis | Glomerulus | Mutant STING | Mutant STING induces glomerulosclerosis [157]. | non-PRRs |
| Pulmonary diseases | ||||
| Lung inflammation | Monocytes, macrophages & dendritic cells | dsDNA | SiO2―dsDNA―cGAS―STING―IFN I pathway drives inflammation [161]. | cGAS |
| Silica particles | Macrophages & fibroblasts | dsDNA | SiO2―dsDNA―cGAS―STING―IRF3/NF-κB―macrophage polarization―lung fibroblast proliferation [160]. | cGAS |
| Lung injury | Lung epithelial cells & neutrophils | dsDNA from NETs | NETs―cGAS―STING pathway promotes inflammatory injury [164,169,170]. | cGAS |
| Acute lung injury | Lung epithelial cells | mtDNA | Mitophagy―mtDNA―cGAS―STING―IFN I/IL-6―ALI [162,171]. | cGAS |
| Neurodegenerative diseases | ||||
| Alzheimer’s disease | Microglia | Aβ, tau and APOE (Apolipoprotein E) ε4-DNA | Aβ, tau and APOE (Apolipoprotein E) ε4-DNA―cGAS―STING―TBK1―IRF3/NF-κB―neuroinflammation and phospho-tau levels [214,215]. | cGAS |
| Alzheimer’s disease | Astrocytes | DNA | IL-6―STAT3 regulates cGAS―STING―neuroinflammation [216]. | cGAS |
| Parkinson’s disease | Astrocytes | mtDNA | cGAS―STING―YY1―LCN2―neurodegeneration [185]. | cGAS |
| Parkinson’s disease | Microglia | mtDNA | cGAS―STING―IRF7―neuroinflammation in Parkinson’s disease [217]. | cGAS |
| Huntington’s disease | T cells, B cells, Dendritic cells & MEFs | STING destabilization | TOLLIP―STING―IRF3―neuroinflammation [58]. | non-PRRs |
| Traumatic brain injury | Neurons | ER stress induced PERK | PERK―ER stress―STING―TBK1―IRF3―IFN-β―white matter injury [79]. | non-PRRs |
| Autoimmune diseases | ||||
| Spontaneous lupus-like inflammatory disease | Systemic tissue | dsDNA induced by mutant TREX1 | Mutant TREX1-dsDNA―cGAS―STING―spontaneous lupus-like inflammatory [190]. | cGAS |
| Familial chilblain lupus | Dendritic cells | Mutant STING | Mutant STING―IFN I―familial chilblain lupus [63,218]. | non-PRRs |
| Autoimmune and autoinflammatory diseases | Murine embryonic fibroblasts | STIM1 deletion or mutation | STIM1 deletion or mutation―STING―IFN―inflammation [77] | non-PRRs |
| Niemann-Pick disease type C | Various cells (e.g., monocytes, macrophages, neurons, fibroblasts) | STING trafficking | NPC1 deficiency―SREBP2 activation―STING―inflammation [86] | non-PRRs |
| COPA syndrome | Various cells (e.g., dendritic cells, macrophages, neurons, fibroblasts) | STING trafficking | COPA mutant―STING stay and palmitoylation―TBKI―inflammation [90,91] | non-PRRs |
| Behçet’s syndrome | Endothelial cells, neutrophils | dsDNA | TNF-α upregulated IFI16―STING―TBK1―apoptosis of intestinal epithelial cells [35]. | IFI16 |
5. Current Therapeutics to Target STING Signaling
5.1. Inhibiting cGAS Enzymatic Activity
5.2. cGAS-dsDNA Binding Inhibitors
5.3. Other cGAS Inhibitors
5.4. STING Antagonists Targeting the CDN-Binding Site
5.5. Targeting STING Phosphorylation Sites
5.6. Targeting STING Palmitoylation Sites
5.7. Targeting STING Cys292 Sites
5.8. Targeting STING Degradation
5.9. Attenuating STING Trafficking
5.10. Clinical Status and Translational Challenges
6. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AC | Aortic Constriction |
| ACBD3 | Acyl-CoA Binding Domain Protein 3 |
| ACSL4 | Acyl-coenzyme A (CoA) Synthetase Long-chain Family Member 4 |
| ADAR1 | Adenosine Deaminase Acting on RNA 1 |
| AGS | Aicardi-Goutières Syndrome |
| AIM2 | Absent in Melanoma 2 |
| ALI | Acute Lung Injury |
| ALS | Amyotrophic Lateral Sclerosis |
| AMP | Adenosine Monophosphate |
| Ang II | Angiotensin II |
| AP | Acute Pancreatitis |
| APOE | Apolipoprotein E |
| ARDS | Acute Respiratory Distress Syndrome |
| ATM | Ataxia Telangiectasia Mutated |
| ATR | ATM and Rad3-Related |
| ATP | Adenosine Triphosphate |
| BSN | Bilateral Striatal Necrosis |
| CDN | Cyclic Dinucleotide |
| COP II | Coat Protein Complex II |
| CTT | C-Terminal Tail |
| CYLD | Cylindromatosis Lysine 63 Deubiquitinase |
| cGAMP | Cyclic GMP-AMP |
| cGAS | Cyclic GMP-AMP Synthase |
| COPA | Coatomer Subunit α |
| DAMP | Damage-Associated Molecular Pattern |
| DDX41 | DEAD-Box Helicase 41 |
| DCs | Dendritic Cells |
| DNA-PK | DNA-Dependent Protein Kinase |
| DNase | Deoxyribonuclease |
| DOT1L | Disruptor of Telomeric Silencing 1-Like |
| dsDNA | Double-Stranded DNA |
| DSS | Dextran Sulfate Sodium |
| eIF3f | Eukaryotic Translation Initiation Factor 3 Subunit F |
| ER | Endoplasmic Reticulum |
| ERGIC | ER-Golgi Intermediate Compartment |
| ESCRT | Endosomal Sorting Complex Required for Transport |
| FASN | Fatty Acid Synthase |
| GS | Gelsevirine |
| GTP | Guanosine Triphosphate |
| hnRNPA2B1 | Heterogeneous Nuclear Ribonucleoprotein A2/B1 |
| HSC | Hepatic Stellate Cell |
| HSV-1 | Herpes Simplex Virus Type 1 |
| HSV-2 | Herpes Simplex Virus Type 2 |
| ICAM-1 | Intercellular adhesion molecule 1 |
| IFI16 | Interferon Gamma Inducible Protein 16 |
| IFIH1 | Interferon Induced with Helicase C Domain 1 |
| IFN | Interferon |
| IFNAR1 | Interferon Alpha/Beta Receptor 1 |
| IFNAR2 | Interferon Alpha/Beta Receptor 2 |
| IKK | IκB Kinase |
| IL-1β | Interleukin-1 Beta |
| IL-6 | Interleukin-6 |
| IL-18 | Interleukin-18 |
| ILD | Interstitial Lung Disease |
| iNOS | Inducible Nitric Oxide Synthase |
| iRhom2 | Inactive Rhomboid Protein 2 |
| IRF3 | Interferon Regulatory Factor 3 |
| IRF9 | Interferon Regulatory Factor 9 |
| ISG | Interferon-Stimulated Gene |
| Jak1 | Janus Kinase 1 |
| JNK1 | c-Jun N-Terminal Kinase 1 |
| K48 | Lysine 48-Linked Ubiquitination |
| LAMP1 | Lysosomal Associated Membrane Protein 1 |
| LCN2 | Lipocalin 2 |
| LSM11 | U7 small nuclear RNA associated protein |
| LPS | Lipopolysaccharide |
| LXR | Liver X Receptor |
| MDA5 | Melanoma Differentiation-Associated Protein 5 |
| MEF | Mouse Embryonic Fibroblasts |
| mtDNA | Mitochondrial DNA |
| MXC | Meloxicam |
| MyD88 | Myeloid Differentiation Primary Response 88 |
| NAFLD | Non-Alcoholic Fatty Liver Disease |
| NEMO | NF-κB Essential Modulator |
| NET | Neutrophil Extracellular Trap |
| NF-κB | Nuclear Factor Kappa B |
| NLRP3 | NLR Family Pyrin Domain Containing 3 |
| NO2-cLA | Nitro-Conjugated Linoleic Acid |
| NO2-OA | Nitro-Oleic Acid |
| NPC1 | Niemann-Pick Type C1 |
| NSCLC | Non-Small Cell Lung Cancer |
| p-IRF3 | Phosphorylated IRF3 |
| p-tau | Phosphorylated Tau |
| PAMP | Pathogen-Associated Molecular Pattern |
| PBMC | Peripheral Blood Mononuclear Cell |
| PERK | Protein Kinase R-like ER Kinase |
| PIKK | Phosphoinositide-3-Kinase-Related Kinase |
| PLK1 | Polo-Like Kinase 1 |
| PM2.5 | Particulate Matter ≤2.5 μm |
| PRRs | Pattern Recognition Receptors |
| PROTAC | Proteolysis-Targeting Chimera |
| PtdIns(3)P | phosphatidylinositol-3-phosphate |
| RIG-I | Retinoic Acid-Inducible Gene I |
| RLR | RIG-I-Like Receptor |
| RNF5 | Ring Finger Protein 5 |
| RNF90 | Ring Finger Protein 90 |
| RNU7-1 | U7 Small Nuclear 1 |
| ROS | Reactive Oxygen Species |
| SAVI | STING-Associated Vasculopathy with Onset in Infancy |
| SAMHD1 | SAM and HD Domain Containing Protein 1 |
| SCAP | SREBP Cleavage-Activating Protein |
| SLE | Systemic Lupus Erythematosus |
| SMPDL3A | Sphingomyelin Phosphodiesterase Acid-Like 3A |
| SREBP2 | Sterol Regulatory Element-Binding Protein 2 |
| STAT | Signal Transducer and Activator of Transcription |
| STEEP | STING ER Exit Protein |
| STIM1 | Stromal Interaction Molecule 1 |
| STING | Stimulator of Interferon Genes |
| TBK1 | TANK-Binding Kinase 1 |
| TCR | T Cell Receptor |
| TLR | Toll-Like Receptor |
| TNF-α | Tumor Necrosis Factor Alpha |
| TOLLIP | Toll-Interacting Protein |
| TREX1 | Three Prime Repair Exonuclease 1 |
| TRIF | TIR-Domain-Containing Adapter-Inducing Interferon-β |
| TRIM13 | Tripartite Motif Containing 13 |
| TRIM21 | Tripartite Motif Containing 21 |
| TRIM29 | Tripartite Motif Containing 29 |
| TRIM30α | Tripartite Motif Containing 30 Alpha |
| Tyk2 | Tyrosine Kinase 2 |
| USP18 | Ubiquitin-Specific Peptidase 18 |
| USP20 | Ubiquitin-Specific Peptidase 20 |
| V155M | Valine 155 to Methionine Mutation |
| WDR5 | WD Repeat Domain 5 |
| XBP1 | X-Box Binding Protein 1 |
| YY1 | Yin Yang 1 |
References
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Vivier, E.; Malissen, B. Innate and adaptive immunity: Specificities and signaling hierarchies revisited. Nat. Immunol. 2005, 6, 17–21. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Ma, M.; Jiang, W.; Zhou, R. DAMPs and DAMP-sensing receptors in inflammation and diseases. Immunity 2024, 57, 752–771. [Google Scholar] [CrossRef]
- Lin, H.; Cao, X. Nuclear innate sensors for nucleic acids in immunity and inflammation. Immunol. Rev. 2020, 297, 162–173. [Google Scholar] [CrossRef]
- Chauvin, S.D.; Stinson, W.A.; Platt, D.J.; Poddar, S.; Miner, J.J. Regulation of cGAS and STING signaling during inflammation and infection. J. Biol. Chem. 2023, 299, 104866. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chiu, Y.H.; Chen, Z.J. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 2014, 54, 289–296. [Google Scholar] [CrossRef]
- Unterholzner, L.; Keating, S.E.; Baran, M.; Horan, K.A.; Jensen, S.B.; Sharma, S.; Sirois, C.M.; Jin, T.; Latz, E.; Xiao, T.S.; et al. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010, 11, 997–1004. [Google Scholar] [CrossRef]
- Zhang, Z.; Yuan, B.; Bao, M.; Lu, N.; Kim, T.; Liu, Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat. Immunol. 2011, 12, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.J.; Mansur, D.S.; Peters, N.E.; Ren, H.; Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012, 1, e00047. [Google Scholar] [CrossRef]
- Wang, L.; Wen, M.; Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019, 365, eaav0758. [Google Scholar] [CrossRef]
- Sun, W.; Li, Y.; Chen, L.; Chen, H.; You, F.; Zhou, X.; Zhou, Y.; Zhai, Z.; Chen, D.; Jiang, Z. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. USA 2009, 106, 8653–8658. [Google Scholar] [CrossRef]
- Zhong, B.; Yang, Y.; Li, S.; Wang, Y.Y.; Li, Y.; Diao, F.; Lei, C.; He, X.; Zhang, L.; Tien, P.; et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 2008, 29, 538–550. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Qin, D.; Zhao, C.; Chai, L.; Yu, Z.; Wang, W.; Tong, L.; Lv, L.; Wang, Y.; Rehwinkel, J.; et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 2020, 21, 727–735. [Google Scholar] [CrossRef]
