A Retrospective Study in Colorectal Adenocarcinoma Uncovers the Potential of Circ-CCT3 as a Predictor of Tumor Recurrence
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Total RNA Extraction and Reverse Transcription
2.3. Relative Circ-CCT3 Quantification, Using Real-Time Quantitative Polymerase Chain Reaction (qPCR)
2.4. Biostatistical Analysis
3. Results
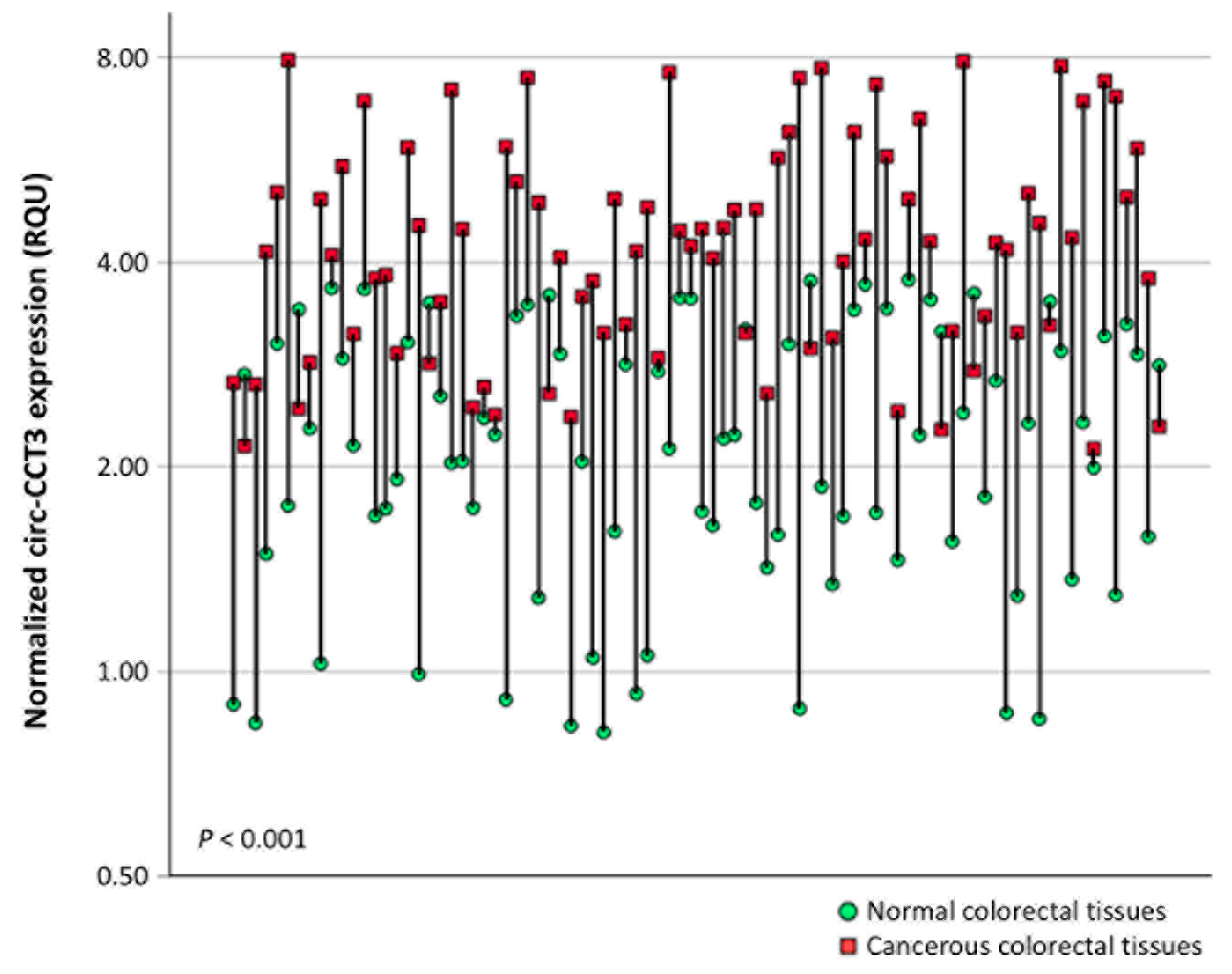
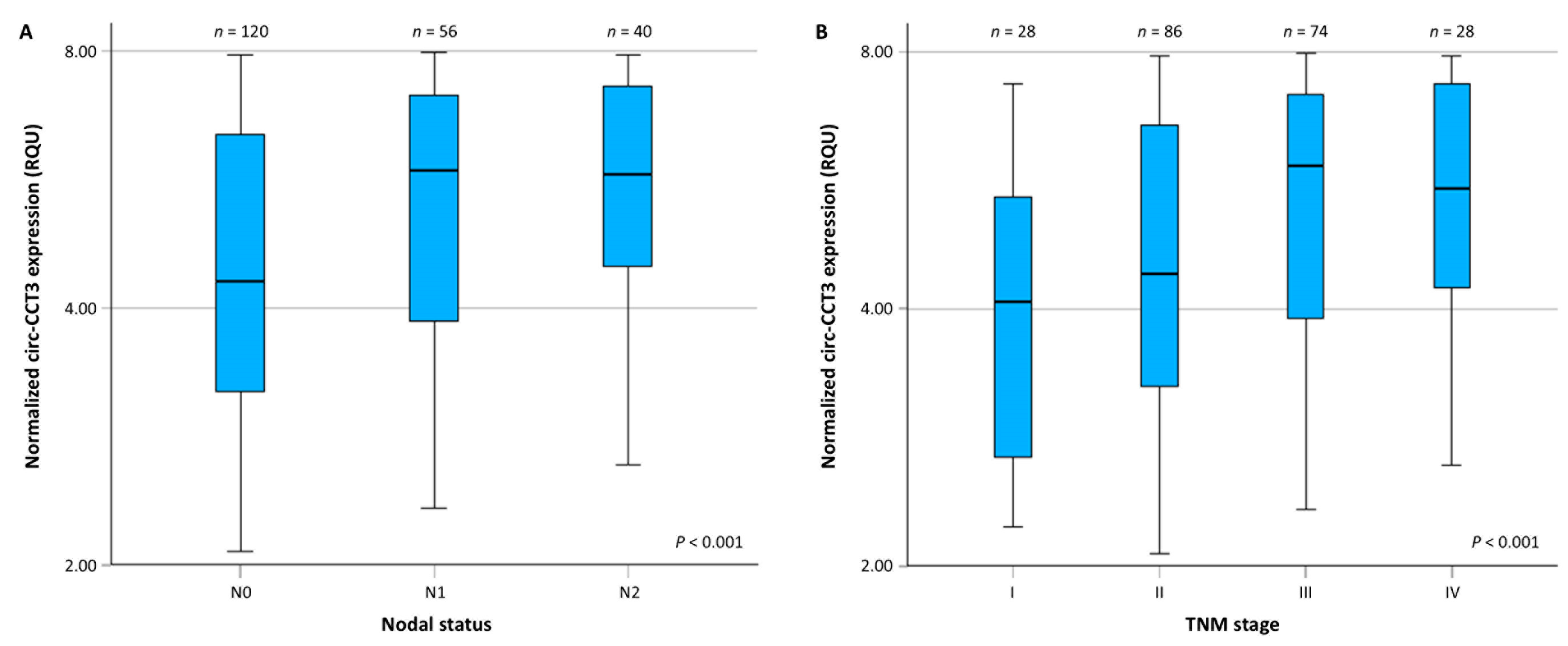
3.1. Circ-CCT3 Expression Is Overexpressed in Colorectal Adenocarcinoma, Compared to Adjacent Non-Cancerous Tissues
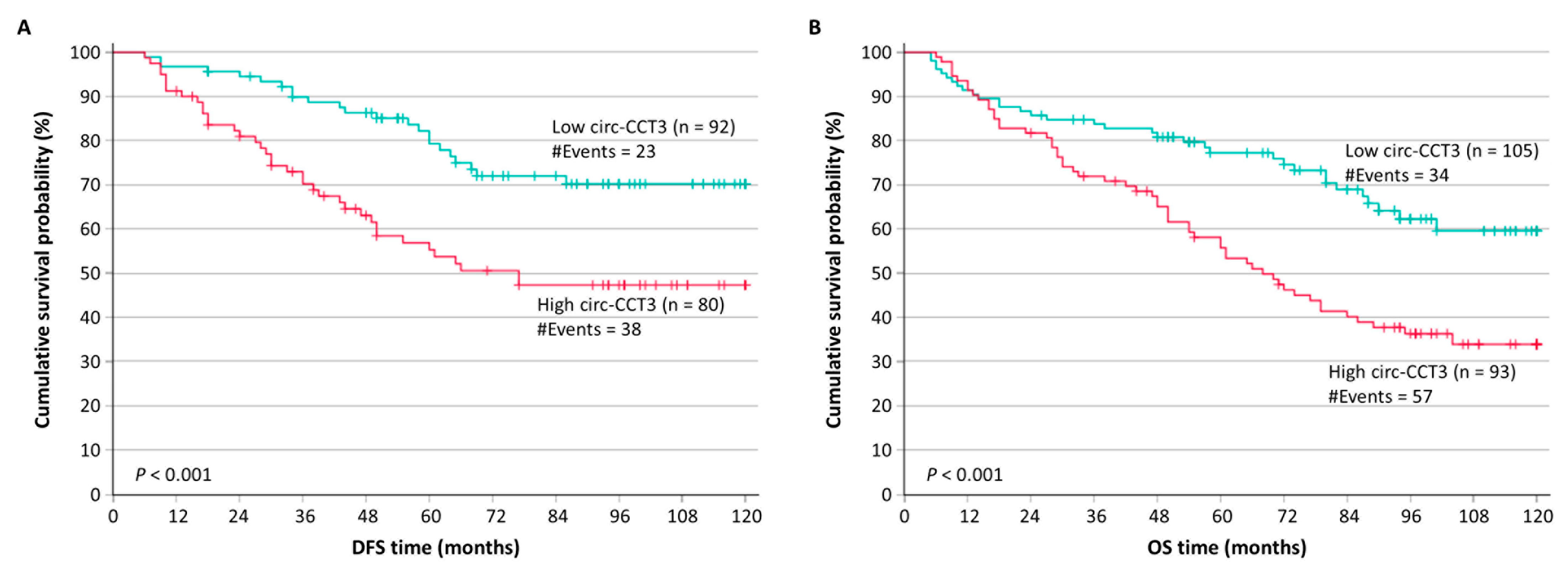
3.2. Circ-CCT3 Overexpression Predicts Poor DFS and OS in Colorectal Adenocarcinoma
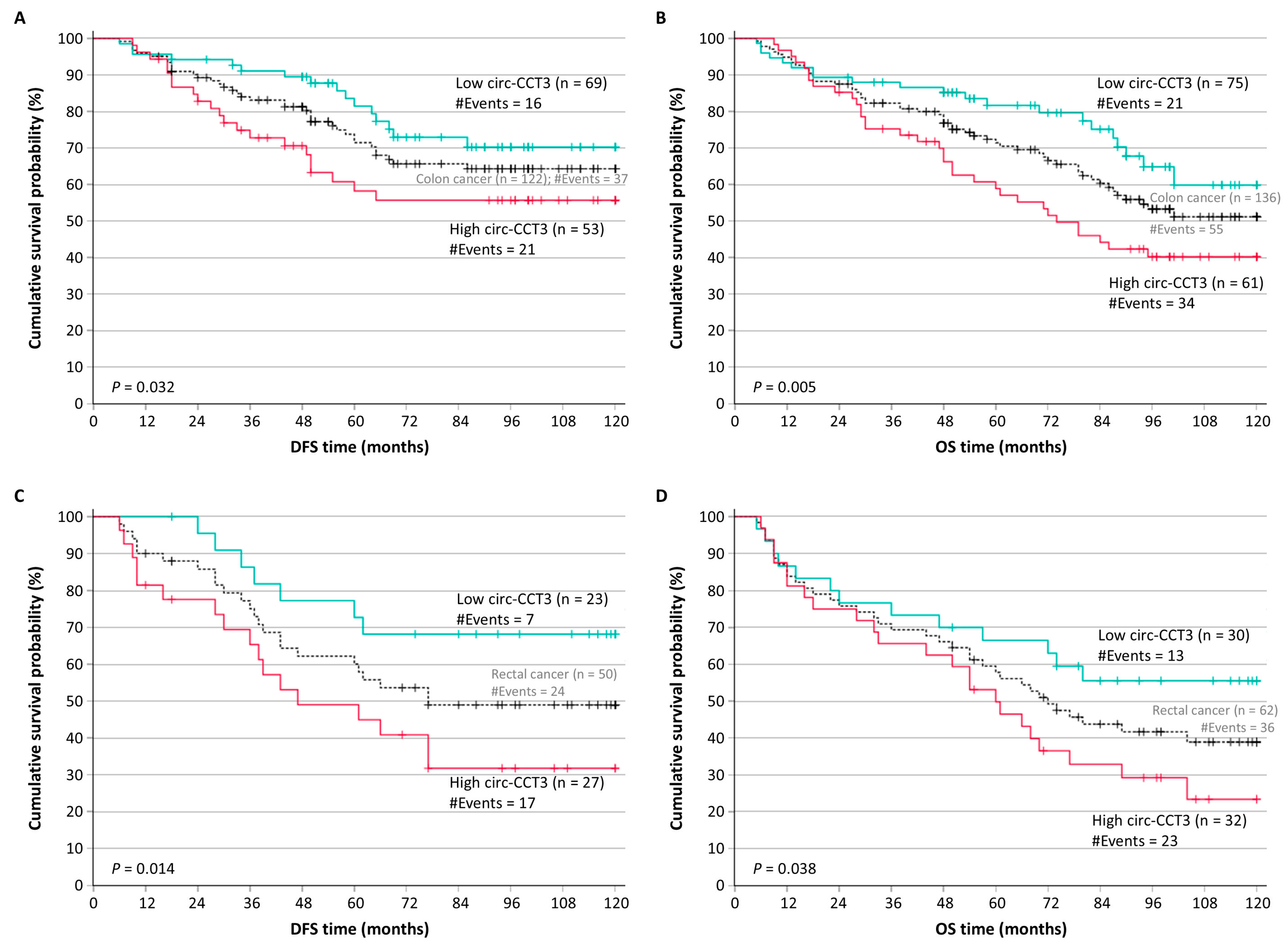
3.3. Circ-CCT3 Overexpression Predicts Short-Term Relapse in Colorectal Adenocarcinoma, Independently of Other Established Prognosticators
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, S.; Zheng, R.; Li, J.; Zeng, H.; Li, L.; Chen, R.; Sun, K.; Han, B.; Bray, F.; Wei, W.; et al. Global, regional, and national lifetime risks of developing and dying from gastrointestinal cancers in 185 countries: A population-based systematic analysis of GLOBOCAN. Lancet Gastroenterol. Hepatol. 2024, 9, 229–237. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef]
- Fadlallah, H.; El Masri, J.; Fakhereddine, H.; Youssef, J.; Chemaly, C.; Doughan, S.; Abou-Kheir, W. Colorectal cancer: Recent advances in management and treatment. World J. Clin. Oncol. 2024, 15, 1136–1156. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, A.S.; Agbaje, K.; Adesina, S.K.; Olajubutu, O. Colorectal Cancer: Disease Process, Current Treatment Options, and Future Perspectives. Pharmaceutics 2023, 15, 2620. [Google Scholar] [CrossRef]