- Valeri, E.; Breggion, S.; Barzaghi, F.; Abou Alezz, M.; Crivicich, G.; Pagani, I.; Forneris, F.; Sartirana, C.; Costantini, M.; Costi, S.; et al. A novel STING variant triggers endothelial toxicity and SAVI disease. J. Exp. Med. 2024, 221, e20232167. [Google Scholar] [CrossRef]
- Liu, S.; Yang, B.; Hou, Y.; Cui, K.; Yang, X.; Li, X.; Chen, L.; Liu, S.; Zhang, Z.; Jia, Y.; et al. The mechanism of STING autoinhibition and activation. Mol. Cell 2023, 83, 1502–1518.e10. [Google Scholar] [CrossRef]
- Fang, R.; Jiang, Q.; Jia, X.; Jiang, Z. ARMH3-mediated recruitment of PI4KB directs Golgi-to-endosome trafficking and activation of the antiviral effector STING. Immunity 2023, 56, 500–515.e6. [Google Scholar] [CrossRef]
- Zhang, Y.; Fu, Y.; Qiang, L.; Zhao, M.; Lu, Z.; Zhao, Z.; Chen, G.; Lei, Z.; Chai, Q.; Ge, P.; et al. Spatiotemporal Regulation of STING Activity by Linear Ubiquitination Governs Antiviral Immunity. Adv. Sci. 2025, 12, e2417660. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, C. Regulation of cGAS-STING signalling and its diversity of cellular outcomes. Nat. Rev. Immunol. 2025, 25, 425–444. [Google Scholar] [CrossRef] [PubMed]
- Lanng, K.R.B.; Lauridsen, E.L.; Jakobsen, M.R. The balance of STING signaling orchestrates immunity in cancer. Nat. Immunol. 2024, 25, 1144–1157. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Mei, J.; Guo, H.; Zhu, J.; Wang, C. Intervention of cGAS-STING signaling in sterile inflammatory diseases. J. Mol. Cell Biol. 2022, 14, mjac005. [Google Scholar] [CrossRef]
- Wu, J.; Sun, L.; Chen, X.; Du, F.; Shi, H.; Chen, C.; Chen, Z.J. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 2013, 339, 826–830. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]
- Du, M.; Chen, Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science 2018, 361, 704–709. [Google Scholar] [CrossRef]
- Zhao, B.; Du, F.; Xu, P.; Shu, C.; Sankaran, B.; Bell, S.L.; Liu, M.; Lei, Y.; Gao, X.; Fu, X.; et al. A conserved PLPLRT/SD motif of STING mediates the recruitment and activation of TBK1. Nature 2019, 569, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.C.; Chen, Z.J. Structural basis of STING binding with and phosphorylation by TBK1. Nature 2019, 567, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Motwani, M.; Pesiridis, S.; Fitzgerald, K.A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 2019, 20, 657–674. [Google Scholar] [CrossRef]
- Corrales, L.; Matson, V.; Flood, B.; Spranger, S.; Gajewski, T.F. Innate immune signaling and regulation in cancer immunotherapy. Cell Res. 2017, 27, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, T.; Bodda, C.; Krapp, C.; Zhang, B.C.; Christensen, M.H.; Sun, C.; Reinert, L.; Cai, Y.; Jensen, S.B.; Skouboe, M.K.; et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018, 37, e97858. [Google Scholar] [CrossRef]
- Zierhut, C.; Funabiki, H. Regulation and Consequences of cGAS Activation by Self-DNA. Trends Cell Biol. 2020, 30, 594–605. [Google Scholar] [CrossRef]
- Zheng, J.; Mo, J.; Zhu, T.; Zhuo, W.; Yi, Y.; Hu, S.; Yin, J.; Zhang, W.; Zhou, H.; Liu, Z. Comprehensive elaboration of the cGAS-STING signaling axis in cancer development and immunotherapy. Mol. Cancer 2020, 19, 133. [Google Scholar] [CrossRef]
- Andreeva, L.; Hiller, B.; Kostrewa, D.; Lässig, C.; de Oliveira Mann, C.C.; Jan Drexler, D.; Maiser, A.; Gaidt, M.; Leonhardt, H.; Hornung, V.; et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein-DNA ladders. Nature 2017, 549, 394–398. [Google Scholar] [CrossRef]
- Jønsson, K.L.; Laustsen, A.; Krapp, C.; Skipper, K.A.; Thavachelvam, K.; Hotter, D.; Egedal, J.H.; Kjolby, M.; Mohammadi, P.; Prabakaran, T.; et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017, 8, 14391. [Google Scholar] [CrossRef]
- Almine, J.F.; O’Hare, C.A.; Dunphy, G.; Haga, I.R.; Naik, R.J.; Atrih, A.; Connolly, D.J.; Taylor, J.; Kelsall, I.R.; Bowie, A.G.; et al. IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat. Commun. 2017, 8, 14392. [Google Scholar] [CrossRef]
- Li, S.; Xu, H.; Song, M.; Shaw, B.I.; Li, Q.J.; Kirk, A.D. IFI16-STING-NF-κB signaling controls exogenous mitochondrion-induced endothelial activation. Am. J. Transplant. 2022, 22, 1578–1592. [Google Scholar] [CrossRef]
- Dunphy, G.; Flannery, S.M.; Almine, J.F.; Connolly, D.J.; Paulus, C.; Jønsson, K.L.; Jakobsen, M.R.; Nevels, M.M.; Bowie, A.G.; Unterholzner, L. Non-canonical Activation of the DNA Sensing Adaptor STING by ATM and IFI16 Mediates NF-κB Signaling after Nuclear DNA Damage. Mol. Cell 2018, 71, 745–760.e5. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Yan, L.; Liu, N.; Xu, M.; Cai, H. IFI16 promotes cervical cancer progression by upregulating PD-L1 in immunomicroenvironment through STING-TBK1-NF-kB pathway. Biomed. Pharmacother. 2020, 123, 109790. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.F.; She, C.H.; Hou, C.C.; Ji, D.N.; Hu, D.; Zou, J.; Shen, Y.; Jian, L.L.; Cai, J.F.; Ye, J.F.; et al. PLK1-activating IFI16-STING-TBK1 pathway induces apoptosis of intestinal epithelial cells in patients with intestinal Behçet’s syndrome. FEBS J. 2024, 291, 3432–3453. [Google Scholar] [CrossRef]
- Kim, H.; Kim, H.; Feng, Y.; Li, Y.; Tamiya, H.; Tocci, S.; Ronai, Z.A. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci. Transl. Med. 2020, 12, eaaz5683. [Google Scholar] [CrossRef]
- Li, Y.; Gao, Y.; Jiang, X.; Cheng, Y.; Zhang, J.; Xu, L.; Liu, X.; Huang, Z.; Xie, C.; Gong, Y. SAMHD1 silencing cooperates with radiotherapy to enhance anti-tumor immunity through IFI16-STING pathway in lung adenocarcinoma. J. Transl. Med. 2022, 20, 628. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Hristova, D.B.; Oliveira, M.; Wagner, E.; Melcher, A.; Harrington, K.J.; Belot, A.; Ferguson, B.J. DNA-PKcs is required for cGAS/STING-dependent viral DNA sensing in human cells. iScience 2024, 27, 108760. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Song, J.; Song, Y.; Zhang, Y.; Yang, J.; Zhang, P.; Zhang, D.; Chen, D.; Sun, Q. Ku proteins promote DNA binding and condensation of cyclic GMP-AMP synthase. Cell Rep. 2022, 40, 111310. [Google Scholar] [CrossRef]
- Burleigh, K.; Maltbaek, J.H.; Cambier, S.; Green, R.; Gale, M., Jr.; James, R.C.; Stetson, D.B. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol. 2020, 5, eaba4219. [Google Scholar] [CrossRef] [PubMed]
- Parvatiyar, K.; Zhang, Z.; Teles, R.M.; Ouyang, S.; Jiang, Y.; Iyer, S.S.; Zaver, S.A.; Schenk, M.; Zeng, S.; Zhong, W.; et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 2012, 13, 1155–1161. [Google Scholar] [CrossRef]
- Omura, H.; Oikawa, D.; Nakane, T.; Kato, M.; Ishii, R.; Ishitani, R.; Tokunaga, F.; Nureki, O. Structural and Functional Analysis of DDX41: A bispecific immune receptor for DNA and cyclic dinucleotide. Sci. Rep. 2016, 6, 34756. [Google Scholar] [CrossRef]
- Lee, K.G.; Kim, S.S.; Kui, L.; Voon, D.C.; Mauduit, M.; Bist, P.; Bi, X.; Pereira, N.A.; Liu, C.; Sukumaran, B.; et al. Bruton’s tyrosine kinase phosphorylates DDX41 and activates its binding of dsDNA and STING to initiate type 1 interferon response. Cell Rep. 2015, 10, 1055–1065. [Google Scholar] [CrossRef]
- Singh, R.S.; Vidhyasagar, V.; Yang, S.; Arna, A.B.; Yadav, M.; Aggarwal, A.; Aguilera, A.N.; Shinriki, S.; Bhanumathy, K.K.; Pandey, K.; et al. DDX41 is required for cGAS-STING activation against DNA virus infection. Cell Rep. 2022, 39, 110856. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, X.; Wang, Z.; Liu, P.; Hou, Y.; Xu, Y.; Su, H.; Koci, M.D.; Yin, H.; Zhang, C. NF-κB activation enhances STING signaling by altering microtubule-mediated STING trafficking. Cell Rep. 2023, 42, 112185. [Google Scholar] [CrossRef]
- Wu, X.; Yu, N.; Ye, Z.; Gu, Y.; Zhang, C.; Chen, M.; Wang, K. Inhibition of cGAS-STING pathway alleviates neuroinflammation-induced retinal ganglion cell death after ischemia/reperfusion injury. Cell Death Dis. 2023, 14, 615. [Google Scholar] [CrossRef] [PubMed]
- Mowat, C.; Schiller, D.; Baker, K. NLRP3 activation promotes cGAS/STING signaling and antitumor immunity by colorectal cancer cells. Cancer Immunol. Immunother. 2025, 74, 238. [Google Scholar] [CrossRef]
- Zhang, R.; Yang, W.; Zhu, H.; Zhai, J.; Xue, M.; Zheng, C. NLRC4 promotes the cGAS-STING signaling pathway by facilitating CBL-mediated K63-linked polyubiquitination of TBK1. J. Med. Virol. 2023, 95, e29013. [Google Scholar] [CrossRef]
- Mangan, M.S.; Latz, E. NLRC3 puts the brakes on STING. Immunity 2014, 40, 305–306. [Google Scholar] [CrossRef]
- Luo, Y.; Chang, L.; Ji, Y.; Liang, T. ER: A critical hub for STING signaling regulation. Trends Cell Biol. 2024, 34, 865–881. [Google Scholar] [CrossRef]
- Ji, Y.; Luo, Y.; Wu, Y.; Sun, Y.; Zhao, L.; Xue, Z.; Sun, M.; Wei, X.; He, Z.; Wu, S.A.; et al. SEL1L-HRD1 endoplasmic reticulum-associated degradation controls STING-mediated innate immunity by limiting the size of the activable STING pool. Nat. Cell Biol. 2023, 25, 726–739. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.W.; Li, S.; Li, C.; Lian, H.; Yang, Q.; Zhong, B.; Shu, H.B. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat. Immunol. 2016, 17, 1057–1066. [Google Scholar] [CrossRef]
- Dobbs, N.; Burnaevskiy, N.; Chen, D.; Gonugunta, V.K.; Alto, N.M.; Yan, N. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe 2015, 18, 157–168. [Google Scholar] [CrossRef]
- Pokatayev, V.; Yang, K.; Tu, X.; Dobbs, N.; Wu, J.; Kalb, R.G.; Yan, N. Homeostatic regulation of STING protein at the resting state by stabilizer TOLLIP. Nat. Immunol. 2020, 21, 158–167. [Google Scholar] [CrossRef]
- Siedel, H.; Roers, A.; Rösen-Wolff, A.; Luksch, H. Type I interferon-independent T cell impairment in a Tmem173 N153S/WT mouse model of STING associated vasculopathy with onset in infancy (SAVI). Clin. Immunol. 2020, 216, 108466. [Google Scholar] [CrossRef] [PubMed]
- Bouis, D.; Kirstetter, P.; Arbogast, F.; Lamon, D.; Delgado, V.; Jung, S.; Ebel, C.; Jacobs, H.; Knapp, A.M.; Jeremiah, N.; et al. Severe combined immunodeficiency in stimulator of interferon genes (STING) V154M/wild-type mice. J. Allergy Clin. Immunol. 2019, 143, 712–725.e5. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, Z.; Fan, F.; Zhou, J.; Hu, Z.; Wang, Q.; Wang, X.; Zeng, Q.; Zhang, Y.; Qiu, J.; et al. Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation. Cell Discov. 2022, 8, 133. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Torreggiani, S.; Kahle, D.; Rumsey, D.G.; Wright, B.L.; Montes-Cano, M.A.; Silveira, L.F.; Alehashemi, S.; Mitchell, J.; Aue, A.G.; et al. Case Report: Novel SAVI-Causing Variants in STING1 Expand the Clinical Disease Spectrum and Suggest a Refined Model of STING Activation. Front. Immunol. 2021, 12, 636225. [Google Scholar] [CrossRef]