- Krul, M.F.; Elferink, M.A.G.; Kok, N.F.M.; Dekker, E.; Lansdorp-Vogelaar, I.; Meijer, G.A.; Nagtegaal, I.D.; Breekveldt, E.C.H.; Ruers, T.J.M.; van Leerdam, M.E.; et al. Initial Impact of National CRC Screening on Incidence and Advanced Colorectal Cancer. Clin. Gastroenterol. Hepatol. 2023, 21, 797–807.e3. [Google Scholar] [CrossRef] [PubMed]
- Koncina, E.; Haan, S.; Rauh, S.; Letellier, E. Prognostic and Predictive Molecular Biomarkers for Colorectal Cancer: Updates and Challenges. Cancers 2020, 12, 319. [Google Scholar] [CrossRef]
- Sepulveda, A.R.; Hamilton, S.R.; Allegra, C.J.; Grody, W.; Cushman-Vokoun, A.M.; Funkhouser, W.K.; Kopetz, S.E.; Lieu, C.; Lindor, N.M.; Minsky, B.D.; et al. Molecular Biomarkers for the Evaluation of Colorectal Cancer: Guideline From the American Society for Clinical Pathology, College of American Pathologists, Association for Molecular Pathology, and the American Society of Clinical Oncology. J. Clin. Oncol. 2017, 35, 1453–1486. [Google Scholar] [CrossRef]
- Liu, C.; Jin, M.; Wang, S.; Han, W.; Zhao, Q.; Wang, Y.; Xu, C.; Diao, L.; Yin, Y.; Peng, C.; et al. Pathway and mechanism of tubulin folding mediated by TRiC/CCT along its ATPase cycle revealed using cryo-EM. Commun. Biol. 2023, 6, 531. [Google Scholar] [CrossRef]
- Ghozlan, H.; Cox, A.; Nierenberg, D.; King, S.; Khaled, A.R. The TRiCky Business of Protein Folding in Health and Disease. Front. Cell Dev. Biol. 2022, 10, 906530. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Song, P.; Liu, X.; Ma, C.; Zheng, M.; Ren, X.; Wang, R.; Liu, W.; Lu, Z.; Li, J. Insights into the roles and driving forces of CCT3 in human tumors. Front. Pharmacol. 