- König, N.; Fiehn, C.; Wolf, C.; Schuster, M.; Cura Costa, E.; Tüngler, V.; Alvarez, H.A.; Chara, O.; Engel, K.; Goldbach-Mansky, R.; et al. Familial chilblain lupus due to a gain-of-function mutation in STING. Ann. Rheum. Dis. 2017, 76, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Shindo, R.; Kuchitsu, Y.; Mukai, K.; Taguchi, T. The activity of disease-causative STING variants can be suppressed by wild-type STING through heterocomplex formation. Front. Cell Dev. Biol. 2022, 10, 1037999. [Google Scholar] [CrossRef] [PubMed]
- David, C.; Frémond, M.L. Lung Inflammation in STING-Associated Vasculopathy with Onset in Infancy (SAVI). Cells 2022, 11, 318. [Google Scholar] [CrossRef]
- Xie, T.; Ruzanov, M.; Critton, D.; Merselis, L.; Naglich, J.; Sack, J.S.; Zhang, P.; Xie, C.; Tredup, J.; Stine, L.B.; et al. Orthosteric STING inhibition elucidates molecular correction of SAVI STING. Nat. Commun. 2025, 16, 5695. [Google Scholar] [CrossRef]
- Ergun, S.L.; Fernandez, D.; Weiss, T.M.; Li, L. STING Polymer Structure Reveals Mechanisms for Activation, Hyperactivation, and Inhibition. Cell 2019, 178, 290–301.e10. [Google Scholar] [CrossRef]
- Liu, Y.; Jesus, A.A.; Marrero, B.; Yang, D.; Ramsey, S.E.; Sanchez, G.A.M.; Tenbrock, K.; Wittkowski, H.; Jones, O.Y.; Kuehn, H.S.; et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 2014, 371, 507–518. [Google Scholar] [CrossRef]
- Keskitalo, S.; Haapaniemi, E.; Einarsdottir, E.; Rajamaki, K.; Heikkila, H.; Ilander, M.; Poyhonen, M.; Morgunova, E.; Hokynar, K.; Lagstrom, S.; et al. Novel TMEM173 Mutation and the Role of Disease Modifying Alleles. Front. Immunol. 2019, 10, 2770. [Google Scholar] [CrossRef]
- Staels, F.; Betrains, A.; Doubel, P.; Willemsen, M.; Cleemput, V.; Vanderschueren, S.; Corveleyn, A.; Meyts, I.; Sprangers, B.; Crow, Y.J.; et al. Adult-Onset ANCA-Associated Vasculitis in SAVI: Extension of the Phenotypic Spectrum, Case Report and Review of the Literature. Front. Immunol. 2020, 11, 575219. [Google Scholar] [CrossRef]
- Tang, X.; Xu, H.; Zhou, C.; Peng, Y.; Liu, H.; Liu, J.; Li, H.; Yang, H.; Zhao, S. STING-Associated Vasculopathy with Onset in Infancy in Three Children with New Clinical Aspect and Unsatisfactory Therapeutic Responses to Tofacitinib. J. Clin. Immunol. 2020, 40, 114–122. [Google Scholar] [CrossRef]
- Motwani, M.; Pawaria, S.; Bernier, J.; Moses, S.; Henry, K.; Fang, T.; Burkly, L.; Marshak-Rothstein, A.; Fitzgerald, K.A. Hierarchy of clinical manifestations in SAVI N153S and V154M mouse models. Proc. Natl. Acad. Sci. USA 2019, 116, 7941–7950. [Google Scholar] [CrossRef]
- Wu, J.; Chen, Y.J.; Dobbs, N.; Sakai, T.; Liou, J.; Miner, J.J.; Yan, N. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 2019, 216, 867–883. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Wang, Y. STING is an essential regulator of heart inflammation and fibrosis in mice with pathological cardiac hypertrophy via endoplasmic reticulum (ER) stress. Biomed. Pharmacother. 2020, 125, 110022. [Google Scholar] [CrossRef]
- Ren, J.; Bi, Y.; Sowers, J.R.; Hetz, C.; Zhang, Y. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat. Rev. Cardiol. 2021, 18, 499–521. [Google Scholar] [CrossRef]
- Chen, X.; Shi, C.; He, M.; Xiong, S.; Xia, X. Endoplasmic reticulum stress: Molecular mechanism and therapeutic targets. Signal Transduct. Target. Ther. 2023, 8, 352. [Google Scholar] [CrossRef]
- Li, Y.; Lin, H.; Tang, H.; Zhu, K.; Zhou, Z.; Zeng, Z.; Pan, B.; Chen, Z. The STING-IRF3 Signaling Pathway, Mediated by Endoplasmic Reticulum Stress, Contributes to Impaired Myocardial Autophagic Flux After Ischemia/Reperfusion. J. Cardiovasc. Pharmacol. 2023, 82, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; He, X.; Wang, J.; Wang, H.; Li, T.; Wen, S.; Ren, Z.; Cai, N.; Yang, J.; Li, M.; et al. Hyperlipidemia induces proinflammatory responses by activating STING pathway through IRE1alpha-XBP1 in retinal endothelial cells. J. Nutr. Biochem. 2023, 112, 109213. [Google Scholar] [CrossRef] [PubMed]
- Sen, T.; Saha, P.; Gupta, R.; Foley, L.M.; Jiang, T.; Abakumova, O.S.; Hitchens, T.K.; Sen, N. Aberrant ER Stress Induced Neuronal-IFNbeta Elicits White Matter Injury Due to Microglial Activation and T-Cell Infiltration after TBI. J. Neurosci. 2020, 40, 424–446. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Zhou, K.; Zhao, M.; Xiang, L.; Mei, T.; Xu, W.; Shang, B.; Liu, X.; Lai, Y.; Lin, M.; et al. TCF7L2 promotes ER stress signaling in diabetic retinopathy. Exp. Eye Res. 2022, 221, 109142. [Google Scholar] [CrossRef]
- Taguchi, T.; Mukai, K.; Takaya, E.; Shindo, R. STING Operation at the ER/Golgi Interface. Front. Immunol. 2021, 12, 646304. [Google Scholar] [CrossRef]
- Wu, J.; Yan, N. STIM1 moonlights as an anchor for STING. Nat. Immunol. 2019, 20, 112–114. [Google Scholar] [CrossRef]
- Srikanth, S.; Woo, J.S.; Wu, B.; El-Sherbiny, Y.M.; Leung, J.; Chupradit, K.; Rice, L.; Seo, G.J.; Calmettes, G.; Ramakrishna, C.; et al. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat. Immunol. 2019, 20, 152–162. [Google Scholar] [CrossRef]
- Zhang, B.C.; Nandakumar, R.; Reinert, L.S.; Huang, J.; Laustsen, A.; Gao, Z.L.; Sun, C.L.; Jensen, S.B.; Troldborg, A.; Assil, S.; et al. STEEP mediates STING ER exit and activation of signaling. Nat. Immunol. 2020, 21, 868–879. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, B.; Zhuang, Y.; Du, S.; Zeng, Z. Single High-Dose Irradiation-Induced iRhom2 Upregulation Promotes Macrophage Antitumor Activity Through cGAS/STING Signaling. Int. J. Radiat. Oncol. Biol. Phys. 2023, 116, 1150–1162. [Google Scholar] [CrossRef]
- Chu, T.T.; Tu, X.; Yang, K.; Wu, J.; Repa, J.J.; Yan, N. Tonic prime-boost of STING signalling mediates Niemann-Pick disease type C. Nature 2021, 596, 570–575. [Google Scholar] [CrossRef]
- Huang, X.; Yao, Y.; Hou, X.; Wei, L.; Rao, Y.; Su, Y.; Zheng, G.; Ruan, X.Z.; Li, D.; Chen, Y. Macrophage SCAP Contributes to Metaflammation and Lean NAFLD by Activating STING-NF-κB Signaling Pathway. Cell Mol. Gastroenterol. Hepatol. 2022, 14, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Motani, K.; Saito-Tarashima, N.; Nishino, K.; Yamauchi, S.; Minakawa, N.; Kosako, H. The Golgi-resident protein ACBD3 concentrates STING at ER-Golgi contact sites to drive export from the ER. Cell Rep. 2022, 41, 111868. [Google Scholar] [CrossRef] [PubMed]
- Luteijn, R.D.; van Terwisga, S.R.; Ver Eecke, J.E.; Onia, L.; Zaver, S.A.; Woodward, J.J.; Wubbolts, R.W.; Raulet, D.H.; van Kuppeveld, F.J.M. The activation of the adaptor protein STING depends on its interactions with the phospholipid PI4P. Sci. Signal 2024, 17, eade3643. [Google Scholar] [CrossRef]
- Delafontaine, S.; Iannuzzo, A.; Bigley, T.M.; Mylemans, B.; Rana, R.; Baatsen, P.; Poli, M.C.; Rymen, D.; Jansen, K.; Mekahli, D.; et al. Heterozygous mutations in the C-terminal domain of COPA underlie a complex autoin fl ammatory syndrome. J. Clin. Invest. 2024, 134, e163604. [Google Scholar] [CrossRef] [PubMed]
- Mukai, K.; Ogawa, E.; Uematsu, R.; Kuchitsu, Y.; Kiku, F.; Uemura, T.; Waguri, S.; Suzuki, T.; Dohmae, N.; Arai, H.; et al. Homeostatic regulation of STING by retrograde membrane traffic to the ER. Nat. Commun. 2021, 12, 61. [Google Scholar] [CrossRef]
- Zhang, L.; Wei, N.; Cui, Y.; Hong, Z.; Liu, X.; Wang, Q.; Li, S.; Liu, H.; Yu, H.; Cai, Y.; et al. The deubiquitinase CYLD is a specific checkpoint of the STING antiviral signaling pathway. PLoS Pathog. 2018, 14, e1007435. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, M.X.; Zhang, Q.; Zhu, G.F.; Yuan, L.; Zhang, D.E.; Zhu, Q.; Yao, J.; Shu, H.B.; Zhong, B. USP18 recruits USP20 to promote innate antiviral response through deubiquitinating STING/MITA. Cell Res. 2016, 26, 1302–1319. [Google Scholar] [CrossRef] [PubMed]
- Balka, K.R.; Venkatraman, R.; Saunders, T.L.; Shoppee, A.; Pang, E.S.; Magill, Z.; Homman-Ludiye, J.; Huang, C.; Lane, R.M.; York, H.M.; et al. Termination of STING responses is mediated via ESCRT-dependent degradation. EMBO J. 2023, 42, e112712. [Google Scholar] [CrossRef] [PubMed]
- Kuchitsu, Y.; Mukai, K.; Uematsu, R.; Takaada, Y.; Shinojima, A.; Shindo, R.; Shoji, T.; Hamano, S.; Ogawa, E.; Sato, R.; et al. STING signalling is terminated through ESCRT-dependent microautophagy of vesicles originating from recycling endosomes. Nat. Cell Biol. 2023, 25, 453–466. [Google Scholar] [CrossRef]
- Gentili, M.; Liu, B.; Papanastasiou, M.; Dele-Oni, D.; Schwartz, M.A.; Carlson, R.J.; Al’Khafaji, A.M.; Krug, K.; Brown, A.; Doench, J.G.; et al. ESCRT-dependent STING degradation inhibits steady-state and cGAMP-induced signalling. Nat. Commun. 2023, 14, 611. [Google Scholar] [CrossRef]
- Frémond, M.L.; Hadchouel, A.; Berteloot, L.; Melki, I.; Bresson, V.; Barnabei, L.; Jeremiah, N.; Belot, A.; Bondet, V.; Brocq, O.; et al. Overview of STING-Associated Vasculopathy with Onset in Infancy (SAVI) Among 21 Patients. J. Allergy Clin. Immunol. Pract. 2021, 9, 803–818.e11. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Berard, R.; Al Rasheed, A.; Aladba, B.; Kranzusch, P.J.; Henderlight, M.; Grom, A.; Kahle, D.; Torreggiani, S.; Aue, A.G.; et al. A novel STING1 variant causes a recessive form of STING-associated vasculopathy with onset in infancy (SAVI). J. Allergy Clin. Immunol. 2020, 146, 1204–1208.e6. [Google Scholar] [CrossRef]
- Wang, J.; Dai, M.; Cui, Y.; Hou, G.; Deng, J.; Gao, X.; Liao, Z.; Liu, Y.; Meng, Y.; Wu, L.; et al. Association of Abnormal Elevations in IFIT3 With Overactive Cyclic GMP-AMP Synthase/Stimulator of Interferon Genes Signaling in Human Systemic Lupus Erythematosus Monocytes. Arthritis Rheumatol. 2018, 70, 2036–2045. [Google Scholar] [CrossRef]
- Kato, Y.; Park, J.; Takamatsu, H.; Konaka, H.; Aoki, W.; Aburaya, S.; Ueda, M.; Nishide, M.; Koyama, S.; Hayama, Y.; et al. Apoptosis-derived membrane vesicles drive the cGAS-STING pathway and enhance type I IFN production in systemic lupus erythematosus. Ann. Rheum. Dis. 2018, 77, 1507–1515. [Google Scholar] [CrossRef]