2022, 13, 1005855. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, L.; Wang, J.; Bao, X.; Huang, J.; Qiu, Y.; Wang, T.; Yu, H. Repurposing cyclovirobuxine D as a novel inhibitor of colorectal cancer progression via modulating the CCT3/YAP axis. Br. J. Pharmacol. 2024, 181, 4348–4368. [Google Scholar] [CrossRef]
- Nibbe, R.K.; Markowitz, S.; Myeroff, L.; Ewing, R.; Chance, M.R. Discovery and scoring of protein interaction subnetworks discriminative of late stage human colon cancer. Mol. Cell Proteomics 2009, 8, 827–845. [Google Scholar] [CrossRef]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.C. CircRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef]
- Liu, W.; Lu, Y.; Yan, X.; Lu, Q.; Sun, Y.; Wan, X.; Li, Y.; Zhao, J.; Li, Y.; Jiang, G. Current understanding on the role of CCT3 in cancer research. Front. Oncol. 2022, 12, 961733. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y.; Li, H.; Peng, S.; Tan, H.; Huang, Z. Circular RNA circ-CCT3 promotes bortezomib resistance in multiple myeloma via modulating miR-223-3p/BRD4 axis. Anticancer. Drugs 2022, 33, e145–e154. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jiang, Y.; Lu, J.; Peng, C.; Ling, Z.; Chen, Y.; Chen, D.; Tong, R.; Zheng, S.; Wu, J. m(6)A-modification regulated circ-CCT3 acts as the sponge of miR-378a-3p to promote hepatocellular carcinoma progression. Epigenetics 2023, 18, 2204772. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, T.; Ding, G.; Hao, L.; Zhang, B.; Yu, J.; Pang, Y.; Geng, F.; Zhan, L.; Zhou, M.; et al. Circular RNA circ-CCT3 promotes hepatocellular carcinoma progression by regulating the miR-1287-5p/TEAD1/PTCH1/LOX axis. Mol. Med. Rep. 2021, 23, 375. [Google Scholar] [CrossRef]
- Li, J.; Lu, R.; Yang, K.; Sun, Q. circCCT3 Enhances Invasion and Epithelial-Mesenchymal Transition (EMT) of Non-Small-Cell Lung Cancer (NSCLC) via the miR-107/Wnt/FGF7 Axis. J. Oncol. 2022, 2022, 7020774. [Google Scholar] [CrossRef]
- Hou, J.P.; Men, X.B.; Yang, L.Y.; Han, E.K.; Han, C.Q.; Liu, L.B. CircCCT3 Acts as a Sponge of miR-613 to Promote Tumor Growth of Pancreatic Cancer Through Regulating VEGFA/VEGFR2 Signaling. Balkan Med. J. 2021, 38, 229–238. [Google Scholar] [CrossRef]
- Papatsirou, M.; Kontos, C.K.; Ntanasis-Stathopoulos, I.; Malandrakis, P.; Sideris, D.C.; Fotiou, D.; Liacos, C.I.; Gavriatopoulou, M.; Kastritis, E.; Dimopoulos, M.A.; et al. Exploring the molecular biomarker utility of circCCT3 in multiple myeloma: A favorable prognostic indicator, particularly for R-ISS II patients. Hemasphere 2024, 8, e34. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, Y.; Wang, X.; Cao, G.; Bu, W.; Wang, X.; Fang, Z.; Xu, Y.; Dong, M.; Tao, Q. circCCT3 Modulates Vascular Endothelial Growth Factor A and Wnt Signaling to Enhance Colorectal Cancer Metastasis Through Sponging miR-613. DNA Cell Biol. 2020, 39, 118–125. [Google Scholar] [CrossRef]
- Luo, L.; Xie, Q.; Wu, Y.; Li, P.; Qin, F.; Liao, D.; Wang, K. Circular RNA CCT3 is a unique molecular marker in bladder cancer. BMC Cancer 2023, 23, 977. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, H.; Liu, Y.; Wang, Y.; Hu, H.; Wang, G. Molecular subtyping in colorectal cancer: A bridge to personalized therapy (Review). Oncol. Lett. 2023, 25, 230. [Google Scholar] [CrossRef]