- Sun, Y.; Aliyari, S.R.; Parvatiyar, K.; Wang, L.; Zhen, A.; Sun, W.; Han, X.; Zhang, A.; Kato, E.; Shi, H.; et al. STING directly interacts with PAR to promote apoptosis upon acute ionizing radiation-mediated DNA damage. Cell Death Differ. 2025, 32, 1167–1179. [Google Scholar] [CrossRef]
- Luo, W.; Wang, Y.; Zhang, L.; Ren, P.; Zhang, C.; Li, Y.; Azares, A.R.; Zhang, M.; Guo, J.; Ghaghada, K.B.; et al. Critical Role of Cytosolic DNA and Its Sensing Adaptor STING in Aortic Degeneration, Dissection, and Rupture. Circulation 2020, 141, 42–66. [Google Scholar] [CrossRef]
- Chen, D.; Tong, J.; Yang, L.; Wei, L.; Stolz, D.B.; Yu, J.; Zhang, J.; Zhang, L. PUMA amplifies necroptosis signaling by activating cytosolic DNA sensors. Proc. Natl. Acad. Sci. USA 2018, 115, 3930–3935. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, J.; Liu, Q.; Li, X.; Li, S.; Chen, J.; Hong, Z.; Wu, X.; Zhao, Y.; Ren, J. mtDNA-STING pathway promotes necroptosis-dependent enterocyte injury in intestinal ischemia reperfusion. Cell Death Dis. 2020, 11, 1050. [Google Scholar] [CrossRef]
- Kim, B.R.; Kim, B.J.; Kook, Y.H.; Kim, B.J. Mycobacterium abscessus infection leads to enhanced production of type 1 interferon and NLRP3 inflammasome activation in murine macrophages via mitochondrial oxidative stress. PLoS Pathog. 2020, 16, e1008294. [Google Scholar] [CrossRef]
- Dai, E.; Han, L.; Liu, J.; Xie, Y.; Zeh, H.J.; Kang, R.; Bai, L.; Tang, D. Ferroptotic damage promotes pancreatic tumorigenesis through a TMEM173/STING-dependent DNA sensor pathway. Nat. Commun. 2020, 11, 6339. [Google Scholar] [CrossRef]
- de Jesus, A.A. STING-Associated Vasculopathy with Onset in Infancy (SAVI). In Encyclopedia of Medical Immunology: Immunodeficiency Diseases; Orange, J.S., Chinen, J., Eds.; Springer: New York, NY, USA, 2020; pp. 609–614. [Google Scholar]
- Song, P.; Yang, W.; Lou, K.F.; Dong, H.; Zhang, H.; Wang, B.; Chen, D. UNC13D inhibits STING signaling by attenuating its oligomerization on the endoplasmic reticulum. EMBO Rep. 2022, 23, e55099. [Google Scholar] [CrossRef] [PubMed]
- Prabakaran, T.; Troldborg, A.; Kumpunya, S.; Alee, I.; Marinković, E.; Windross, S.J.; Nandakumar, R.; Narita, R.; Zhang, B.C.; Carstensen, M.; et al. A STING antagonist modulating the interaction with STIM1 blocks ER-to-Golgi trafficking and inhibits lupus pathology. EBioMedicine 2021, 66, 103314. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Cui, D.; Guo, J.; Ma, X.; Wang, J.; Guo, L.; Li, T.; Yang, M.; Huang, G.; Xu, A.; et al. STING COPII ER Export Trafficking and Signaling Primed by Phosphorylation Switches. Adv. Sci. 2025, 12, e03660. [Google Scholar] [CrossRef] [PubMed]
- Ran, Y.; Xiong, M.G.; Xu, Z.S.; Luo, W.W.; Wang, S.Y.; Wang, Y.Y. YIPF5 Is Essential for Innate Immunity to DNA Virus and Facilitates COPII-Dependent STING Trafficking. J. Immunol. 2019, 203, 1560–1570. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.Y.; Wang, X.L.; Li, Z.C.; Zhang, C.; Xu, X.; Cui, B.J.; Tian, M.Z.; Zhou, C.J.; Xu, N.; Wu, Y.; et al. Grass carp reovirus VP4 manipulates TOLLIP to degrade STING for inhibition of IFN production. J. Virol. 2025, 99, e0158324. [Google Scholar] [CrossRef]
- Shi, G.; Liu, L.; Cao, Y.; Ma, G.; Zhu, Y.; Xu, J.; Zhang, X.; Li, T.; Mi, L.; Jia, H.; et al. Inhibition of neutrophil extracellular trap formation ameliorates neuroinflammation and neuronal apoptosis via STING-dependent IRE1α/ASK1/JNK signaling pathway in mice with traumatic brain injury. J. Neuroinflamm. 2023, 20, 222. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef]
- Zhou, S.; Cheng, F.; Zhang, Y.; Su, T.; Zhu, G. Engineering and Delivery of cGAS-STING Immunomodulators for the Immunotherapy of Cancer and Autoimmune Diseases. Acc. Chem. Res. 2023, 56, 2933–2943. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Wu, J.; Wu, Y.T.; Du, F.; Aroh, C.; Yan, N.; Sun, L.; Chen, Z.J. Cyclic GMP-AMP synthase is an innate immune sensor of HIV and other retroviruses. Science 2013, 341, 903–906. [Google Scholar] [CrossRef]
- Ritchie, C.; Carozza, J.A.; Li, L. Biochemistry, Cell Biology, and Pathophysiology of the Innate Immune cGAS-cGAMP-STING Pathway. Annu. Rev. Biochem. 2022, 91, 599–628. [Google Scholar] [CrossRef] [PubMed]
- Hopfner, K.P.; Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 2020, 21, 501–521. [Google Scholar] [CrossRef]
- Abe, T.; Harashima, A.; Xia, T.; Konno, H.; Konno, K.; Morales, A.; Ahn, J.; Gutman, D.; Barber, G.N. STING recognition of cytoplasmic DNA instigates cellular defense. Mol. Cell 2013, 50, 5–15. [Google Scholar] [CrossRef]
- Decout, A.; Katz, J.D.; Venkatraman, S.; Ablasser, A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat. Rev. Immunol. 2021, 21, 548–569. [Google Scholar] [CrossRef]
- Stetson, D.B.; Ko, J.S.; Heidmann, T.; Medzhitov, R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 2008, 134, 587–598. [Google Scholar] [CrossRef]
- Mohr, L.; Toufektchan, E.; von Morgen, P.; Chu, K.; Kapoor, A.; Maciejowski, J. ER-directed TREX1 limits cGAS activation at micronuclei. Mol. Cell 2021, 81, 724–738.e9. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Gutman, D.; Saijo, S.; Barber, G.N. STING manifests self DNA-dependent inflammatory disease. Proc. Natl. Acad. Sci. USA 2012, 109, 19386–19391. [Google Scholar] [CrossRef]
- Wang, X.; Hu, R.; Song, Z.; Zhao, H.; Pan, Z.; Feng, Y.; Yu, Y.; Han, Q.; Zhang, J. Sorafenib combined with STAT3 knockdown triggers ER stress-induced HCC apoptosis and cGAS-STING-mediated anti-tumor immunity. Cancer Lett. 2022, 547, 215880. [Google Scholar] [CrossRef]
- Ablasser, A.; Chen, Z.J. cGAS in action: Expanding roles in immunity and inflammation. Science 2019, 363, eaat8657. [Google Scholar] [CrossRef]
- Peng, Y.; Yang, Y.; Li, Y.; Shi, T.; Xu, N.; Liu, R.; Luan, Y.; Yao, Y.; Yin, C. Mitochondrial (mt)DNA-cyclic GMP-AMP synthase (cGAS)-stimulator of interferon genes (STING) signaling promotes pyroptosis of macrophages via interferon regulatory factor (IRF)7/IRF3 activation to aggravate lung injury during severe acute pancreatitis. Cell. Mol. Biol. Lett. 2024, 29, 61. [Google Scholar] [CrossRef]
- Zhao, Q.; Wei, Y.; Pandol, S.J.; Li, L.; Habtezion, A. STING Signaling Promotes Inflammation in Experimental Acute Pancreatitis. Gastroenterology 2018, 154, 1822–1835.e2. [Google Scholar] [CrossRef]
- Yan, K.; Yang, J.; Qian, Q.; Xu, D.; Liu, H.; Wei, L.; Li, M.; Xu, W. Pathogenic Role of an IL-23/γδT17/Neutrophil Axis in Coxsackievirus B3-Induced Pancreatitis. J. Immunol. 2019, 203, 3301–3312. [Google Scholar] [CrossRef]
- Kuhl, N.; Linder, A.; Philipp, N.; Nixdorf, D.; Fischer, H.; Veth, S.; Kuut, G.; Xu, T.T.; Theurich, S.; Carell, T.; et al. STING agonism turns human T cells into interferon-producing cells but impedes their functionality. EMBO Rep. 2023, 24, e55536. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Zhao, C.; Tai, Y.; Li, B.; Lan, T.; Lai, E.; Dai, W.; Guo, Y.; Gan, C.; Kostallari, E.; et al. STING mediates hepatocyte pyroptosis in liver fibrosis by Epigenetically activating the NLRP3 inflammasome. Redox Biol. 2023, 62, 102691. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, M.; Wang, X.; Bu, Q.; Wang, Q.; Su, W.; Li, L.; Zhou, H.; Lu, L. XBP1 deficiency promotes hepatocyte pyroptosis by impairing mitophagy to activate mtDNA-cGAS-STING signaling in macrophages during acute liver injury. Redox Biol. 2022, 52, 102305. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Li, H.; Ma, L.; Zhou, J.; Guo, X.; Woo, S.L.; Pei, Y.; Knight, L.R.; Deveau, M.; Chen, Y.; et al. Expression of STING Is Increased in Liver Tissues From Patients With NAFLD and Promotes Macrophage-Mediated Hepatic Inflammation and Fibrosis in Mice. Gastroenterology 2018, 155, 1971–1984.e4. [Google Scholar] [CrossRef]
- Shmuel-Galia, L.; Humphries, F.; Lei, X.; Ceglia, S.; Wilson, R.; Jiang, Z.; Ketelut-Carneiro, N.; Foley, S.E.; Pechhold, S.; Houghton, J.; et al. Dysbiosis exacerbates colitis by promoting ubiquitination and accumulation of the innate immune adaptor STING in myeloid cells. Immunity 2021, 54, 1137–1153.e8. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.; Ren, J.; Li, J. Microbial DNA recognition by cGAS-STING and other sensors in dendritic cells in inflammatory bowel diseases. Inflamm. Bowel Dis. 2015, 21, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Son, S.; Oliveira, S.C.; Barber, G.N. STING-Dependent Signaling Underlies IL-10 Controlled Inflammatory Colitis. Cell Rep. 2017, 21, 3873–3884. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Ji, Y.; Gao, H.; Gomes Dos Reis, F.C.; Bandyopadhyay, G.; Jin, Z.; Ly, C.; Chang, Y.J.; Zhang, D.; Kumar, D.; et al. CRIg(+) Macrophages Prevent Gut Microbial DNA-Containing Extracellular Vesicle-Induced Tissue Inflammation and Insulin Resistance. Gastroenterology 2021, 160, 863–874. [Google Scholar] [CrossRef]
- Cai, Y.; Li, S.; Yang, Y.; Duan, S.; Fan, G.; Bai, J.; Zheng, Q.; Gu, Y.; Li, X.; Liu, R. Intestinal epithelial damage-derived mtDNA activates STING-IL12 axis in dendritic cells to promote colitis. Theranostics 2024, 14, 4393–4410. [Google Scholar] [CrossRef]
- Petrasek, J.; Iracheta-Vellve, A.; Csak, T.; Satishchandran, A.; Kodys, K.; Kurt-Jones, E.A.; Fitzgerald, K.A.; Szabo, G. STING-IRF3 pathway links endoplasmic reticulum stress with hepatocyte apoptosis in early alcoholic liver disease. Proc. Natl. Acad. Sci. USA 2013, 110, 16544–16549. [Google Scholar] [CrossRef]
- Iracheta-Vellve, A.; Petrasek, J.; Gyongyosi, B.; Satishchandran, A.; Lowe, P.; Kodys, K.; Catalano, D.; Calenda, C.D.; Kurt-Jones, E.A.; Fitzgerald, K.A.; et al. Endoplasmic Reticulum Stress-induced Hepatocellular Death Pathways Mediate Liver Injury and Fibrosis via Stimulator of Interferon Genes. J. Biol. Chem. 2016, 291, 26794–26805. [Google Scholar] [CrossRef]
- Cao, D.J.; Schiattarella, G.G.; Villalobos, E.; Jiang, N.; May, H.I.; Li, T.; Chen, Z.J.; Gillette, T.G.; Hill, J.A. Cytosolic DNA Sensing Promotes Macrophage Transformation and Governs Myocardial Ischemic Injury. Circulation 2018, 137, 2613–2634. [Google Scholar] [CrossRef] [PubMed]