- Chakrabarti, S.; Peterson, C.Y.; Sriram, D.; Mahipal, A. Early stage colon cancer: Current treatment standards, evolving paradigms, and future directions. World J. Gastrointest. Oncol. 2020, 12, 808–832. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Shi, Q.; Smyrk, T.C.; Thibodeau, S.N.; Dienstmann, R.; Guinney, J.; Bot, B.M.; Tejpar, S.; Delorenzi, M.; Goldberg, R.M.; et al. Molecular markers identify subtypes of stage III colon cancer associated with patient outcomes. Gastroenterology 2015, 148, 88–99. [Google Scholar] [CrossRef]
- Weng, W.; Wei, Q.; Toden, S.; Yoshida, K.; Nagasaka, T.; Fujiwara, T.; Cai, S.; Qin, H.; Ma, Y.; Goel, A. Circular RNA ciRS-7-A Promising Prognostic Biomarker and a Potential Therapeutic Target in Colorectal Cancer. Clin. Cancer Res. 2017, 23, 3918–3928. [Google Scholar] [CrossRef]
- Lin, J.; Cai, D.; Li, W.; Yu, T.; Mao, H.; Jiang, S.; Xiao, B. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin. Biochem. 2019, 74, 60–68. [Google Scholar] [CrossRef]
- Kokoropoulos, P.; Christodoulou, S.; Tsiakanikas, P.; Vassiliu, P.; Kontos, C.K.; Arkadopoulos, N. Overexpression of Circular PRMT1 Transcripts in Colorectal Adenocarcinoma Predicts Recurrence and Poor Overall Survival. Int. J. Mol. Sci. 2025, 26, 6683. [Google Scholar] [CrossRef] [PubMed]
- de Assis, J.V.; Coutinho, L.A.; Oyeyemi, I.T.; Oyeyemi, O.T.; Grenfell, R. Diagnostic and therapeutic biomarkers in colorectal cancer: A review. Am. J. Cancer Res. 2022, 12, 661–680. [Google Scholar] [PubMed]