- Rech, L.; Abdellatif, M.; Pottler, M.; Stangl, V.; Mabotuwana, N.; Hardy, S.; Rainer, P.P. Small molecule STING inhibition improves myocardial infarction remodeling. Life Sci. 2022, 291, 120263. [Google Scholar] [CrossRef]
- Mao, Y.; Luo, W.; Zhang, L.; Wu, W.; Yuan, L.; Xu, H.; Song, J.; Fujiwara, K.; Abe, J.I.; LeMaire, S.A.; et al. STING-IRF3 Triggers Endothelial Inflammation in Response to Free Fatty Acid-Induced Mitochondrial Damage in Diet-Induced Obesity. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 920–929. [Google Scholar] [CrossRef]
- Xie, S.; Su, E.; Song, X.; Xue, J.; Yu, P.; Zhang, B.; Liu, M.; Jiang, H. GSDME in Endothelial Cells: Inducing Vascular Inflammation and Atherosclerosis via Mitochondrial Damage and STING Pathway Activation. Biomedicines 2023, 11, 2579. [Google Scholar] [CrossRef]
- Wu, B.; Xu, M.M.; Fan, C.; Feng, C.L.; Lu, Q.K.; Lu, H.M.; Xiang, C.G.; Bai, F.; Wang, H.Y.; Wu, Y.W.; et al. STING inhibitor ameliorates LPS-induced ALI by preventing vascular endothelial cells-mediated immune cells chemotaxis and adhesion. Acta Pharmacol. Sin. 2022, 43, 2055–2066. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; You, Y.; Shang, F.F.; Wang, X.; Huang, B.; Zhao, B.; Lv, D.; Yang, S.; Xie, M.; Kong, L.; et al. iNOS aggravates pressure overload-induced cardiac dysfunction via activation of the cytosolic-mtDNA-mediated cGAS-STING pathway. Theranostics 2023, 13, 4229–4246. [Google Scholar] [CrossRef]
- Pham, P.T.; Fukuda, D.; Nishimoto, S.; Kim-Kaneyama, J.R.; Lei, X.F.; Takahashi, Y.; Sato, T.; Tanaka, K.; Suto, K.; Kawabata, Y.; et al. STING, a cytosolic DNA sensor, plays a critical role in atherogenesis: A link between innate immunity and chronic inflammation caused by lifestyle-related diseases. Eur. Heart J. 2021, 42, 4336–4348. [Google Scholar] [CrossRef] [PubMed]
- Audu, C.O.; Melvin, W.J.; Joshi, A.D.; Wolf, S.J.; Moon, J.Y.; Davis, F.M.; Barrett, E.C.; Mangum, K.D.; Deng, H.; Xing, X.; et al. Macrophage-specific inhibition of the histone demethylase JMJD3 decreases STING and pathologic inflammation in diabetic wound repair. Cell. Mol. Immunol. 2022, 19, 1251–1262. [Google Scholar] [CrossRef] [PubMed]
- de Cevins, C.; Delage, L.; Batignes, M.; Riller, Q.; Luka, M.; Remaury, A.; Sorin, B.; Fali, T.; Masson, C.; Hoareau, B.; et al. Single-cell RNA-sequencing of PBMCs from SAVI patients reveals disease-associated monocytes with elevated integrated stress response. Cell Rep. Med. 2023, 4, 101333. [Google Scholar] [CrossRef]
- Maekawa, H.; Inoue, T.; Ouchi, H.; Jao, T.M.; Inoue, R.; Nishi, H.; Fujii, R.; Ishidate, F.; Tanaka, T.; Tanaka, Y.; et al. Mitochondrial Damage Causes Inflammation via cGAS-STING Signaling in Acute Kidney Injury. Cell Rep. 2019, 29, 1261–1273.e6. [Google Scholar] [CrossRef]
- Chung, K.W.; Dhillon, P.; Huang, S.; Sheng, X.; Shrestha, R.; Qiu, C.; Kaufman, B.A.; Park, J.; Pei, L.; Baur, J.; et al. Mitochondrial Damage and Activation of the STING Pathway Lead to Renal Inflammation and Fibrosis. Cell Metab. 2019, 30, 784–799.e5. [Google Scholar] [CrossRef]
- Yi, B.J.; Wang, C.C.; Li, X.W.; Xu, Y.R.; Ma, X.Y.; Jian, P.A.; Talukder, M.; Li, X.N.; Li, J.L. Lycopene Protects against Atrazine-Induced Kidney STING-Dependent PANoptosis through Stabilizing mtDNA via Interaction with Sam50/PHB1. J. Agric. Food Chem. 2024, 72, 14956–14966. [Google Scholar] [CrossRef]
- Jiao, B.; An, C.; Du, H.; Tran, M.; Yang, D.; Zhao, Y.; Wang, P.; Hu, Z.; Zhou, D.; Wang, Y. Genetic deficiency or pharmacological inhibition of cGAS-STING signalling suppresses kidney inflammation and fibrosis. Br. J. Pharmacol. 2025, 182, 1741–1762. [Google Scholar] [CrossRef]
- Khedr, S.; Dissanayake, L.V.; Alsheikh, A.J.; Zietara, A.; Spires, D.R.; Kerketta, R.; Mathison, A.J.; Urrutia, R.; Palygin, O.; Staruschenko, A. Role of cGAS/STING pathway in aging and sexual dimorphism in diabetic kidney disease. JCI Insight 2024, 10, e174126. [Google Scholar] [CrossRef]
- Davis, S.E.; Khatua, A.K.; Popik, W. Nucleosomal dsDNA Stimulates APOL1 Expression in Human Cultured Podocytes by Activating the cGAS/IFI16-STING Signaling Pathway. Sci. Rep. 2019, 9, 15485. [Google Scholar] [CrossRef]
- Cao, Y.; Chen, X.; Zhu, Z.; Luo, Z.; Hao, Y.; Yang, X.; Feng, J.; Zhang, Z.; Hu, J.; Jian, Y.; et al. STING contributes to lipopolysaccharide-induced tubular cell inflammation and pyroptosis by activating endoplasmic reticulum stress in acute kidney injury. Cell Death Dis. 2024, 15, 217. [Google Scholar] [CrossRef]
- Andrade-Silva, M.; Dhillon, P.; Sanchez-Navarro, A.; Mukhi, D.; Hu, H.; Kolligundla, L.P.; Bergeson, A.; Abedini, A.; Levinsohn, J.; Dumoulin, B.; et al. The critical role of endoplasmic reticulum stress and the stimulator of interferon genes (STING) pathway in kidney fibrosis. Kidney Int. 2025, 107, 302–316. [Google Scholar] [CrossRef]
- Abid, Q.; Best Rocha, A.; Larsen, C.P.; Schulert, G.; Marsh, R.; Yasin, S.; Patty-Resk, C.; Valentini, R.P.; Adams, M.; Baracco, R. APOL1-Associated Collapsing Focal Segmental Glomerulosclerosis in a Patient With Stimulator of Interferon Genes (STING)-Associated Vasculopathy With Onset in Infancy (SAVI). Am. J. Kidney Dis. 2020, 75, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Zhang, J.; Yang, T.; Jiang, L.; Liu, X.; Wang, S.; Wang, X.; Huang, Y.; Wang, H.; Zhang, M.; et al. STING/ACSL4 axis-dependent ferroptosis and inflammation promote hypertension-associated chronic kidney disease. Mol. Ther. 2023, 31, 3084–3103. [Google Scholar] [CrossRef]
- Zhou, Q.; Yi, G.; Chang, M.; Li, N.; Bai, Y.; Li, H.; Yao, S. Activation of Sirtuin3 by honokiol ameliorates alveolar epithelial cell senescence in experimental silicosis via the cGAS-STING pathway. Redox Biol. 2024, 74, 103224. [Google Scholar] [CrossRef]
- Ou, L.; Zhang, P.; Huang, Z.; Cheng, Y.; Miao, Q.; Niu, R.; Hu, Y.; Chen, Y. Targeting STING-mediated pro-inflammatory and pro-fibrotic effects of alveolar macrophages and fibroblasts blunts silicosis caused by silica particles. J. Hazard. Mater. 2023, 458, 131907. [Google Scholar] [CrossRef] [PubMed]
- Benmerzoug, S.; Rose, S.; Bounab, B.; Gosset, D.; Duneau, L.; Chenuet, P.; Mollet, L.; Le Bert, M.; Lambers, C.; Geleff, S.; et al. STING-dependent sensing of self-DNA drives silica-induced lung inflammation. Nat. Commun. 2018, 9, 5226. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liu, E.; Li, Z.; Yan, C.; Zhang, X.; Guan, J.; Zhan, Y.; Zhao, B.; Ding, W. SIRT1-Rab7 axis attenuates NLRP3 and STING activation through late endosomal-dependent mitophagy during sepsis-induced acute lung injury. Int. J. Surg. 2024, 110, 2649–2668. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Wei, W.; Wenyang, J.; Rui, X.; Qing, G. Cytosolic DNA-STING-NLRP3 axis is involved in murine acute lung injury induced by lipopolysaccharide. Clin. Transl. Med. 2020, 10, e228. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Yu, Y.; Qu, M.; Qiu, Z.; Zhang, H.; Miao, C.; Guo, K. Neutrophil extracellular traps contribute to immunothrombosis formation via the STING pathway in sepsis-associated lung injury. Cell Death Discov. 2023, 9, 315. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Guo, T.; Zhou, Y.; Jiang, S.; Zhang, L.; Zhu, Z. STING signaling activation modulates macrophage polarization via CCL2 in radiation-induced lung injury. J. Transl. Med. 2023, 21, 590. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wang, T.; Gu, L.; Xu, Y.; Yang, Z.; Zhu, W.; Zhang, Q.; Luo, J.; Cao, J.; Jiao, Y. USP11 Exacerbates Radiation-Induced Pneumonitis by Activating Endothelial Cell Inflammatory Response via OTUD5-STING Signaling. Int. J. Radiat. Oncol. Biol. Phys. 2024, 119, 1261–1274. [Google Scholar] [CrossRef]
- di Flora, D.C.; Lara, J.P.Z.; Dionizio, A.; Buzalaf, M.A.R. The Dual Role of cGAS-STING Signaling in COVID-19: Implications for Therapy. Cells 2025, 14, 362. [Google Scholar] [CrossRef]
- Berthelot, J.M.; Lioté, F.; Maugars, Y.; Sibilia, J. Lymphocyte Changes in Severe COVID-19: Delayed Over-Activation of STING? Front. Immunol. 2020, 11, 607069. [Google Scholar] [CrossRef]
- Zhao, J.; Zhen, N.; Zhou, Q.; Lou, J.; Cui, W.; Zhang, G.; Tian, B. NETs Promote Inflammatory Injury by Activating cGAS-STING Pathway in Acute Lung Injury. Int. J. Mol. Sci. 2023, 24, 5125. [Google Scholar] [CrossRef]
- Sha, H.X.; Liu, Y.B.; Qiu, Y.L.; Zhong, W.J.; Yang, N.S.; Zhang, C.Y.; Duan, J.X.; Xiong, J.B.; Guan, C.X.; Zhou, Y. Neutrophil extracellular traps trigger alveolar epithelial cell necroptosis through the cGAS-STING pathway during acute lung injury in mice. Int. J. Biol. Sci. 2024, 20, 4713–4730. [Google Scholar] [CrossRef]
- Liu, Q.; Wu, J.; Zhang, X.; Li, X.; Wu, X.; Zhao, Y.; Ren, J. Circulating mitochondrial DNA-triggered autophagy dysfunction via STING underlies sepsis-related acute lung injury. Cell Death Dis. 2021, 12, 673. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, P.; Ablasser, A. Regulation of the cGAS-STING Pathway. Annu. Rev. Immunol. 2025, 43, 667–692. [Google Scholar] [CrossRef]
- Elahi, R.; Hozhabri, S.; Moradi, A.; Siahmansouri, A.; Jahani Maleki, A.; Esmaeilzadeh, A. Targeting the cGAS-STING pathway as an inflammatory crossroad in coronavirus disease 2019 (COVID-19). Immunopharmacol. Immunotoxicol. 2023, 45, 639–649. [Google Scholar] [CrossRef]
- Frémond, M.L.; Crow, Y.J. STING-Mediated Lung Inflammation and Beyond. J. Clin. Immunol. 2021, 41, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Ferecsko, A.S.; Smallwood, M.J.; Moore, A.; Liddle, C.; Newcombe, J.; Holley, J.; Whatmore, J.; Gutowski, N.J.; Eggleton, P. STING-Triggered CNS Inflammation in Human Neurodegenerative Diseases. Biomedicines 2023, 11, 1375. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Ma, G.; Li, X.; Zhao, J.; Zhao, Z.; Zeng, J. Activation of innate immune cGAS-STING pathway contributes to Alzheimer’s pathogenesis in 5xFAD mice. Nat. Aging 2023, 3, 202–212. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 30. [Google Scholar] [CrossRef]
- Cai, Q.; Jeong, Y.Y. Mitophagy in Alzheimer’s Disease and Other Age-Related Neurodegenerative Diseases. Cells 2020, 9, 150. [Google Scholar] [CrossRef] [PubMed]
- Chakravorty, A.; Jetto, C.T.; Manjithaya, R. Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2019, 11, 311. [Google Scholar] [CrossRef]