- Zamanian, M.Y.; Darmadi, D.; Darabi, R.; Fanoukh Aboqader Al-Aouadi, R.; Sadeghi Ivraghi, M.; Küpeli Akkol, E. Biomarkers for colorectal cancer detection: An insight into colorectal cancer and FDA-approved biomarkers. Bioimpacts 2025, 15, 31211. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.N.; Yu, J.H.; Hou, D.; Xie, Y.Q.; Lai, D.M.; Zheng, F.; Yang, B.; Zeng, J.T.; Chen, Y.; Lu, S.H.; et al. Extracellular Vesicle-Packaged circTAX1BP1 from Cancer-Associated Fibroblasts Regulates RNA m6A Modification through Lactylation of VIRMA in Colorectal Cancer Cells. Adv. Sci. 2025, e14008. [Google Scholar] [CrossRef]
- Xu, J.; Liu, J.; Huang, F.; Chao, Z.; Chen, J.; Xu, A.; Zhang, H.; Zhu, W. Research progress of circular RNA and N6-methyladenosine modification in colorectal cancer. Gene 2025, 967, 149684. [Google Scholar] [CrossRef]
- Kerimis, D.; Kontos, C.K.; Christodoulou, S.; Papadopoulos, I.N.; Scorilas, A. Elevated expression of miR-24-3p is a potentially adverse prognostic factor in colorectal adenocarcinoma. Clin. Biochem. 2017, 50, 285–292. [Google Scholar] [CrossRef]
- Kontos, C.K.; Chantzis, D.; Papadopoulos, I.N.; Scorilas, A. Kallikrein-related peptidase 4 (KLK4) mRNA predicts short-term relapse in colorectal adenocarcinoma patients. Cancer Lett. 2013, 330, 106–112. [Google Scholar] [CrossRef]
- Ryuk, J.P.; Choi, G.S.; Park, J.S.; Kim, H.J.; Park, S.Y.; Yoon, G.S.; Jun, S.H.; Kwon, Y.C. Predictive factors and the prognosis of recurrence of colorectal cancer within 2 years after curative resection. Ann. Surg. Treat. Res. 2014, 86, 143–151. [Google Scholar] [CrossRef]
- Abe, S.; Kawai, K.; Nozawa, H.; Sasaki, K.; Murono, K.; Emoto, S.; Ozawa, T.; Yokoyama, Y.; Nagai, Y.; Anzai, H.; et al. Clinical impact of primary tumor sidedness and sex on unresectable post-recurrence survival in resected pathological stage II–III colorectal cancers: A nationwide multicenter retrospective study. BMC Cancer 2022, 22, 486. [Google Scholar] [CrossRef]
- O’Connell, M.J.; Campbell, M.E.; Goldberg, R.M.; Grothey, A.; Seitz, J.F.; Benedetti, J.K.; Andre, T.; Haller, D.G.; Sargent, D.J. Survival following recurrence in stage II and III colon cancer: Findings from the ACCENT data set. J. Clin. Oncol. 2008, 26, 2336–2341. [Google Scholar] [CrossRef]
- Nikolic, N.; Radosavljevic, D.; Gavrilovic, D.; Nikolic, V.; Stanic, N.; Spasic, J.; Cacev, T.; Castellvi-Bel, S.; Cavic, M.; Jankovic, G. Prognostic Factors for Post-Recurrence Survival in Stage II and III Colorectal Carcinoma Patients. Medicina 2021, 57, 1108. [Google Scholar] [CrossRef]
- Pu, H.; Chen, Y.; Shen, R.; Zhang, Y.; Yang, D.; Liu, L.; Dong, X.; Yang, G. Influence of the initial recurrence site on prognosis after radical surgery for colorectal cancer: A retrospective cohort study. World J. Surg. Oncol. 2023, 21, 137. [Google Scholar] [CrossRef]
- Valenzuela, G.; Canepa, J.; Simonetti, C.; Solo de Zaldívar, L.; Marcelain, K.; González-Montero, J. Consensus molecular subtypes of colorectal cancer in clinical practice: A translational approach. World J. Clin. Oncol. 2021, 12, 1000–1008. [Google Scholar] [CrossRef] [PubMed]