- Hou, Y.; Wei, Y.; Lautrup, S.; Yang, B.; Wang, Y.; Cordonnier, S.; Mattson, M.P.; Croteau, D.L.; Bohr, V.A. NAD(+) supplementation reduces neuroinflammation and cell senescence in a transgenic mouse model of Alzheimer’s disease via cGAS-STING. Proc. Natl. Acad. Sci. USA 2021, 118, e2011226118. [Google Scholar] [CrossRef]
- Scopa, C.; Barnada, S.M.; Cicardi, M.E.; Singer, M.; Trotti, D.; Trizzino, M. JUN upregulation drives aberrant transposable element mobilization, associated innate immune response, and impaired neurogenesis in Alzheimer’s disease. Nat. Commun. 2023, 14, 8021. [Google Scholar] [CrossRef]
- Gao, D.; Hao, J.P.; Li, B.Y.; Zheng, C.C.; Miao, B.B.; Zhang, L.; Li, Y.L.; Li, L.; Li, X.J.; Zhang, L. Tetrahydroxy stilbene glycoside ameliorates neuroinflammation for Alzheimer’s disease via cGAS-STING. Eur. J. Pharmacol. 2023, 953, 175809. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Pallares, J.; Garcia-Garrote, M.; Parga, J.A.; Labandeira-Garcia, J.L. Combined cell-based therapy strategies for the treatment of Parkinson’s disease: Focus on mesenchymal stromal cells. Neural Regen. Res. 2023, 18, 478–484. [Google Scholar] [CrossRef]
- Hinkle, J.T.; Patel, J.; Panicker, N.; Karuppagounder, S.S.; Biswas, D.; Belingon, B.; Chen, R.; Brahmachari, S.; Pletnikova, O.; Troncoso, J.C.; et al. STING mediates neurodegeneration and neuroinflammation in nigrostriatal alpha-synucleinopathy. Proc. Natl. Acad. Sci. USA 2022, 119, e2118819119. [Google Scholar] [CrossRef]
- Jiang, S.Y.; Tian, T.; Yao, H.; Xia, X.M.; Wang, C.; Cao, L.; Hu, G.; Du, R.H.; Lu, M. The cGAS-STING-YY1 axis accelerates progression of neurodegeneration in a mouse model of Parkinson’s disease via LCN2-dependent astrocyte senescence. Cell Death Differ. 2023, 30, 2280–2292. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, B.; Sinha, S.C.; Amin, S.; Gan, L. Mechanism and therapeutic potential of targeting cGAS-STING signaling in neurological disorders. Mol. Neurodegener. 2023, 18, 79. [Google Scholar] [CrossRef]
- Li, Q.; Cao, Y.; Dang, C.; Han, B.; Han, R.; Ma, H.; Hao, J.; Wang, L. Inhibition of double-strand DNA-sensing cGAS ameliorates brain injury after ischemic stroke. EMBO Mol. Med. 2020, 12, e11002. [Google Scholar] [CrossRef]
- Sharma, M.; Rajendrarao, S.; Shahani, N.; Ramirez-Jarquin, U.N.; Subramaniam, S. Cyclic GMP-AMP synthase promotes the inflammatory and autophagy responses in Huntington disease. Proc. Natl. Acad. Sci. USA 2020, 117, 15989–15999. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.Q.; Wang, H.; Su, W.M.; Gu, X.J.; Shen, X.F.; Jiang, Z.; Ren, Y.L.; Cao, B.; Li, G.B.; Wang, Y.; et al. TBK1, a prioritized drug repurposing target for amyotrophic lateral sclerosis: Evidence from druggable genome Mendelian randomization and pharmacological verification in vitro. BMC Med. 2024, 22, 96. [Google Scholar] [CrossRef]
- Grieves, J.L.; Fye, J.M.; Harvey, S.; Grayson, J.M.; Hollis, T.; Perrino, F.W. Exonuclease TREX1 degrades double-stranded DNA to prevent spontaneous lupus-like inflammatory disease. Proc. Natl. Acad. Sci. USA 2015, 112, 5117–5122. [Google Scholar] [CrossRef] [PubMed]
- Wolf, C.; Rapp, A.; Berndt, N.; Staroske, W.; Schuster, M.; Dobrick-Mattheuer, M.; Kretschmer, S.; Konig, N.; Kurth, T.; Wieczorek, D.; et al. RPA and Rad51 constitute a cell intrinsic mechanism to protect the cytosol from self DNA. Nat. Commun. 2016, 7, 11752. [Google Scholar] [CrossRef]
- Zhou, W.; Richmond-Buccola, D.; Wang, Q.; Kranzusch, P.J. Structural basis of human TREX1 DNA degradation and autoimmune disease. Nat. Commun. 2022, 13, 4277. [Google Scholar] [CrossRef] [PubMed]
- Gulati, A.; Bale, A.E.; Dykas, D.J.; Bia, M.J.; Danovitch, G.M.; Moeckel, G.W.; Somlo, S.; Dahl, N.K. TREX1 Mutation Causing Autosomal Dominant Thrombotic Microangiopathy and CKD-A Novel Presentation. Am. J. Kidney Dis. 2018, 72, 895–899. [Google Scholar] [CrossRef]
- Wang, Q.; Du, J.; Hua, S.; Zhao, K. TREX1 plays multiple roles in human diseases. Cell. Immunol. 2022, 375, 104527. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Li, T.; Li, X.D.; Chen, X.; Li, Q.Z.; Wight-Carter, M.; Chen, Z.J. Activation of cyclic GMP-AMP synthase by self-DNA causes autoimmune diseases. Proc. Natl. Acad. Sci. USA 2015, 112, E5699–E5705. [Google Scholar] [CrossRef]
- Liu, Y.; Xiao, N.; Du, H.; Kou, M.; Lin, L.; Huang, M.; Zhang, S.; Xu, S.; Li, D.; Chen, Q. Celastrol ameliorates autoimmune disorders in Trex1-deficient mice. Biochem. Pharmacol. 2020, 178, 114090. [Google Scholar] [CrossRef] [PubMed]
- Uggenti, C.; Lepelley, A.; Depp, M.; Badrock, A.P.; Rodero, M.P.; El-Daher, M.T.; Rice, G.I.; Dhir, S.; Wheeler, A.P.; Dhir, A.; et al. cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing. Nat. Genet. 2020, 52, 1364–1372. [Google Scholar] [CrossRef]
- Song, B.; Shiromoto, Y.; Minakuchi, M.; Nishikura, K. The role of RNA editing enzyme ADAR1 in human disease. Wiley Interdiscip. Rev. RNA 2022, 13, e1665. [Google Scholar] [CrossRef]
- de Reuver, R.; Dierick, E.; Wiernicki, B.; Staes, K.; Seys, L.; De Meester, E.; Muyldermans, T.; Botzki, A.; Lambrecht, B.N.; Van Nieuwerburgh, F.; et al. ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep. 2021, 36, 109500. [Google Scholar] [CrossRef]
- Jeremiah, N.; Neven, B.; Gentili, M.; Callebaut, I.; Maschalidi, S.; Stolzenberg, M.C.; Goudin, N.; Frémond, M.L.; Nitschke, P.; Molina, T.J.; et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Investig. 2014, 124, 5516–5520. [Google Scholar] [CrossRef]
- Larkin, B.; Ilyukha, V.; Sorokin, M.; Buzdin, A.; Vannier, E.; Poltorak, A. Cutting Edge: Activation of STING in T Cells Induces Type I IFN Responses and Cell Death. J. Immunol. 2017, 199, 397–402. [Google Scholar] [CrossRef]
- Tu, X.; Chu, T.T.; Jeltema, D.; Abbott, K.; Yang, K.; Xing, C.; Han, J.; Dobbs, N.; Yan, N. Interruption of post-Golgi STING trafficking activates tonic interferon signaling. Nat. Commun. 2022, 13, 6977. [Google Scholar] [CrossRef] [PubMed]
- Lepelley, A.; Martin-Niclós, M.J.; Le Bihan, M.; Marsh, J.A.; Uggenti, C.; Rice, G.I.; Bondet, V.; Duffy, D.; Hertzog, J.; Rehwinkel, J.; et al. Mutations in COPA lead to abnormal trafficking of STING to the Golgi and interferon signaling. J. Exp. Med. 2020, 217, e20200600. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Chong, Z.; Law, C.S.; Mukai, K.; Ho, F.O.; Martinu, T.; Backes, B.J.; Eckalbar, W.L.; Taguchi, T.; Shum, A.K. A defect in COPI-mediated transport of STING causes immune dysregulation in COPA syndrome. J. Exp. Med. 2020, 217, e20201045. [Google Scholar] [CrossRef]
- Deloron, P.; Sexton, J.D.; Bugilimfura, L.; Sezibera, C. Amodiaquine and sulfadoxine-pyrimethamine as treatment for chloroquine-resistant Plasmodium falciparum in Rwanda. Am. J. Trop. Med. Hyg. 1988, 38, 244–248. [Google Scholar] [CrossRef]
- Jeltema, D.; Abbott, K.; Yan, N. STING trafficking as a new dimension of immune signaling. J. Exp. Med. 2023, 220, e20220990. [Google Scholar] [CrossRef]
- Murthy, A.M.V.; Robinson, N.; Kumar, S. Crosstalk between cGAS-STING signaling and cell death. Cell Death Differ. 2020, 27, 2989–3003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Li, J.; Zhao, L.; Yang, Z.; Wu, C.; Liu, Y.; Li, W.; Jin, Z.; Ma, J. Mitochondrial DNA Leakage Promotes Persistent Pancreatic Acinar Cell Injury in Acute Pancreatitis via the cGAS-STING-NF-κB Pathway. Inflammation 2024, 48, 1420–1437. [Google Scholar] [CrossRef]
- Gao, Y.; Mi, N.; Wu, W.; Zhao, Y.; Fan, F.; Liao, W.; Ming, Y.; Guan, W.; Bai, C. Transfer of inflammatory mitochondria via extracellular vesicles from M1 macrophages induces ferroptosis of pancreatic beta cells in acute pancreatitis. J. Extracell. Vesicles 2024, 13, e12410. [Google Scholar] [CrossRef]
- Uthaman, S.; Parvinroo, S.; Mathew, A.P.; Jia, X.; Hernandez, B.; Proctor, A.; Sajeevan, K.A.; Nenninger, A.; Long, M.J.; Park, I.K.; et al. Inhibiting the cGAS-STING Pathway in Ulcerative Colitis with Programmable Micelles. ACS Nano 2024, 18, 12117–12133. [Google Scholar] [CrossRef]
- Kang, J.; Wu, J.; Liu, Q.; Jiang, H.; Li, W.; Li, Y.; Li, X.; Ni, C.; Wu, L.; Liu, M.; et al. FASN regulates STING palmitoylation via malonyl-CoA in macrophages to alleviate sepsis-induced liver injury. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 167299. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, X.; Zheng, L.; Chen, M.; Zhang, Y.; Zhu, R.; Chen, J.; Gu, J.; Yin, Q.; Jiang, H.; et al. Non-canonical STING-PERK pathway dependent epigenetic regulation of vascular endothelial dysfunction via integrating IRF3 and NF-κB in inflammatory response. Acta Pharm. Sin. B 2023, 13, 4765–4784. [Google Scholar] [CrossRef] [PubMed]
- Mitrofanova, A.; Fontanella, A.; Tolerico, M.; Mallela, S.; Molina David, J.; Zuo, Y.; Boulina, M.; Kim, J.J.; Santos, J.; Ge, M.; et al. Activation of Stimulator of IFN Genes (STING) Causes Proteinuria and Contributes to Glomerular Diseases. J. Am. Soc. Nephrol. 2022, 33, 2153–2173. [Google Scholar] [CrossRef]
- Chung, S.; Jeong, J.H.; Park, J.C.; Han, J.W.; Lee, Y.; Kim, J.I.; Mook-Jung, I. Blockade of STING activation alleviates microglial dysfunction and a broad spectrum of Alzheimer’s disease pathologies. Exp. Mol. Med. 2024, 56, 1936–1951. [Google Scholar] [CrossRef] [PubMed]
- Carling, G.K.; Fan, L.; Foxe, N.R.; Norman, K.; Wong, M.Y.; Zhu, D.; Corona, C.; Razzoli, A.; Yu, F.; Yarahmady, A.; et al. Alzheimer’s disease-linked risk alleles elevate microglial cGAS-associated senescence and neurodegeneration in a tauopathy model. Neuron 2024, 112, 3877–3896.e8. [Google Scholar] [CrossRef]
- Liu, M.; Pan, J.; Li, X.; Zhang, X.; Tian, F.; Li, M.; Wu, X.; Zhang, L.; Qin, C. Interleukin-6 deficiency reduces neuroinflammation by inhibiting the STAT3-cGAS-STING pathway in Alzheimer’s disease mice. J. Neuroinflamm. 2024, 21, 282. [Google Scholar] [CrossRef]
- Zhou, S.; Li, T.; Zhang, W.; Wu, J.; Hong, H.; Quan, W.; Qiao, X.; Cui, C.; Qiao, C.; Zhao, W.; et al. The cGAS-STING-interferon regulatory factor 7 pathway regulates neuroinflammation in Parkinson’s disease. Neural Regen. Res. 2025, 20, 2361–2372. [Google Scholar] [CrossRef]
- Yi, C.; Li, Q.; Xiao, J. Familial chilblain lupus due to a novel mutation in TREX1 associated with Aicardi-Goutie’res syndrome. Pediatr. Rheumatol. Online J. 2020, 18, 32. [Google Scholar] [CrossRef]
- Hall, J.; Brault, A.; Vincent, F.; Weng, S.; Wang, H.; Dumlao, D.; Aulabaugh, A.; Aivazian, D.; Castro, D.; Chen, M.; et al. Discovery of PF-06928215 as a high affinity inhibitor of cGAS enabled by a novel fluorescence polarization assay. PLoS ONE 2017, 12, e0184843. [Google Scholar] [CrossRef]
- Wang, X.; Lu, W.; Xia, X.; Zhu, Y.; Ge, C.; Guo, X.; Zhang, N.; Chen, H.; Xu, S. Selenomethionine mitigate PM2.5-induced cellular senescence in the lung via attenuating inflammatory response mediated by cGAS/STING/NF-kappaB pathway. Ecotoxicol. Environ. Saf. 2022, 247, 114266. [Google Scholar] [CrossRef]
- Vincent, J.; Adura, C.; Gao, P.; Luz, A.; Lama, L.; Asano, Y.; Okamoto, R.; Imaeda, T.; Aida, J.; Rothamel, K.; et al. Small molecule inhibition of cGAS reduces interferon expression in primary macrophages from autoimmune mice. Nat. Commun. 2017, 8, 750. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Meng, Y.; Yuan, K.; Wu, Q.; Zhu, S.; Li, Y.; Wu, P.; Zheng, J.; Shi, H. RU.521 mitigates subarachnoid hemorrhage-induced brain injury via regulating microglial polarization and neuroinflammation mediated by the cGAS/STING/NF-kappaB pathway. Cell Commun. Signal. 2023, 21, 264. [Google Scholar] [CrossRef]
- Lama, L.; Adura, C.; Xie, W.; Tomita, D.; Kamei, T.; Kuryavyi, V.; Gogakos, T.; Steinberg, J.I.; Miller, M.; Ramos-Espiritu, L.; et al. Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression. Nat. Commun. 2019, 10, 2261. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Xiao, R.; Zeng, R.; He, E.; Zhang, A. Small molecules targeting cGAS-STING pathway for autoimmune disease. Eur. J. Med. Chem. 2022, 238, 114480. [Google Scholar] [CrossRef]
- Skeldon, A.M.; Wang, L.; Sgarioto, N.; Beveridge, R.E.; Chan, S.; Dorich, S.; Dumais, V.; Fradet, N.; Gaudreault, S.; LeGros, P.; et al. Structural insight into the cGAS active site explains differences between therapeutically relevant species. Commun. Chem. 2025, 8, 88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Xiong, M.; Yuan, X.; Li, M.; Sun, H.; Xu, Y. In Silico Screening-Based Discovery of Novel Inhibitors of Human Cyclic GMP-AMP Synthase: A Cross-Validation Study of Molecular Docking and Experimental Testing. J. Chem. Inf. Model. 2020, 60, 3265–3276. [Google Scholar] [CrossRef]
- Song, J.; Yang, R.R.; Chang, J.; Liu, Y.D.; Lu, C.H.; Chen, L.F.; Guo, H.; Zhang, Y.H.; Fan, Z.S.; Zhou, J.Y.; et al. Discovery and characterization of a novel cGAS covalent inhibitor for the treatment of inflammatory bowel disease. Acta Pharmacol. Sin. 2023, 44, 791–800. [Google Scholar] [CrossRef]
- Padilla-Salinas, R.; Sun, L.; Anderson, R.; Yang, X.; Zhang, S.; Chen, Z.J.; Yin, H. Discovery of Small-Molecule Cyclic GMP-AMP Synthase Inhibitors. J. Org. Chem. 2020, 85, 1579–1600. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lei, S.; Zhou, Z.; Wang, M.; Feng, C.; Gao, X.; Ding, C.; Song, Z.; Tang, W.; Zhang, A. Design, Synthesis, and Pharmacological Evaluation of Spiro[carbazole-3,3′-pyrrolidine] Derivatives as cGAS Inhibitors for Treatment of Acute Lung Injury. J. Med. Chem. 2024, 67, 6268–6291. [Google Scholar] [CrossRef]
- An, J.; Minie, M.; Sasaki, T.; Woodward, J.J.; Elkon, K.B. Antimalarial Drugs as Immune Modulators: New Mechanisms for Old Drugs. Annu. Rev. Med. 2017, 68, 317–330. [Google Scholar] [CrossRef]
- Busto, N.; Garcia, B.; Leal, J.M.; Gaspar, J.F.; Martins, C.; Boggioni, A.; Secco, F. ACMA (9-amino-6-chloro-2-methoxy acridine) forms three complexes in the presence of DNA. Phys. Chem. Chem. Phys. 2011, 13, 19534–19545. [Google Scholar] [CrossRef] [PubMed]
- Tonduti, D.; Fazzi, E.; Badolato, R.; Orcesi, S. Novel and emerging treatments for Aicardi-Goutieres syndrome. Expert Rev. Clin. Immunol. 2020, 16, 189–198. [Google Scholar] [CrossRef] [PubMed]
- An, J.; Woodward, J.J.; Lai, W.; Minie, M.; Sun, X.; Tanaka, L.; Snyder, J.M.; Sasaki, T.; Elkon, K.B. Inhibition of Cyclic GMP-AMP Synthase Using a Novel Antimalarial Drug Derivative in Trex1-Deficient Mice. Arthritis Rheumatol. 2018, 70, 1807–1819. [Google Scholar] [CrossRef]
- Wang, M.; Sooreshjani, M.A.; Mikek, C.; Opoku-Temeng, C.; Sintim, H.O. Suramin potently inhibits cGAMP synthase, cGAS, in THP1 cells to modulate IFN-beta levels. Future Med. Chem. 2018, 10, 1301–1317. [Google Scholar] [CrossRef]
- Green, J.P.; El-Sharkawy, L.Y.; Roth, S.; Zhu, J.; Cao, J.; Leach, A.G.; Liesz, A.; Freeman, S.; Brough, D. Discovery of an inhibitor of DNA-driven inflammation that preferentially targets the AIM2 inflammasome. iScience 2023, 26, 106758. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Cao, A.; Luo, Q.; Chen, D.; Zhao, W.; Xu, J.; Li, Q.; Bu, X.; Quan, J. Development of cyclopeptide inhibitors of cGAS targeting protein-DNA interaction and phase separation. Nat. Commun. 2023, 14, 6132. [Google Scholar] [CrossRef]
- Valencia, D.X.; Spiegelstein, O.; Fradet, N.; Chefson, A.; Cyr, P.; Pike, K.; Stewart, J.; Beveridge, R. ABS115 Afety, Tolerability, Pharmacokinetics and Pharmacodynamics in Healthy Volunteers of VENT-03, a Novel cGAS INHibitor for the Treatment of Systemic Lupus Erythematosus. Ann. Rheum. Dis. 2025, 84, 1541–1542. [Google Scholar] [CrossRef]
- ImmuneSensor Therapeutics, Inc. A Phase I randomized study of IMSB301 in healthy volunteers. ISRCTN Regist. 2024. [Google Scholar] [CrossRef]
- Siu, T.; Altman, M.D.; Baltus, G.A.; Childers, M.; Ellis, J.M.; Gunaydin, H.; Hatch, H.; Ho, T.; Jewell, J.; Lacey, B.M.; et al. Discovery of a Novel cGAMP Competitive Ligand of the Inactive Form of STING. ACS Med. Chem. Lett. 2019, 10, 92–97. [Google Scholar] [CrossRef]
- Li, S.; Hong, Z.; Wang, Z.; Li, F.; Mei, J.; Huang, L.; Lou, X.; Zhao, S.; Song, L.; Chen, W.; et al. The Cyclopeptide Astin C Specifically Inhibits the Innate Immune CDN Sensor STING. Cell Rep. 2018, 25, 3405–3421.e7. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Mei, J.; Li, C.; Bai, G.; Maimaiti, M.; Hu, H.; Yu, W.; Sun, L.; Zhang, L.; Cheng, D.; et al. STING inhibitors target the cyclic dinucleotide binding pocket. Proc. Natl. Acad. Sci. USA 2021, 118, e2105465118. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Bian, H.; Lv, J.; Song, W.; Xing, C.; Hui, C.; Zhang, D.; Zhang, C.; Zhao, L.; Li, Y.; et al. Gelsevirine is a novel STING-specific inhibitor and mitigates STING-related inflammation in sepsis. Front. Immunol. 2023, 14, 1190707. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, Z.; Liu, P.; Wei, X.; Zhang, Z.; Fan, S.; Zhang, L.; Han, F.; Song, Y.; Chu, L.; et al. SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing. Immunity 2023, 56, 2492–2507.e10. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, J.; Gao, Y.; Shu, Z.; Sun, F.; Ma, J.; Zhou, X.; Li, W.; Mao, H.; Lei, X. Discovery and Total Synthesis of Anhydrotuberosin as a STING Antagonist for Treating Autoimmune Diseases. Angew. Chem. Int. Ed. Engl. 2025, 64, e202407641. [Google Scholar] [CrossRef]
- Liu, H.; Ottosen, R.N.; Jennet, K.M.; Svenningsen, E.B.; Kristensen, T.F.; Biltoft, M.; Jakobsen, M.R.; Poulsen, T.B. Macrodiolide Diversification Reveals Broad Immunosuppressive Activity That Impairs the cGAS-STING Pathway. Angew. Chem. Int. Ed. Engl. 2021, 60, 18734–18741. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, M.; Li, X.W.; Mao, J.; Zhu, Y.J.; Wang, N.; Yin, L.H.; Guo, Z.L.; Cai, H.; Li, T.; et al. Meloxicam inhibits STING phosphorylation and alleviates intracellular DNA-mediated autoimmune responses. Cell. Biosci. 2023, 13, 76. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Kang, J.; Jiang, H.; Gong, W.; Chen, L.; Wu, C.; Liu, M.; Wu, X.; Zhao, Y.; et al. 4-octyl itaconate as a metabolite derivative inhibits inflammation via alkylation of STING. Cell Rep. 2023, 42, 112145. [Google Scholar] [CrossRef]
- Hansen, A.L.; Mukai, K.; Schopfer, F.J.; Taguchi, T.; Holm, C.K. STING palmitoylation as a therapeutic target. Cell Mol. Immunol. 2019, 16, 236–241. [Google Scholar] [CrossRef] [PubMed]
- Haag, S.M.; Gulen, M.F.; Reymond, L.; Gibelin, A.; Abrami, L.; Decout, A.; Heymann, M.; van der Goot, F.G.; Turcatti, G.; Behrendt, R.; et al. Targeting STING with covalent small-molecule inhibitors. Nature 2018, 559, 269–273. [Google Scholar] [CrossRef]
- Kobritz, M.; Borjas, T.; Patel, V.; Coppa, G.; Aziz, M.; Wang, P. H151, a Small Molecule Inhibitor of Sting as a Novel Therapeutic in Intestinal Ischemia-Reperfusion Injury. Shock 2022, 58, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.L.; Buchan, G.J.; Rühl, M.; Mukai, K.; Salvatore, S.R.; Ogawa, E.; Andersen, S.D.; Iversen, M.B.; Thielke, A.L.; Gunderstofte, C.; et al. Nitro-fatty acids are formed in response to virus infection and are potent inhibitors of STING palmitoylation and signaling. Proc. Natl. Acad. Sci. USA 2018, 115, E7768–E7775. [Google Scholar] [CrossRef]
- Vinogradova, E.V.; Zhang, X.; Remillard, D.; Lazar, D.C.; Suciu, R.M.; Wang, Y.; Bianco, G.; Yamashita, Y.; Crowley, V.M.; Schafroth, M.A.; et al. An Activity-Guided Map of Electrophile-Cysteine Interactions in Primary Human T Cells. Cell 2020, 182, 1009–1026.e29. [Google Scholar] [CrossRef]
- Su, C.; Cheng, T.; Huang, J.; Zhang, T.; Yin, H. 4-Octyl itaconate restricts STING activation by blocking its palmitoylation. Cell Rep. 2023, 42, 113040. [Google Scholar] [CrossRef]
- Barasa, L.; Chaudhuri, S.; Zhou, J.Y.; Jiang, Z.; Choudhary, S.; Green, R.M.; Wiggin, E.; Cameron, M.; Humphries, F.; Fitzgerald, K.A.; et al. Development of LB244, an Irreversible STING Antagonist. J. Am. Chem. Soc. 2023, 145, 20273–20288. [Google Scholar] [CrossRef]
- Sakamoto, K.M.; Kim, K.B.; Kumagai, A.; Mercurio, F.; Crews, C.M.; Deshaies, R.J. Protacs: Chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. USA 2001, 98, 8554–8559. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Ohoka, N.; Shibata, N.; Inoue, T.; Tsuji, G.; Demizu, Y. Development of STING degrader with double covalent ligands. Bioorg. Med. Chem. Lett. 2024, 102, 129677. [Google Scholar] [CrossRef]
- Liu, J.; Yuan, L.; Ruan, Y.; Deng, B.; Yang, Z.; Ren, Y.; Li, L.; Liu, T.; Zhao, H.; Mai, R.; et al. Novel CRBN-Recruiting Proteolysis-Targeting Chimeras as Degraders of Stimulator of Interferon Genes with In Vivo Anti-Inflammatory Efficacy. J. Med. Chem. 2022, 65, 6593–6611. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Johnson, R.L.; Zhang, Z.; Herring, L.E.; Jiang, G.; Damania, B.; James, L.I.; Liu, P. Development of VHL-recruiting STING PROTACs that suppress innate immunity. Cell. Mol. Life Sci. 2023, 80, 149. [Google Scholar] [CrossRef]
- Zhang, J.; Du, J.; Liu, D.; Zhuo, J.; Chu, L.; Li, Y.; Gao, L.; Xu, M.; Chen, W.; Huang, W.; et al. Polystyrene microplastics induce pulmonary fibrosis by promoting alveolar epithelial cell ferroptosis through cGAS/STING signaling. Ecotoxicol. Environ. Saf. 2024, 277, 116357. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kundu, S.; Suresh Kumar, G. Quinacrine and 9-amino acridine inhibit B-Z and B-H(l) form DNA conformational transitions. DNA Cell Biol. 2011, 30, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Steinhagen, F.; Zillinger, T.; Peukert, K.; Fox, M.; Thudium, M.; Barchet, W.; Putensen, C.; Klinman, D.; Latz, E.; Bode, C. Suppressive oligodeoxynucleotides containing TTAGGG motifs inhibit cGAS activation in human monocytes. Eur. J. Immunol. 2018, 48, 605–611. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y.; Lin, G. Blocking cGAS/STING signaling protects against sepsis-associated acute liver injury. Int. Immunopharmacol. 2022, 113, 109276. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Goldberg, E.; Goldberg, J. ER retention is imposed by COPII protein sorting and attenuated by 4-phenylbutyrate. Elife 2017, 6, e26624. [Google Scholar] [CrossRef] [PubMed]


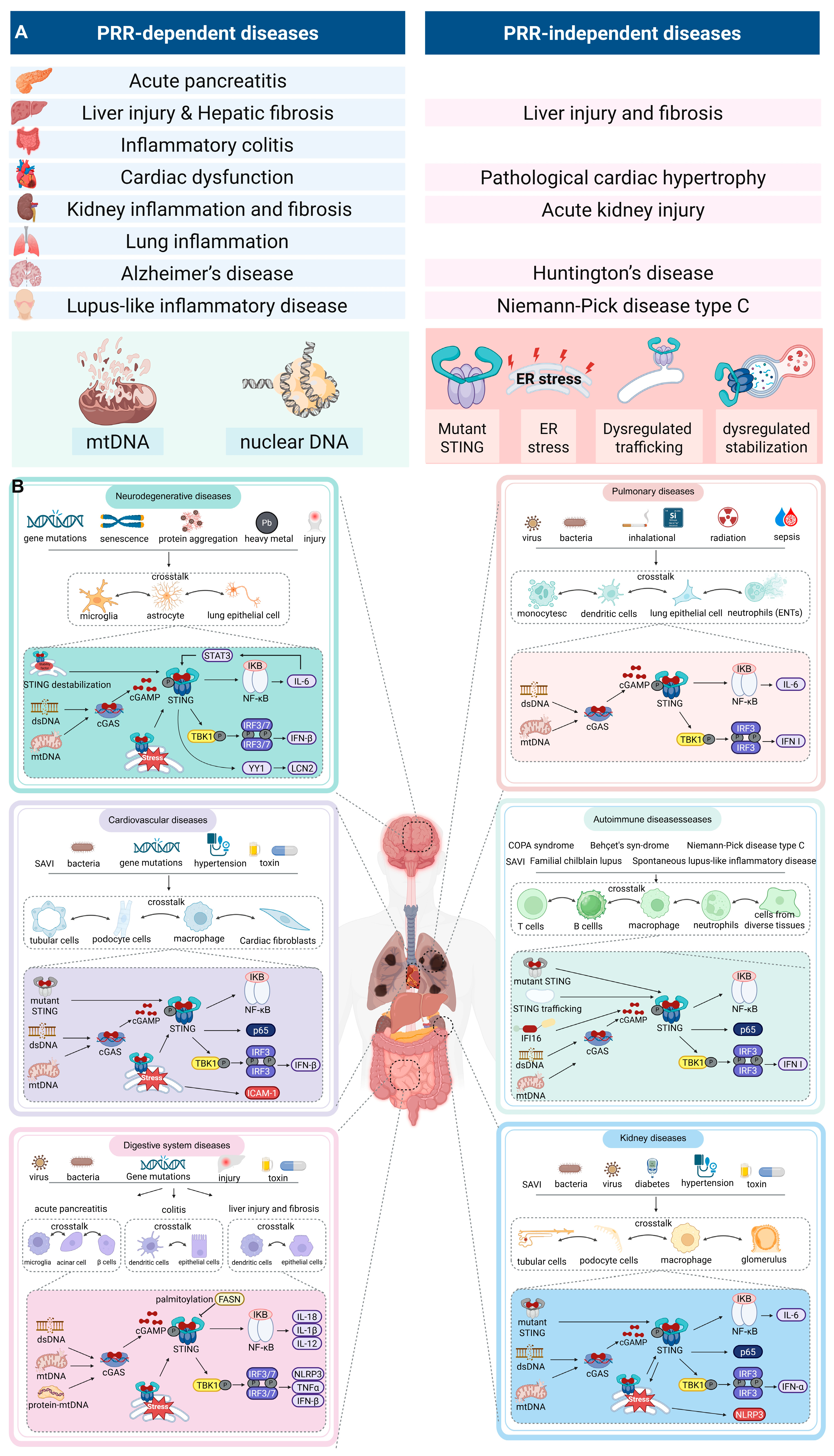

| Drug (Code Name) | Target | Species | Clinical Phase | Indication | Trial Identifier |
|---|---|---|---|---|---|
| VENT-03 | cGAS | Human | Phase 1 | Lupus | EUCT2023-507504-31-00 |
| IMSB-301 | cGAS | Human | Phase 1 | Lupus | ISRCTN90049550 |
| CXA-10 | STING (palmitoylation) | Human | Phase 2 | Pulmonary Arterial Hypertension | NCT04053543 NCT04125745 NCT03449524 |
| Reagent | Type | Effects | Structure | Therapeutics | References |
|---|---|---|---|---|---|
| cGAS catalytic site inhibitors | |||||
| PF-06928215 | Substrate-competitive inhibitor | Inhibiting catalytic site of cGAS (human) |  | Ameliorating PM2.5-induced cellular senescence in the lung | [219,220,226] |
| RU.521 | Substrate-competitive inhibitor | Inhibiting catalytic site of cGAS (mouse) |  | Mitigating subarachnoid hemorrhage-induced brain injury | [221,222] |
| G150 | Non-covalent inhibitor | Inhibiting catalytic site of cGAS (human) |  | Alleviating polystyrene microplastics induce pulmonary fibrosis | [223,259] |
| Compound S3 | Non-covalent inhibitor | Inhibiting catalytic site of cGAS (human) | 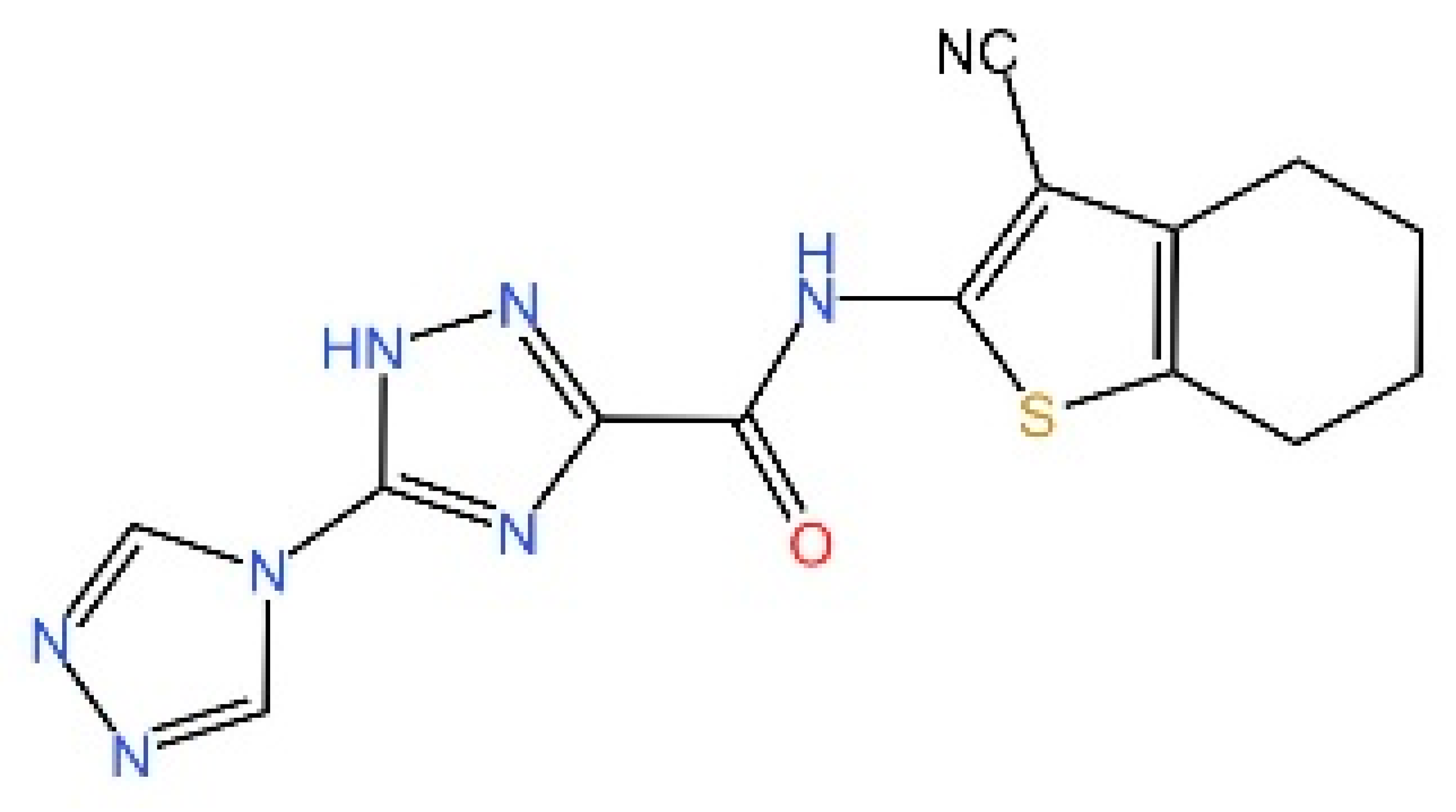 | Treatment of Acute Lung Injury | [226,229] |
| Compound 3 | Covalently modified inhibitor | Inhibiting catalytic site of cGAS (mouse) | 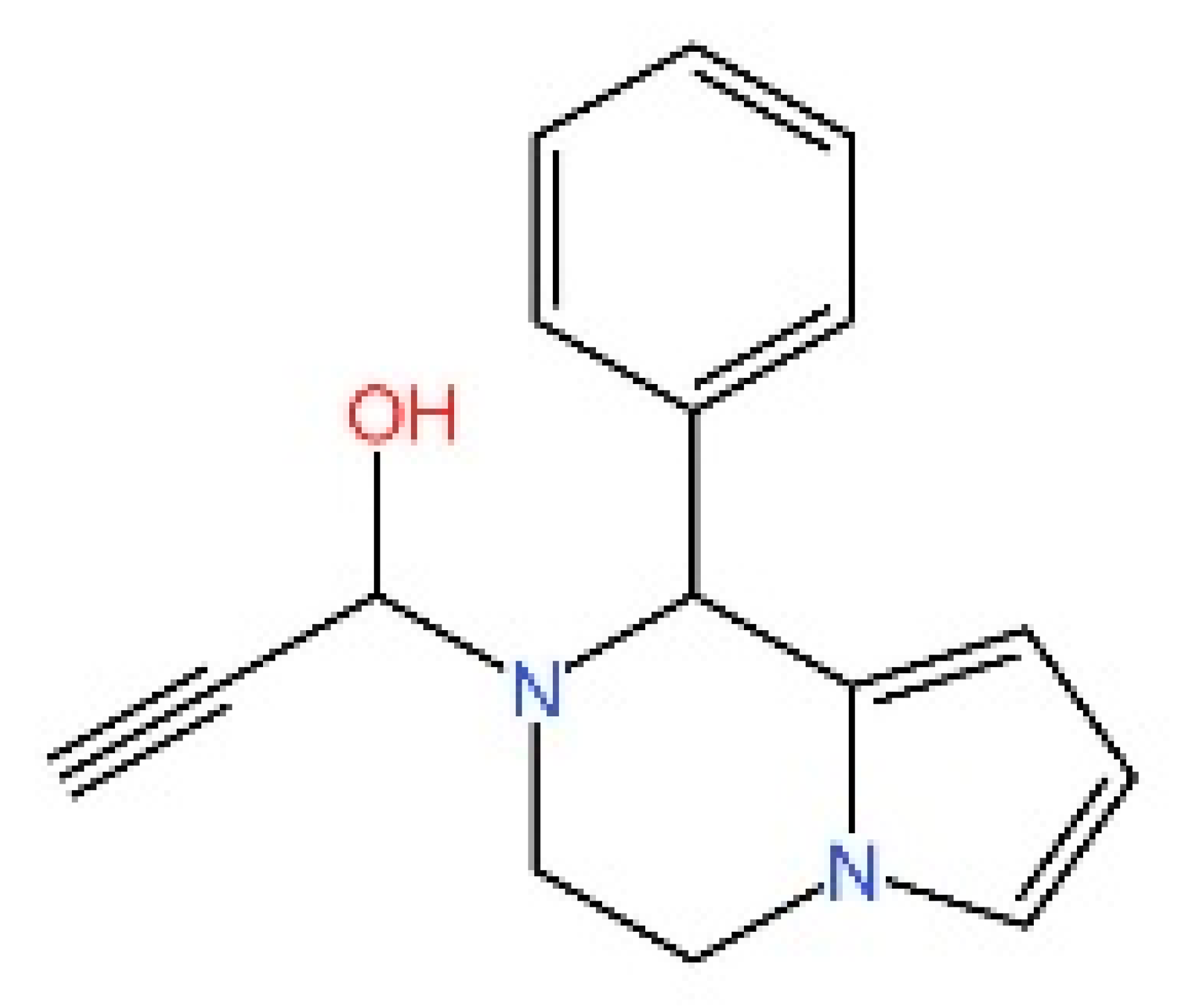 | Alleviating DSS-induced colitis | [227] |
| CU-32 & CU-76 | Substrate-no-competitive inhibitor | Inhibiting catalytic site of cGAS (human) | 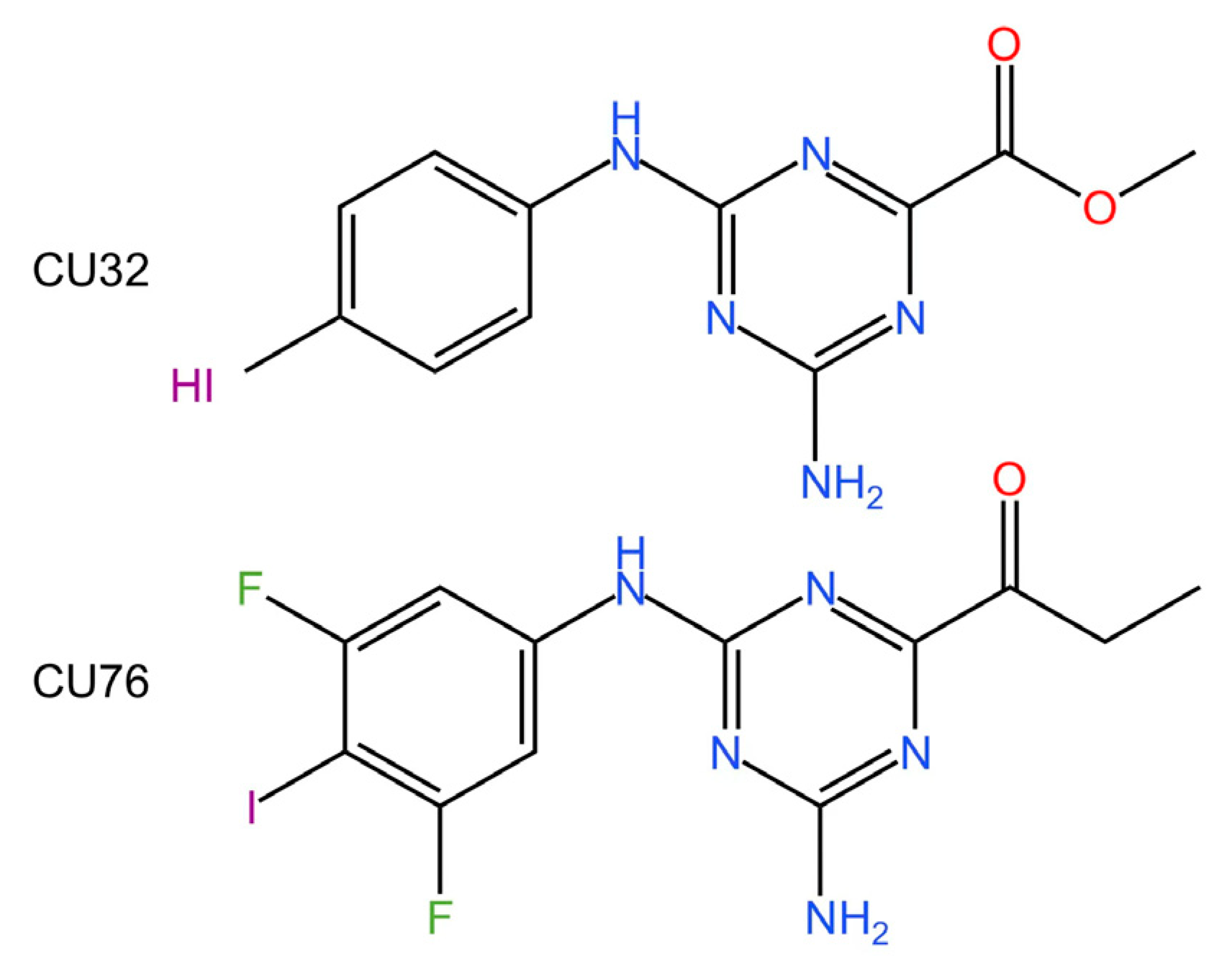 | Reducing IFN-β production in macrophages | [228] |
| Compounds 30d-S | Substrate-competitive inhibitor | Inhibiting catalytic site of cGAS (human) | 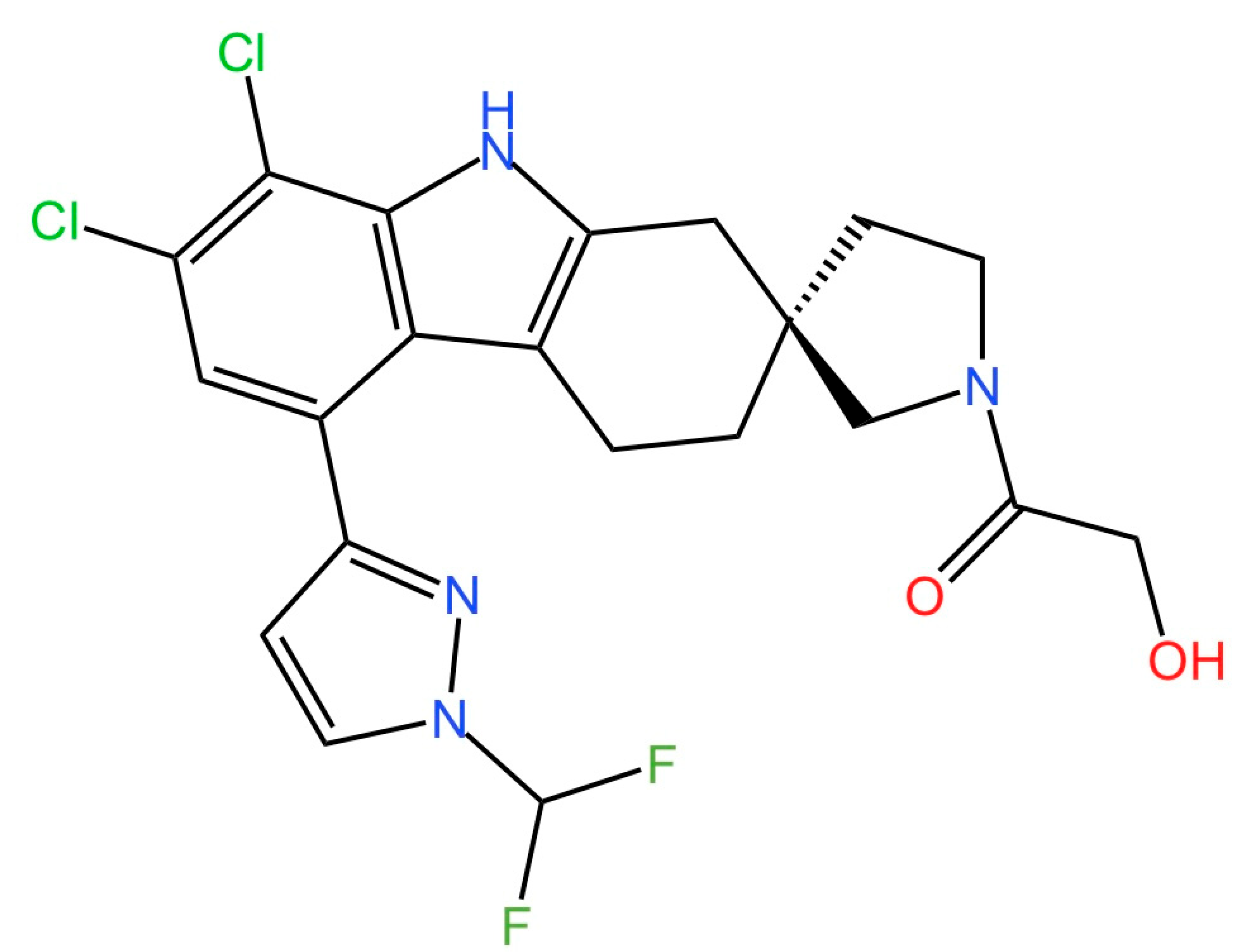 | Alleviating LPS-Induced Acute Lung Injury (ALI) In Vivo. | [229] |
| cGAS-dsDNA banding inhibitors | |||||
| Quinacrine | Binding to dsDNA | Inhibiting cGAS-DNA interaction (human & mouse) |  | Reducing IFN-β production in vitro | [231,260] |
| X6 | Binding to dsDNA | Inhibiting cGAS-DNA interaction (mouse) | 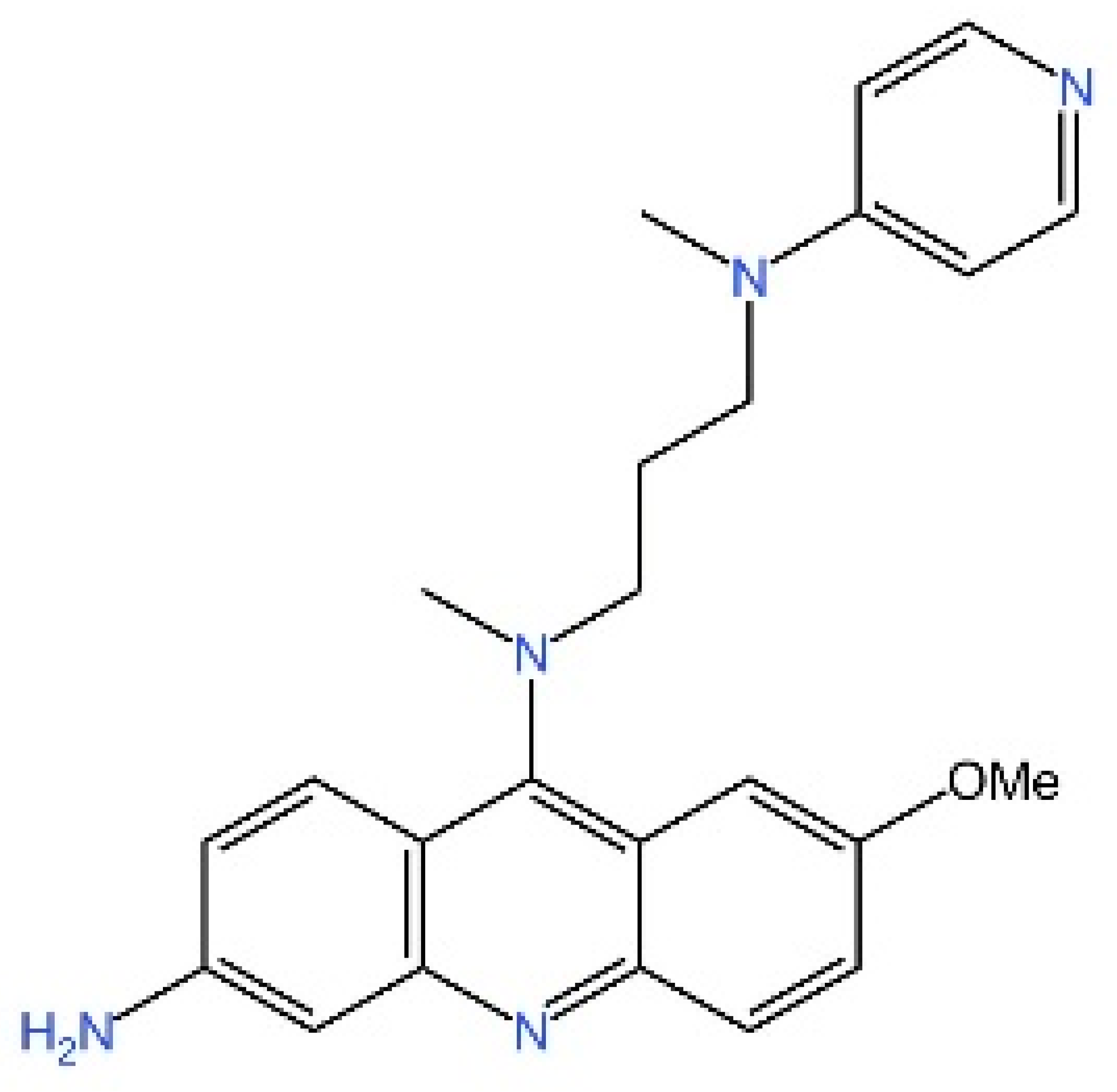 | Inhibiting cGAMP synthesis in Trex1-deficient mice | [233] |
| Suramin | Competitive inhibitor | Inhibiting cGAS-DNA interaction (human) | 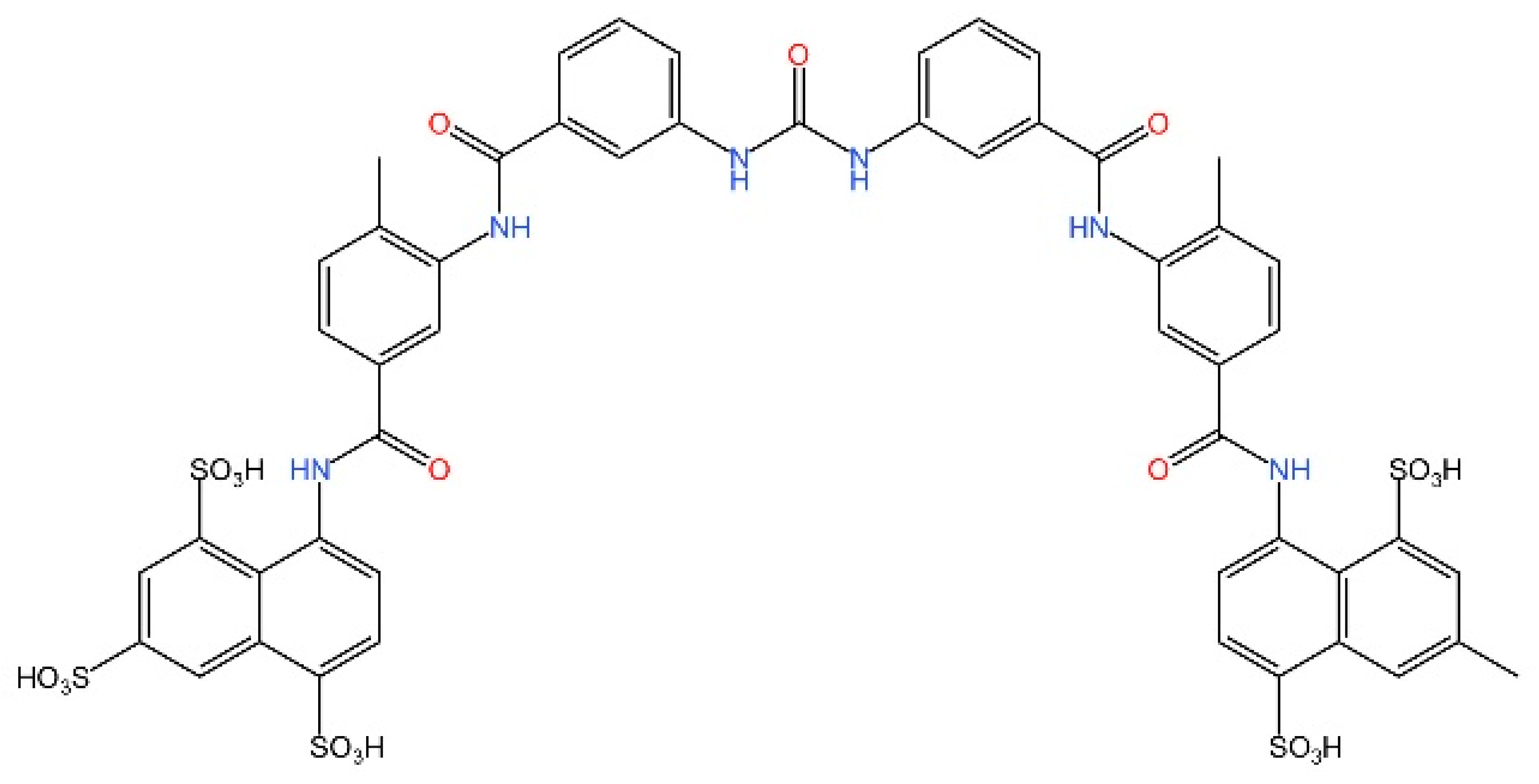 | Reducing IFN-β pro-duction in THP-1 cells | [234] |
| 4-sulfonic calix[6] | Competitive inhibitor | Inhibiting cGAS-DNA interaction/ Inhibiting catalytic site of cGAS (mouse) |  | Preventing AIM2-dependent immunosuppression following stroke. | [235] |
| A151 | Competitive inhibitor | Inhibiting cGAS-DNA interaction (human & mouse) | 5′-TTAGGGTTAGGG TTAGGGTTAGGG-3′ | Inhibiting type I interferon response in TREX1-deficient cells. | [261] |
| XQ2B | Competitive inhibitor | Inhibiting cGAS-DNA interaction (mouse) | 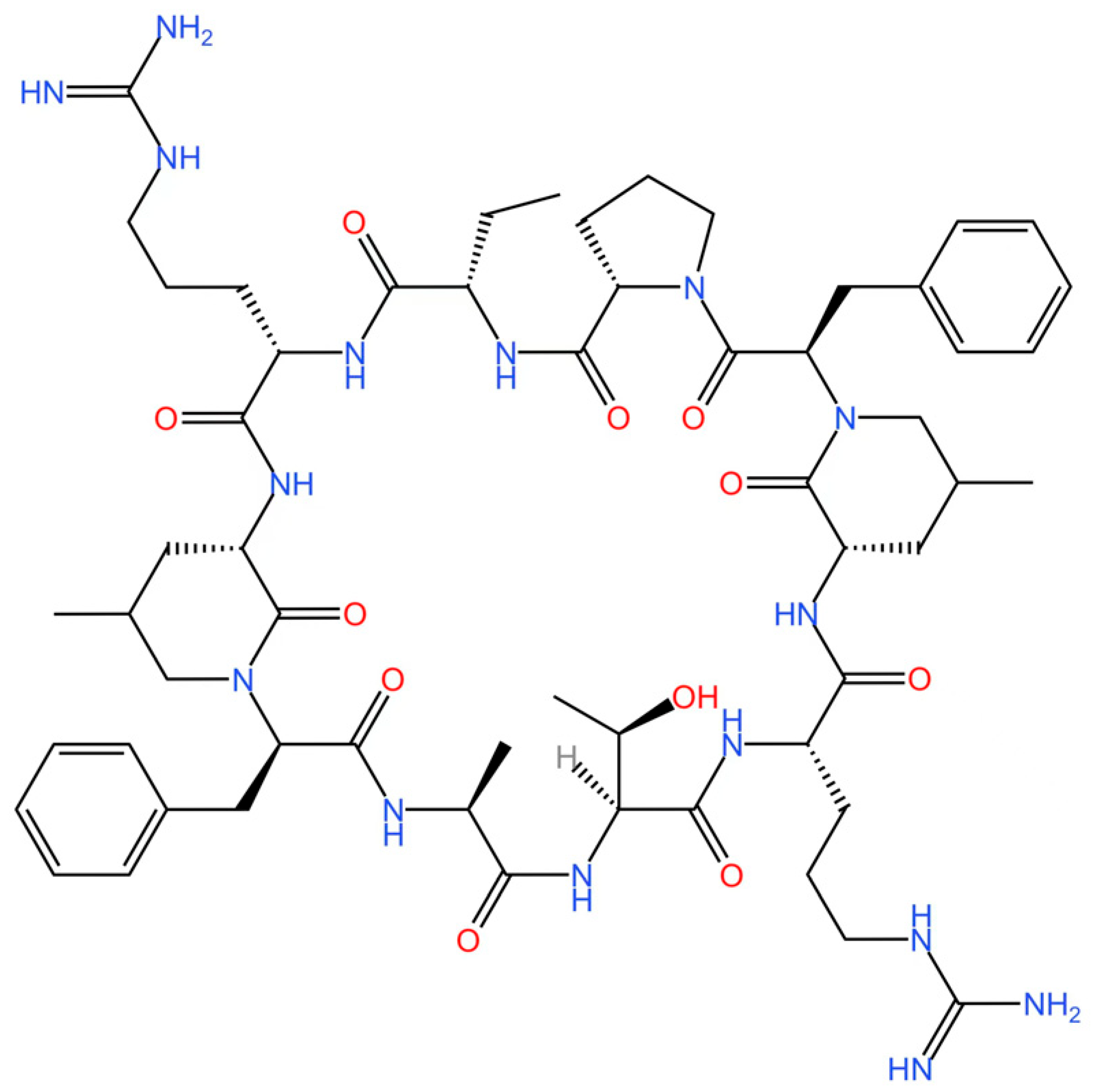 | Inhibiting STING-mediated inflammation induced by Trex1 deficiency and in inhibiting IFN I production during HSV-1 infection. | [236]. |
| STING antagonists targeting the CDN-binding site | |||||
| Compound 18 | cGAMP-competitive inhibitor | Inhibiting CDN-binding site of STING (human) | 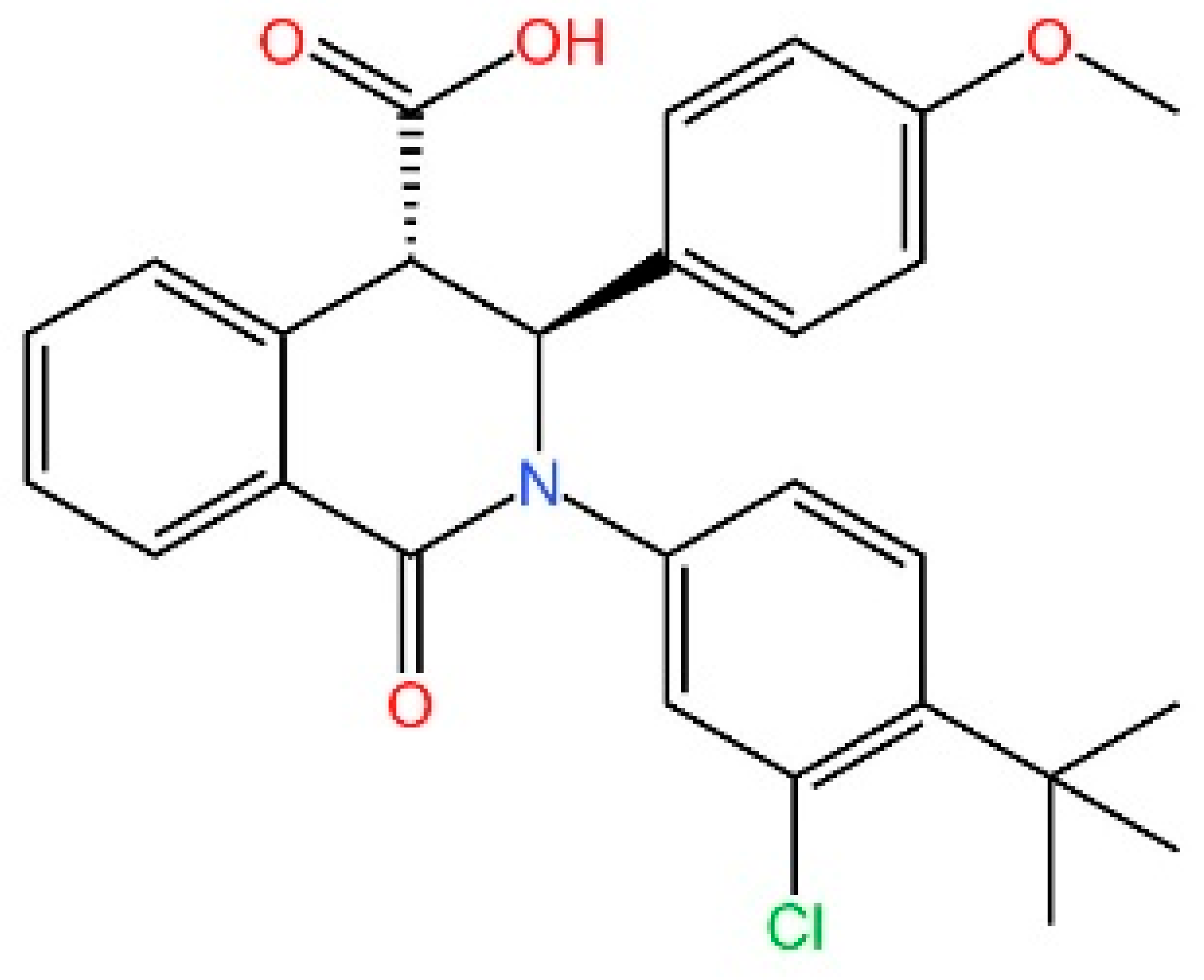 | Inhibiting cGAMP-induced IFN-β secretion. | [239] |
| Astin C | cGAMP-competitive inhibitor | Inhibiting CDN-binding site of STING (mouse & human) | 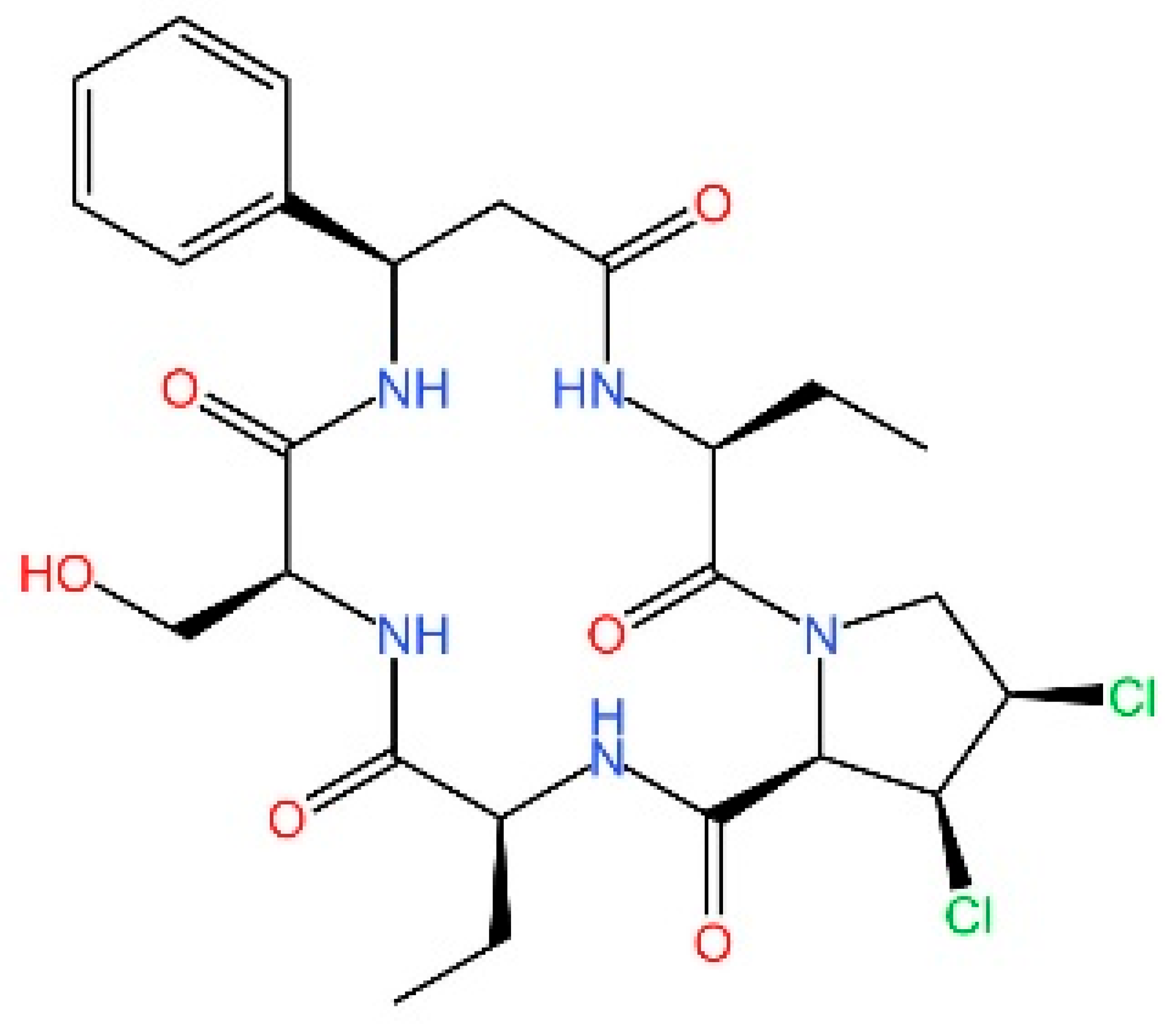 | Attenuating the autoinflammatory responses in Trex1−/− mouse autoimmune disease model. | [240] |
| SN-011 | cGAMP-competitive inhibitor | cGAMP-competitive inhibitor (mouse & human) | 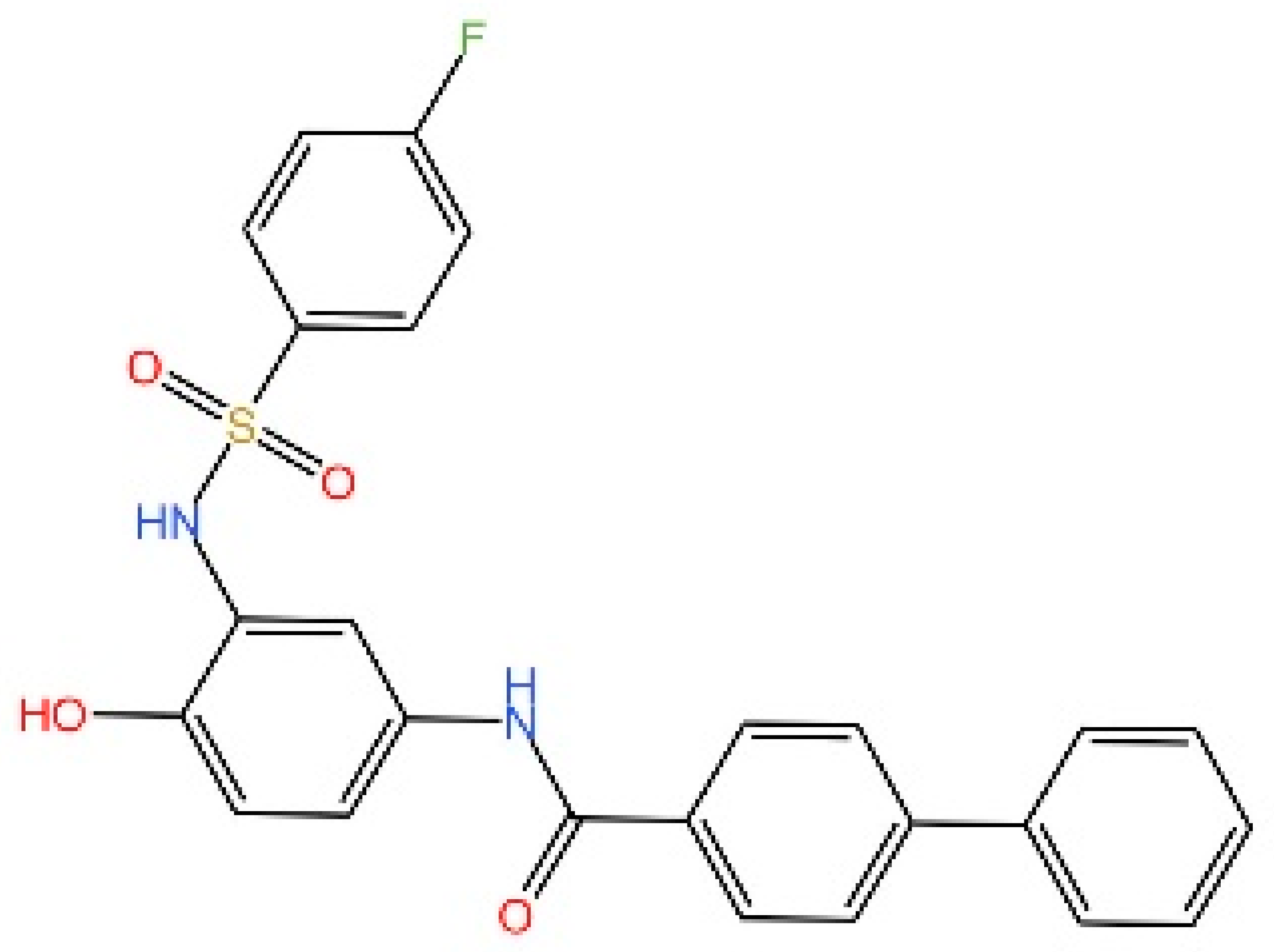 | Suppressing Systemic Inflammation in Trex1−/− mice. | [241] |
| Gelsevirine | cGAMP-competitive inhibitor | cGAMP-competitive inhibitor (human & mouse) |  | Mitigating STING-related inflammation in sepsis. | [242] |
| T0901317 | SMPDL3A agonist | Degradation of cGAMP via LXR-SMPDL3A (mouse & human) | 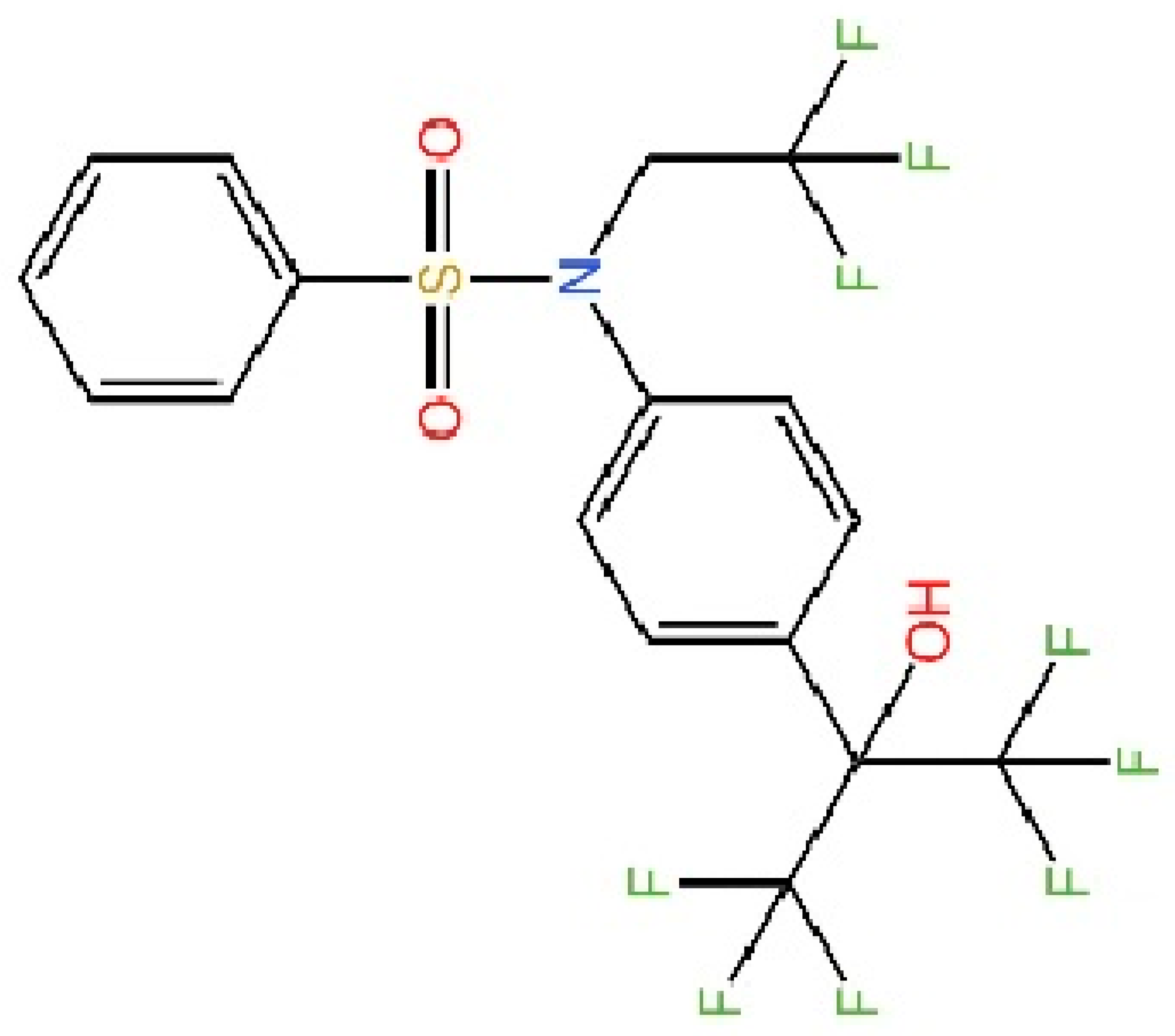 | In the Smpdl3a+/+ mouse model, T0901317 induces an increase in HSV-1 viral load. | [243] |
| Anhydrotuberosin | cGAMP-competitive inhibitor (mouse & human) | 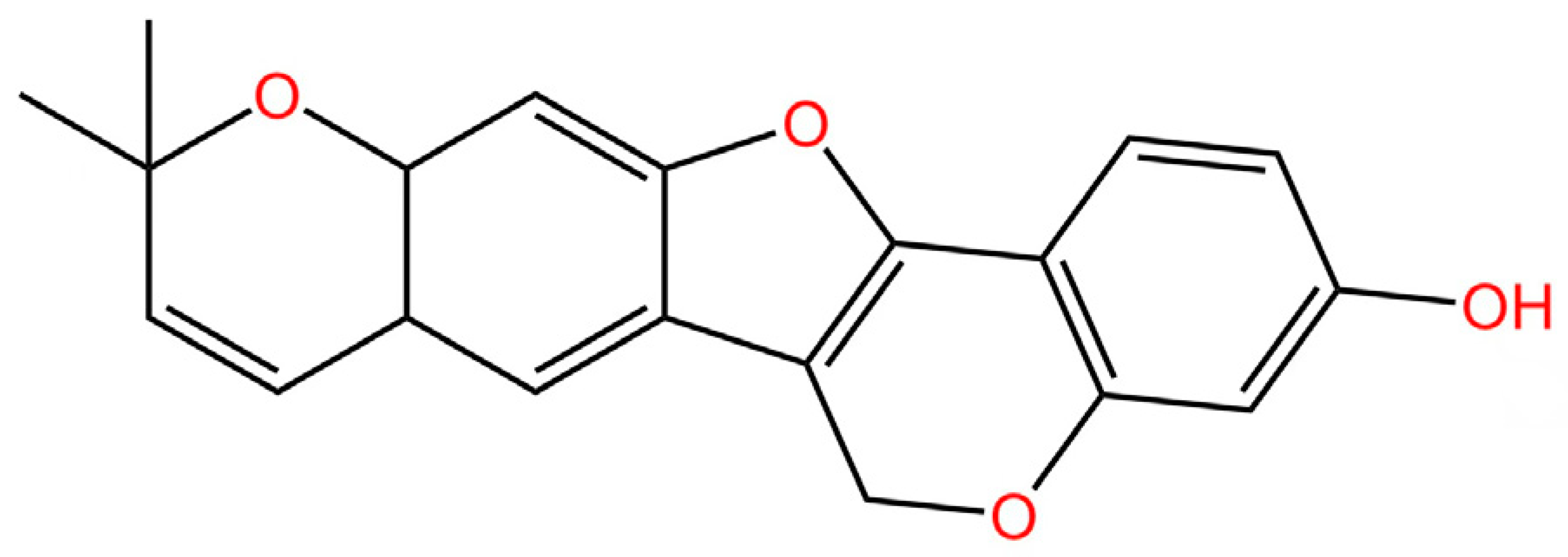 | ATS prevents aberrant STING activation driven by SAVI-related mutations. | [244] | |
| Targeting STING phosphorylation sites | |||||
| Meloxicam | No report | Inhibiting phosphorylation of STING (human & mouse) | 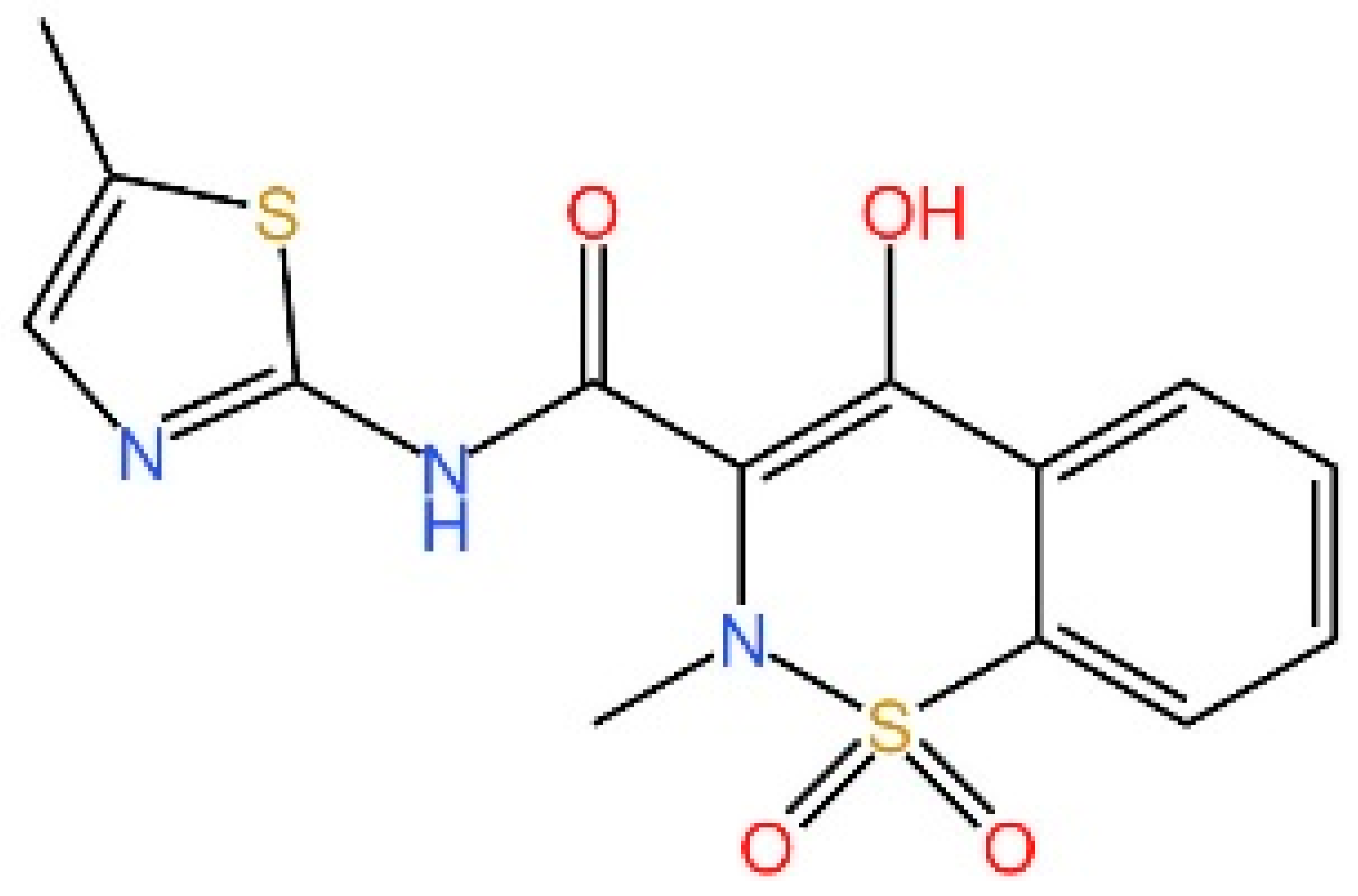 | Promoting the survival in Trex1−/− mouse model for Aicardi-Goutières syndrome | [246] |
| LH531 | Covalently modified inhibitor | Inhibiting phosphorylation of STING (human) | 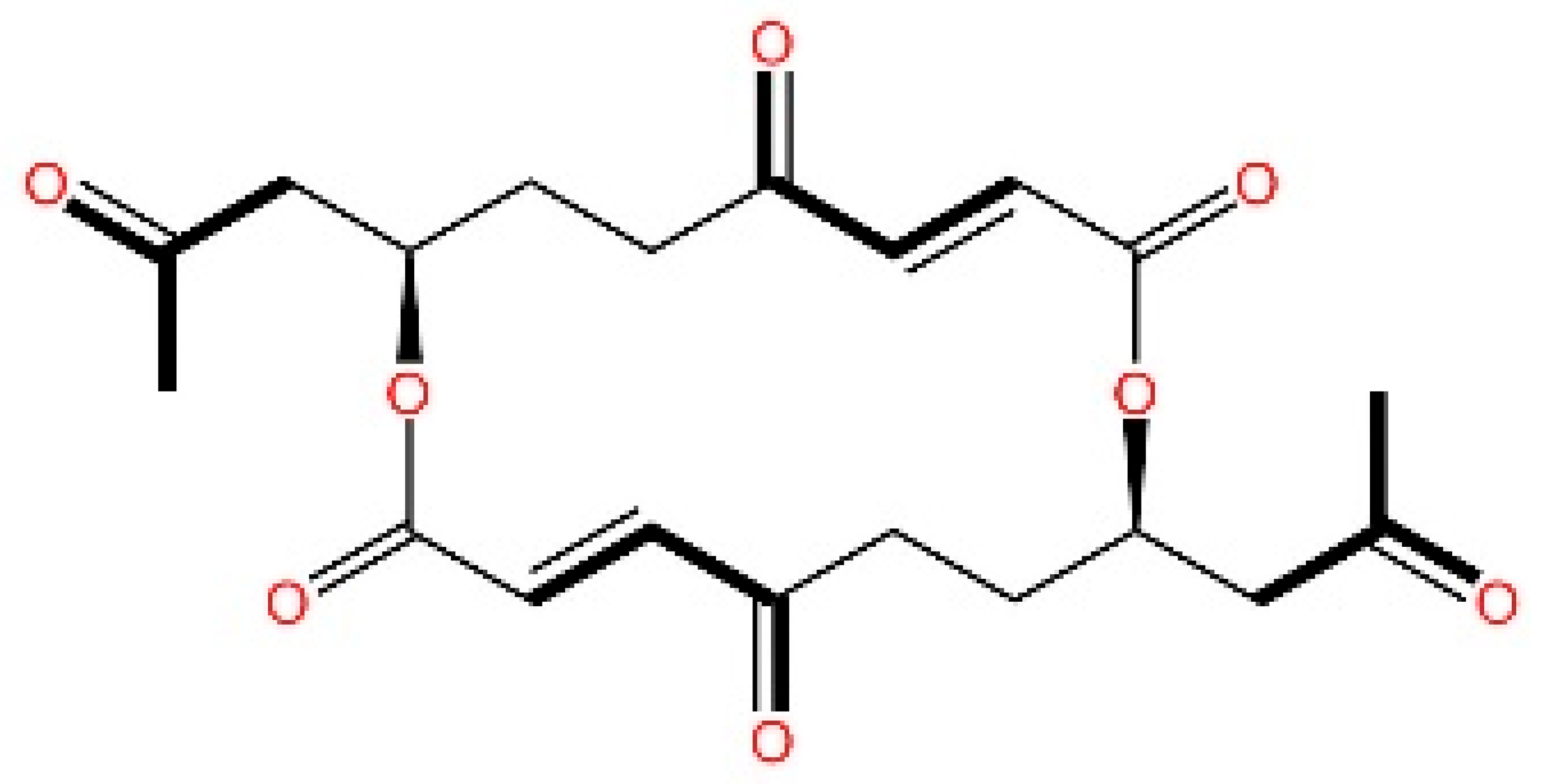 | No report | [245] |
| LH519 | Covalently modified inhibitor | Inhibiting phosphorylation of STING (human) | 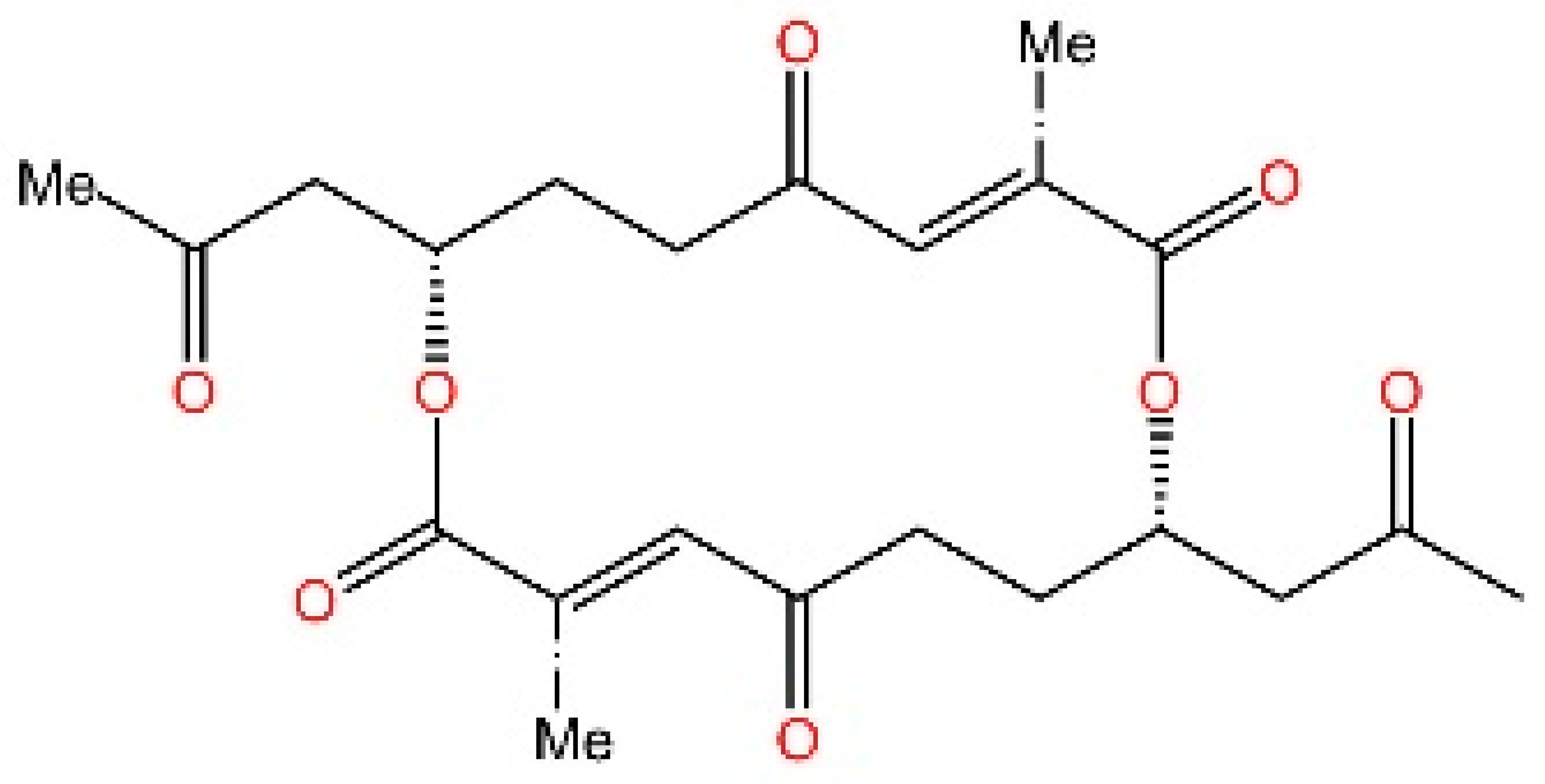 | No report | [245] |
| 4-OI | Covalently modified inhibitor | Inhibiting phosphorylation of STING (human & mouse) | 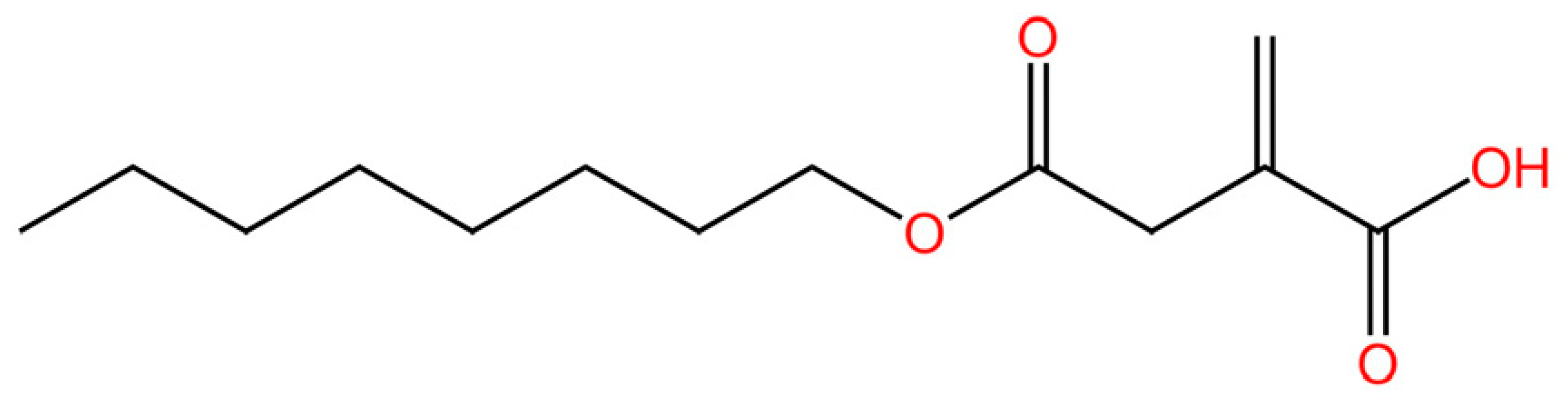 | Alleviate sepsis | [247] |
| Targeting STING palmitoylation sites | |||||
| C-176 | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) |  | Reducing STING agonist mediated elevation of serum levels of type I IFNs and IL-6 in Trex1−/− mice | [249] |
| C-178 | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 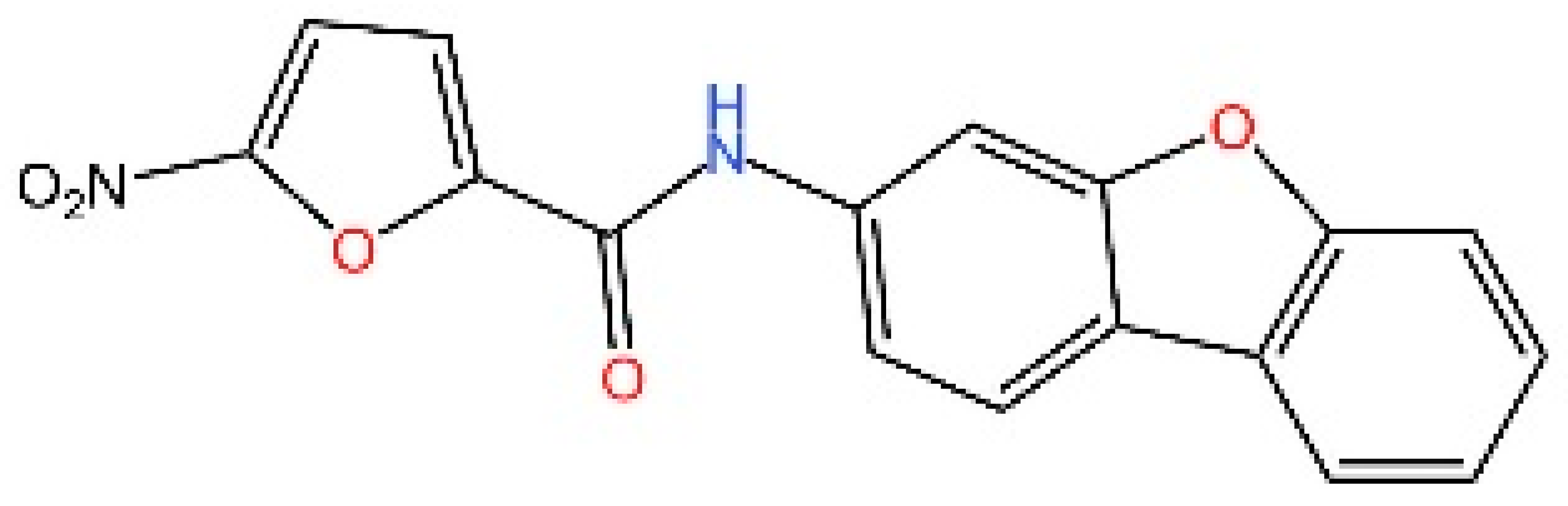 | Suppressing the STING responses elicited by distinct activators | [249] |
| H151 | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 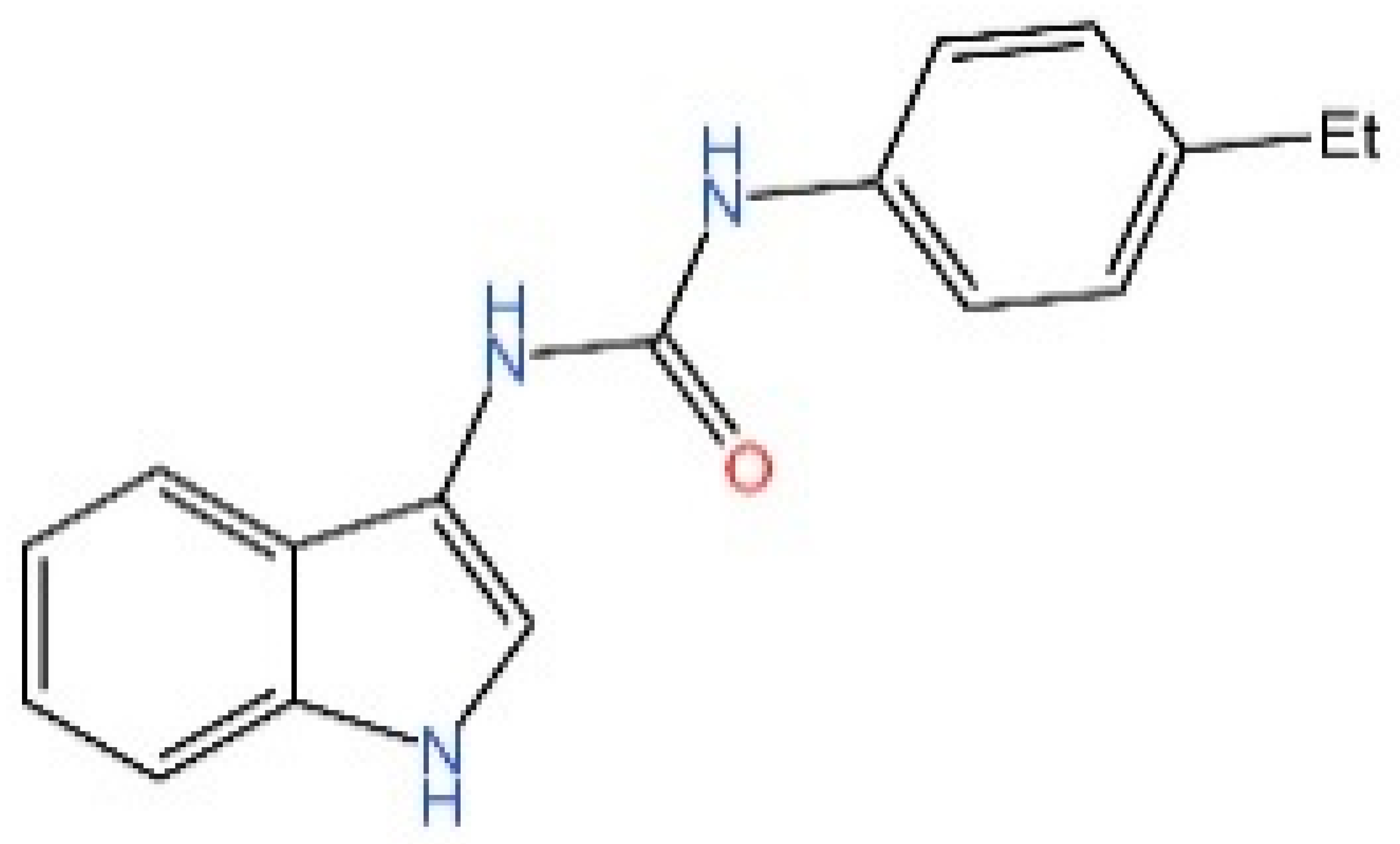 | Reducing IFN-β levels in Trex1−/− mice | [262] |
| NO2-cLA | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 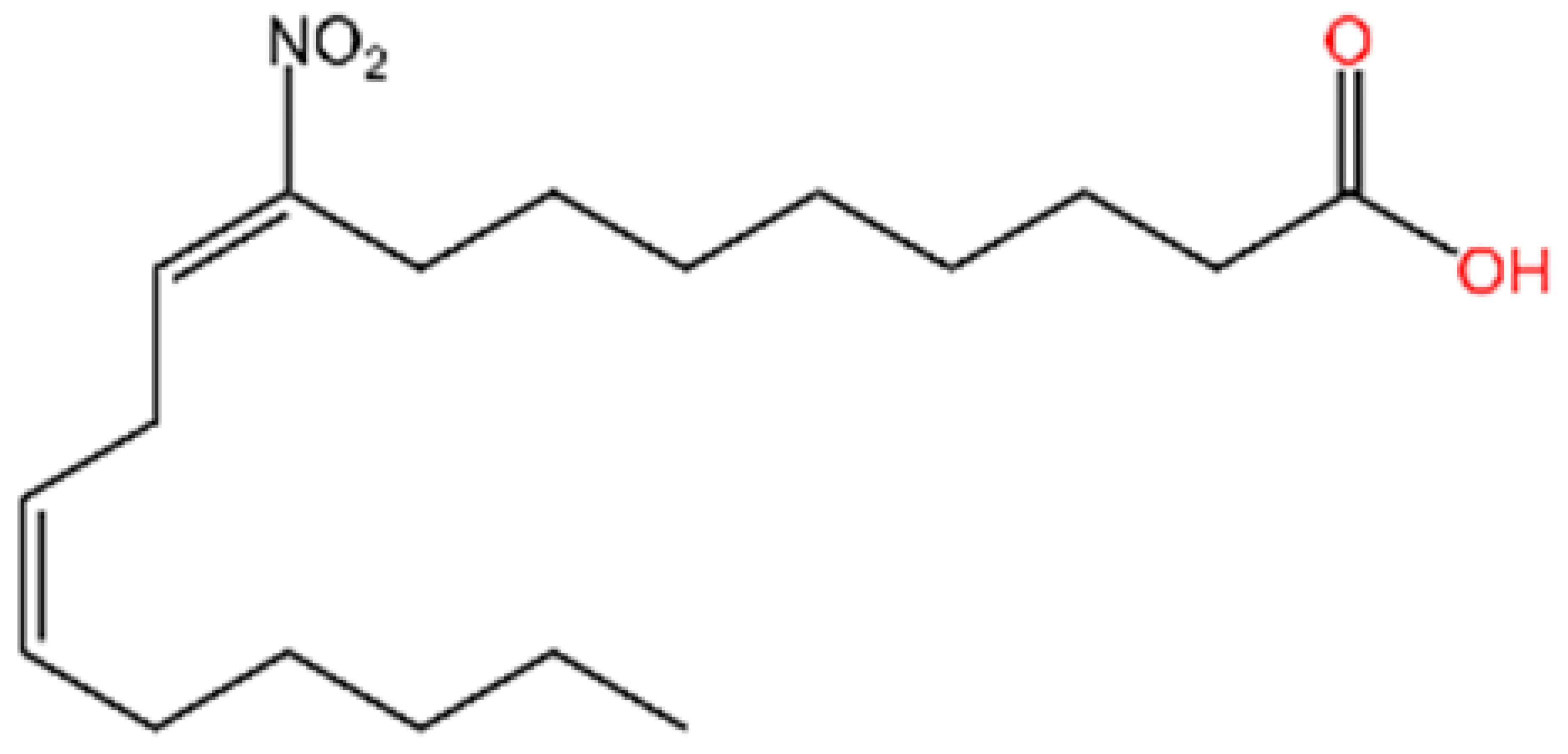 | Reducing of type I IFNs in response to HSV-2 infection | [251] |
| NO2-OA | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 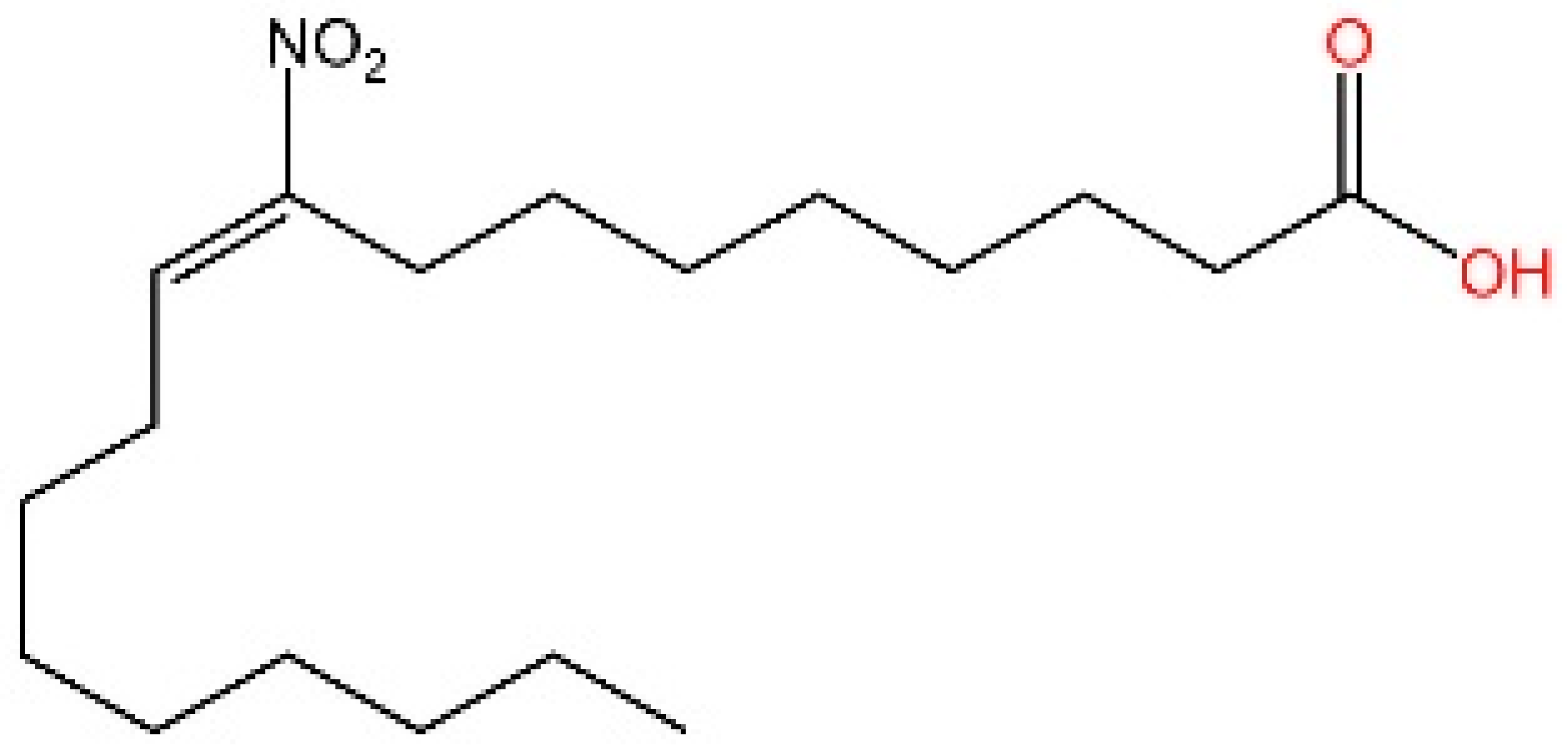 | Inhibiting the increase in pTBK1 in fibroblasts from patients with SAVI | [251] |
| BPK-21 | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 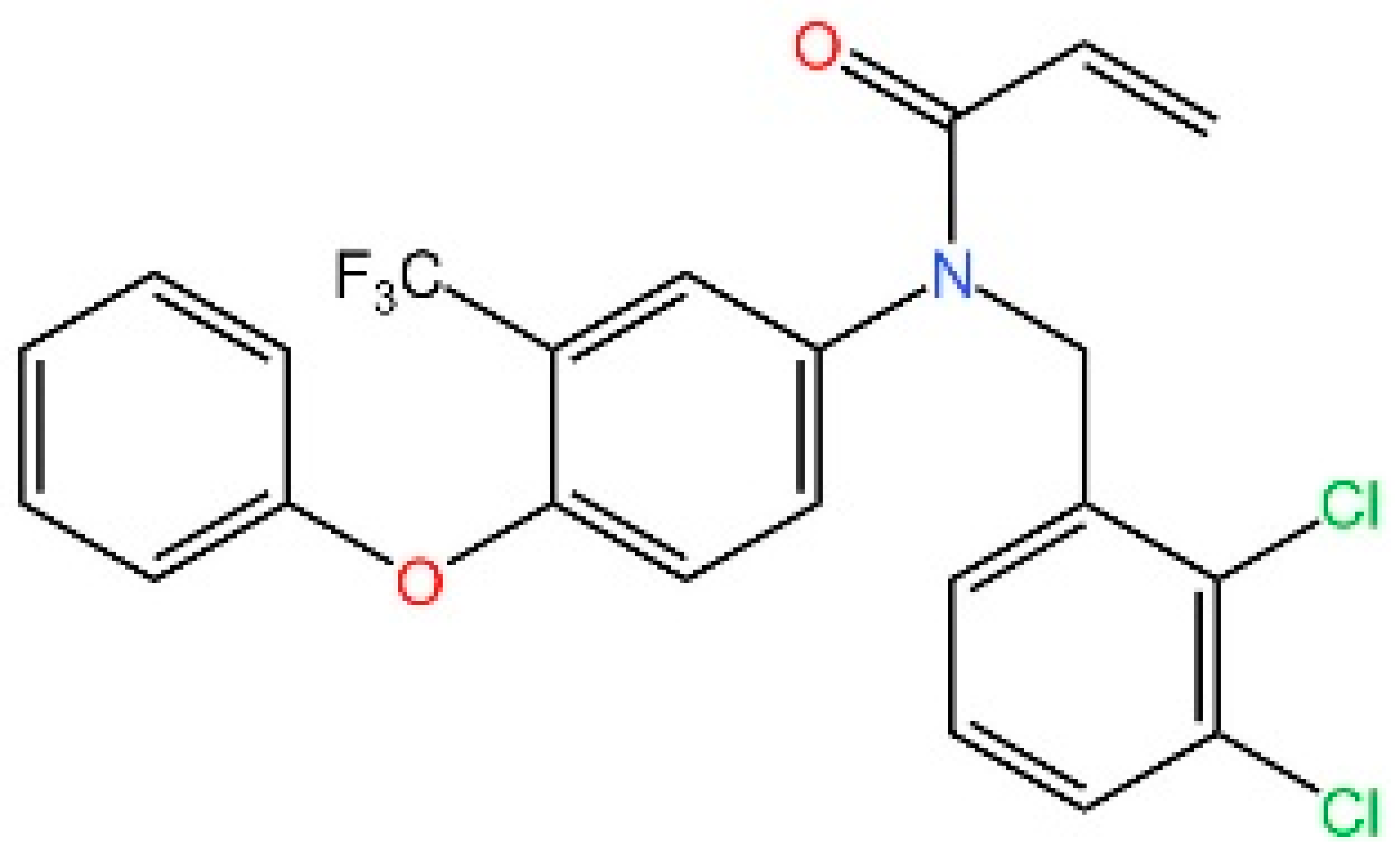 | No report | [252] |
| BPK-25 | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human) | 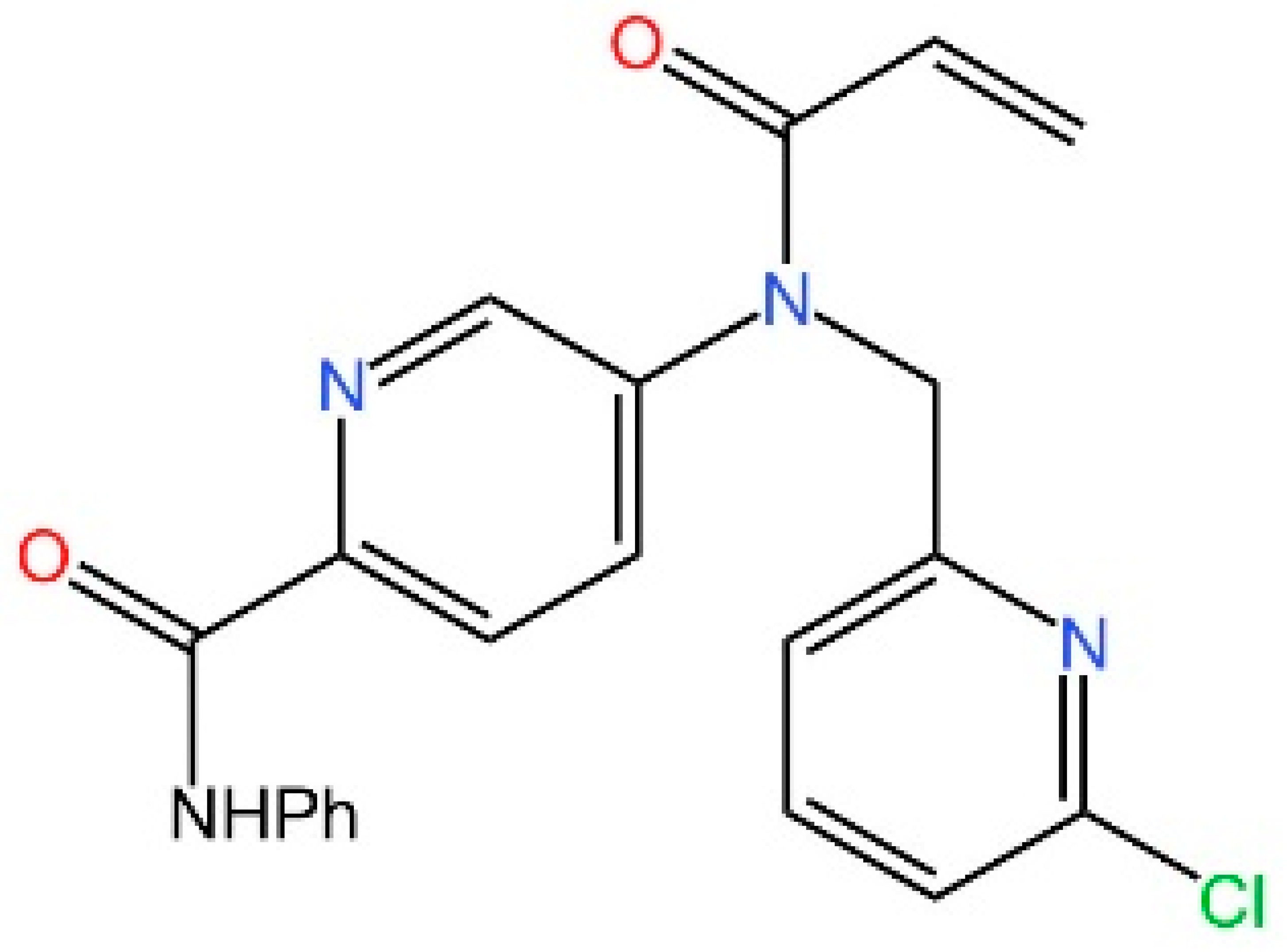 | No report | [252] |
| 4-OI | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 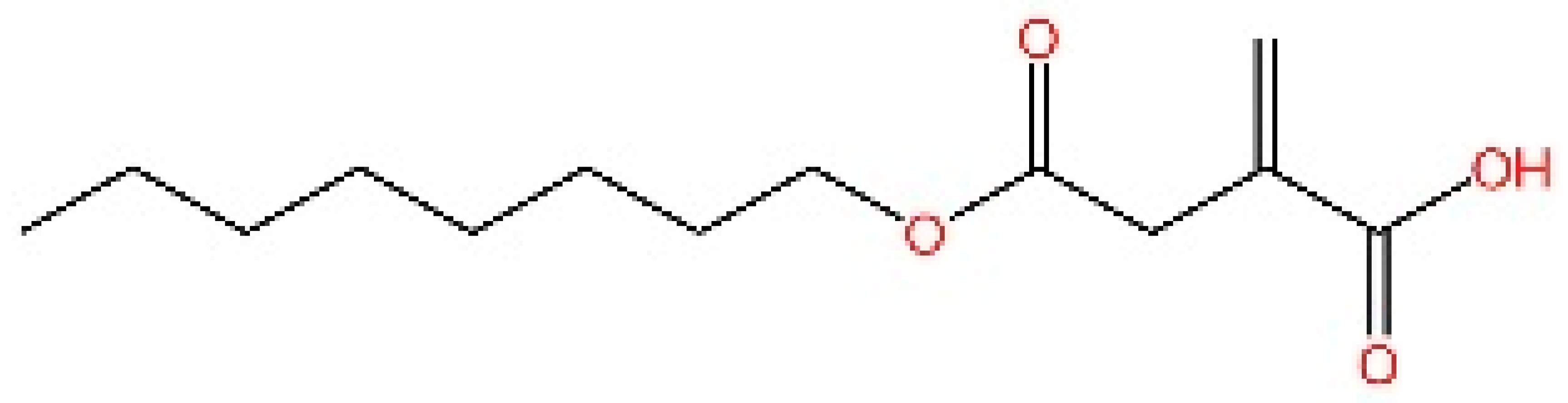 | Restricting the cGAS-STING antiviral (HSV-1) immune response and inflammatory responses in Trex1−/− Raw264.7 cells | [253] |
| NO2-cLA | Covalently modified inhibitor | Inhibiting palmitoylation of STING (human & mouse) | 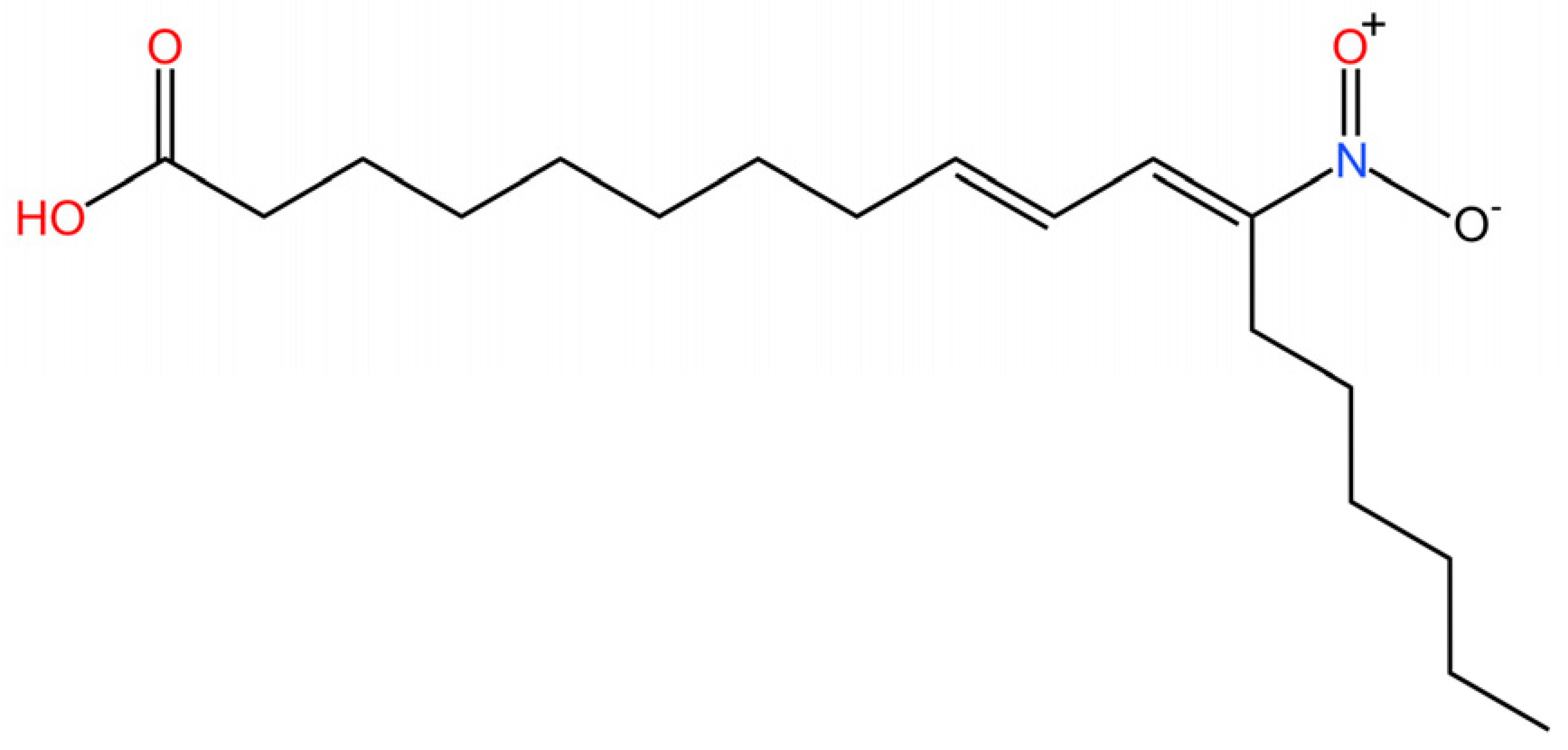 | NO2-FAs inhibit release of IFN I from SAVI fibroblasts. | [251] |
| Targeting STING Cys292 sites | |||||
| LB224 | Covalently modified inhibitor | Inhibiting modification of the Cys292 site of STING (human & mouse) | 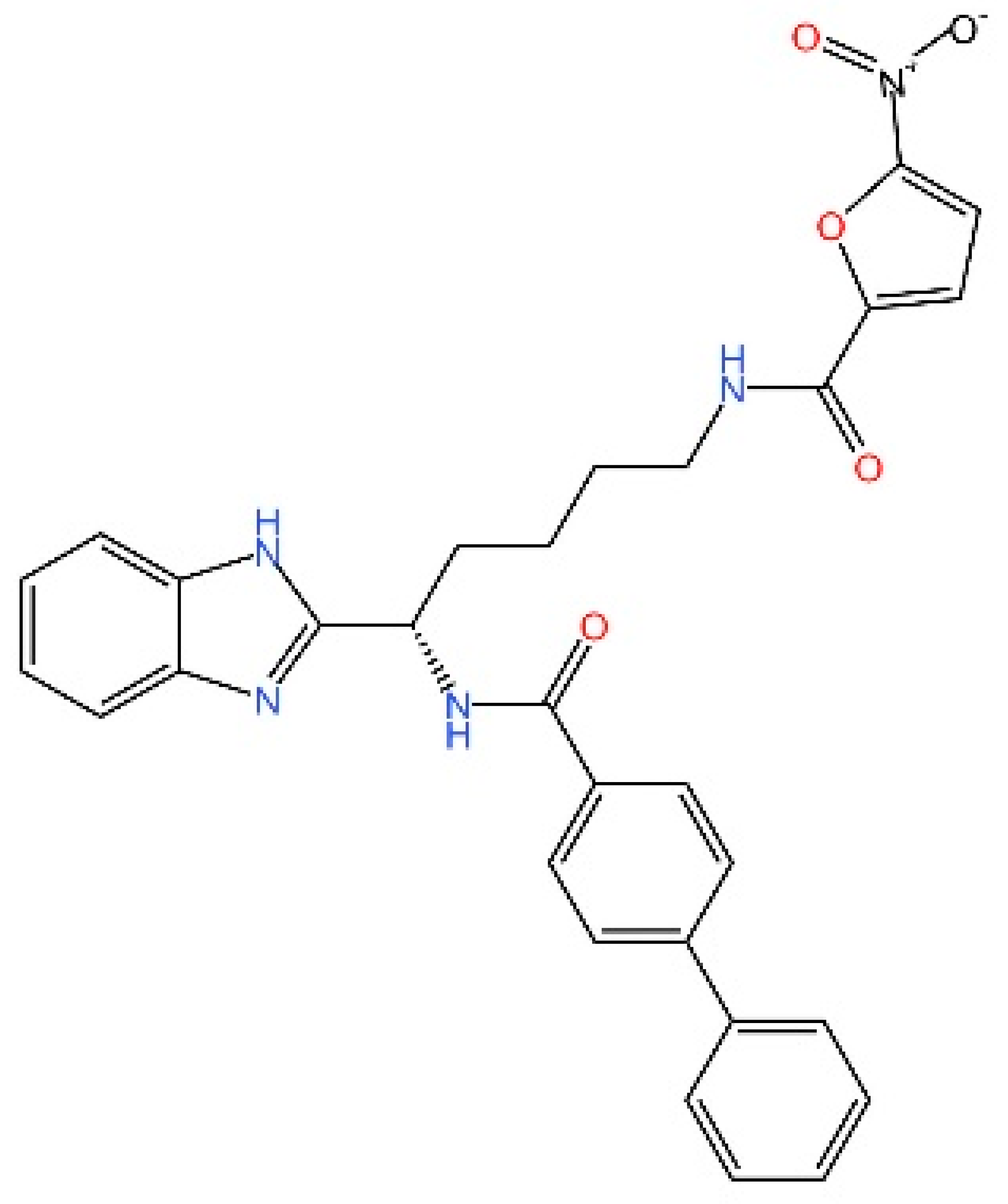 | Reducing serum IFN-β and IL-6 after administration of diABZI in mice | [254] |
| Targeting STING degradation | |||||
| STING Degrader-1 | Ubiquitin–proteasome system | Binding covalently to STING and E3 ligase (human & mouse) | 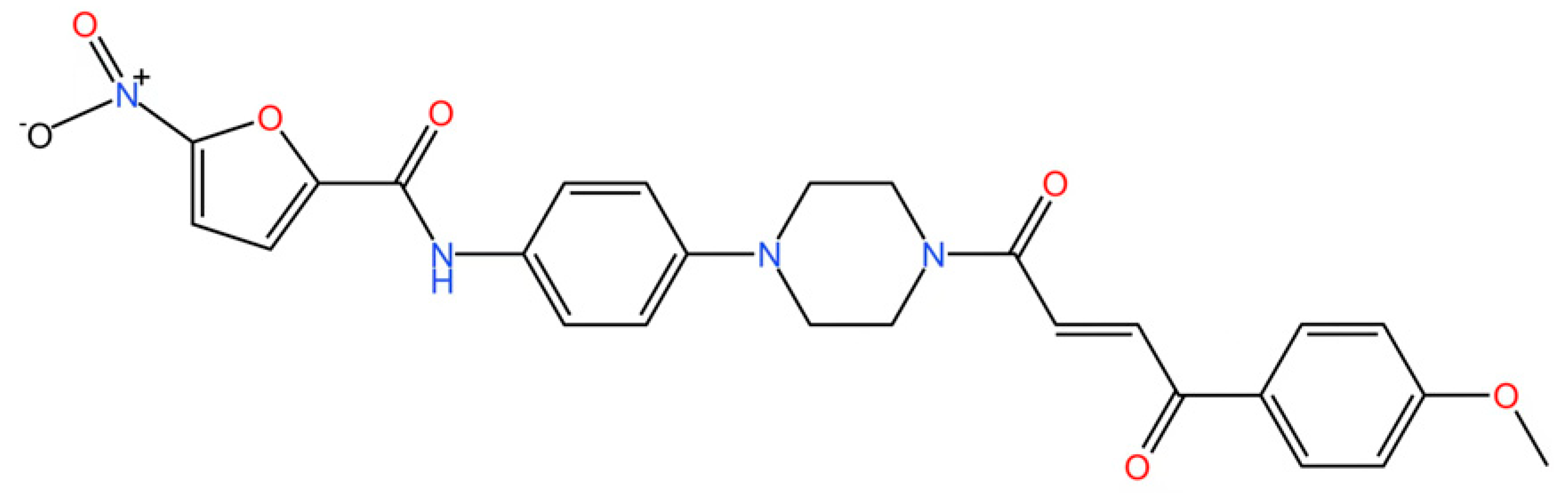 | No report | [256] |
| SP23 | Ubiquitin–proteasome system | Binding covalently to STING and E3 ligase (human & mouse) | 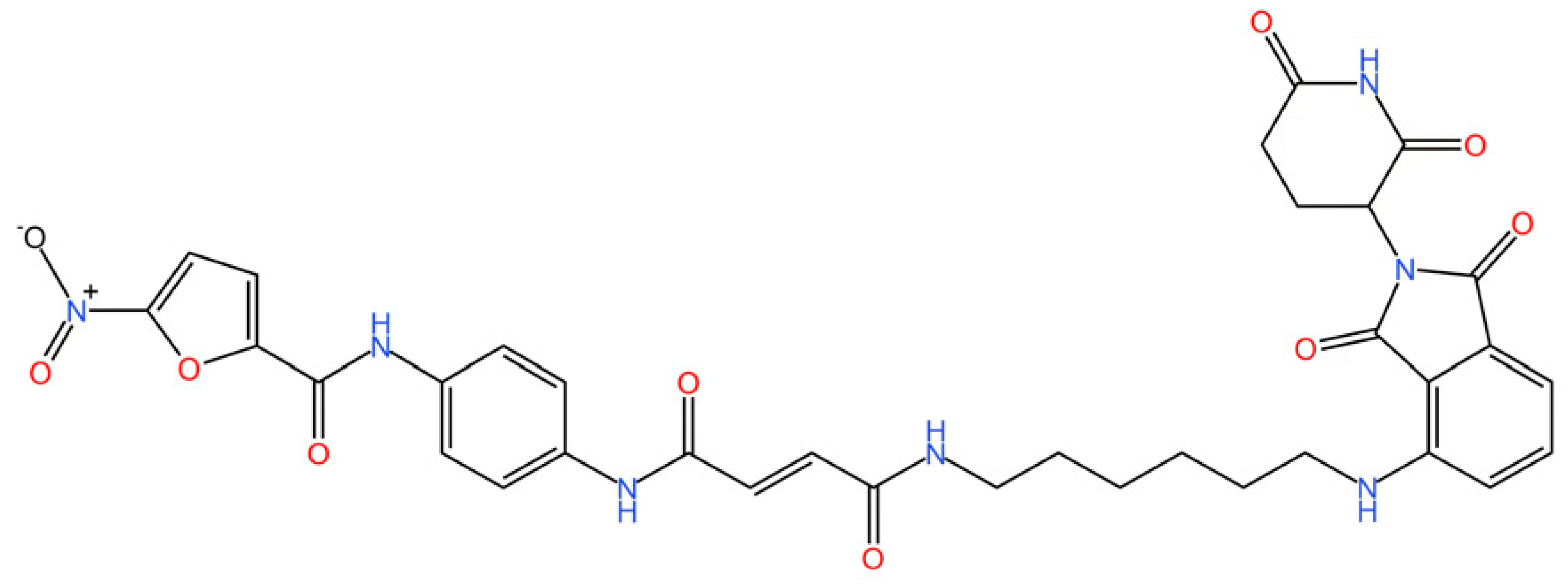 | Alleviating cisplatin-induced acute kidney injury by inhibiting the STING signaling pathway. | [257] |
| UNC9036 | Ubiquitin–proteasome system | Binding covalently to STING and E3 ligase (human & mouse) | 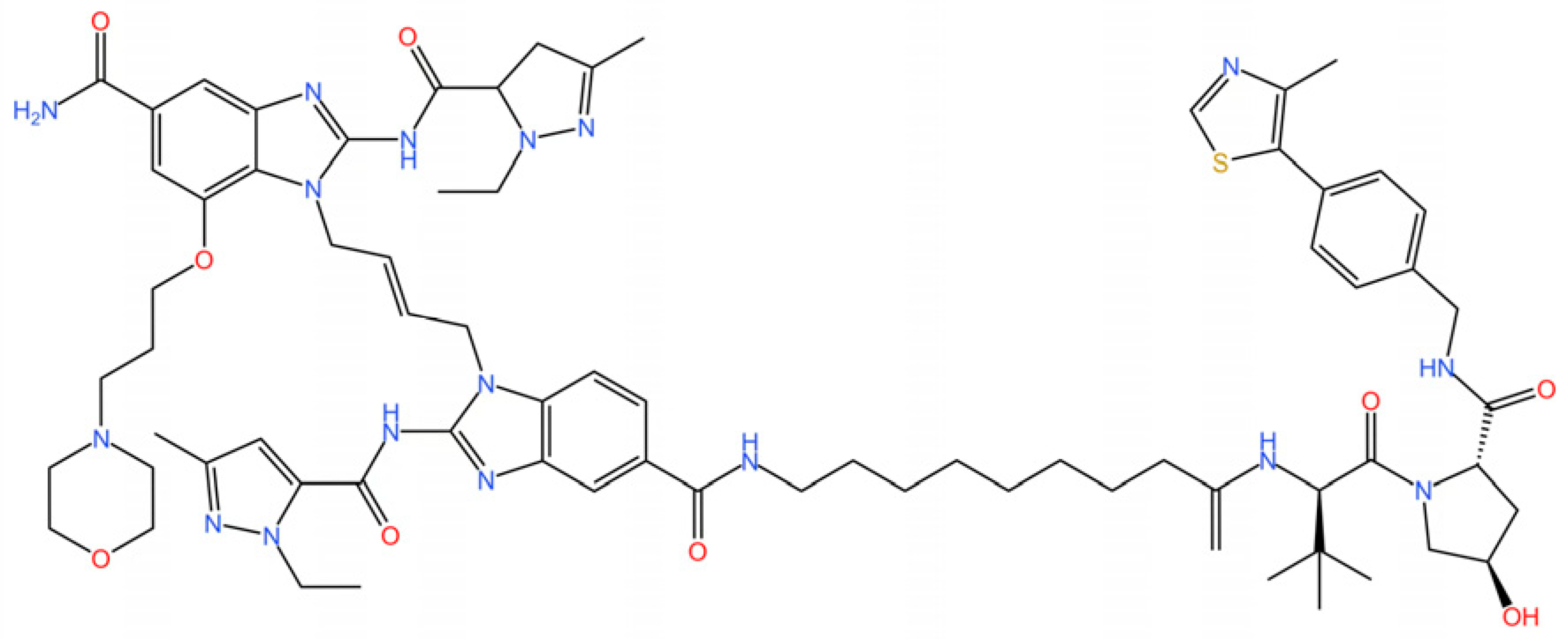 | No report | [258] |
| Targeting STING trafficking | |||||
| 4-phenylcinnamic acid | modulating STING trafficking | Competitive inhibitor |  | Significantly diminishing COPII-mediated vesicular transport of STING, it suppresses inflammatory signaling pathways. | [110,263] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, L.; Li, J.; Zhu, X.; Zhou, L.; Sun, Z.; Zhang, Z.; Xu, W.; Song, Y. PRRs-Dependent and Independent Mechanisms of STING Signaling in Inflammatory and Autoimmune Diseases. Biomedicines 2025, 13, 2533. https://doi.org/10.3390/biomedicines13102533
Xu L, Li J, Zhu X, Zhou L, Sun Z, Zhang Z, Xu W, Song Y. PRRs-Dependent and Independent Mechanisms of STING Signaling in Inflammatory and Autoimmune Diseases. Biomedicines. 2025; 13(10):2533. https://doi.org/10.3390/biomedicines13102533
Chicago/Turabian StyleXu, Le, Jingrou Li, Xingchen Zhu, Liting Zhou, Zhirong Sun, Zhipeng Zhang, Wei Xu, and Yahui Song. 2025. "PRRs-Dependent and Independent Mechanisms of STING Signaling in Inflammatory and Autoimmune Diseases" Biomedicines 13, no. 10: 2533. https://doi.org/10.3390/biomedicines13102533
APA StyleXu, L., Li, J., Zhu, X., Zhou, L., Sun, Z., Zhang, Z., Xu, W., & Song, Y. (2025). PRRs-Dependent and Independent Mechanisms of STING Signaling in Inflammatory and Autoimmune Diseases. Biomedicines, 13(10), 2533. https://doi.org/10.3390/biomedicines13102533