- Lugli, A.; Kirsch, R.; Ajioka, Y.; Bosman, F.; Cathomas, G.; Dawson, H.; El Zimaity, H.; Fléjou, J.F.; Hansen, T.P.; Hartmann, A.; et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod. Pathol. 2017, 30, 1299–1311. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Yang, M.; Fang, H.; Zhang, B.; Ma, Y.; Li, Y.; Liu, Y.; Cheng, Z.; Zhao, Y.; Si, Z.; et al. Tailoring a Functional Synthetic Microbial Community Alleviates Fusobacterium nucleatum-infected Colorectal Cancer via Ecological Control. Adv. Sci. 2025, 12, e14232. [Google Scholar] [CrossRef] [PubMed]
- Kanda, S.; Watanabe, K.; Nakamura, S.; Narimatsu, H. Association between socioeconomic background and cancer: An ecological study using cancer registry and various community socioeconomic status indicators in Kanagawa, Japan. PLoS ONE 2025, 20, e0326895. [Google Scholar] [CrossRef]

| Assay | Target | Sequence (5′ → 3′) | Ta (°C) |
|---|---|---|---|
| Pre-amplification | circ-CCT3 | TCTGCTCGTCTTCCAACATC | 58 |
| GCTCAGGATTATCTGGAAGACC | |||
| GAPDH mRNA | CCACATCGCTCAGACACCAT | 60 | |
| TGACAAGCTTCCCGTTCTCA | |||
| Real-time qPCR | circ-CCT3 | TACCCAGTCTTCCATCAACTGG | 60 |
| ACACAGGTGCCATCGGAAAC | |||
| GAPDH mRNA | ATGGGGAAGGTGAAGGTCG | 60 | |
| GGGTCATTGATGGCAACAATATC |
| Variable | Number of Patients (%) |
|---|---|
| Gender | |
| Male | 113 (52.3%) |
| Female | 103 (47.7%) |
| Histological grade | |
| I | 19 (8.8%) |
| II | 166 (76.8%) |
| III | 31 (14.4%) |
| T (tumor invasion) | |
| T1 | 6 (2.8%) |
| T2 | 25 (11.6%) |
| T3 | 134 (62.0%) |
| T4 | 51 (23.6%) |
| N (nodal status) | |
| N0 | 120 (55.6%) |
| N1 | 56 (25.9%) |
| N2 | 40 (18.5%) |
| M (distant metastasis) | |
| M0 | 188 (87.0%) |
| M1 | 28 (13.0%) |
| TNM stage | |
| I | 28 (13.0%) |
| II | 86 (39.7%) |
| III | 74 (34.3%) |
| IV | 28 (13.0%) |
| Radiotherapy | |
| No | 169 (83.7%) |
| Yes | 33 (16.3%) |
| Chemotherapy | |
| No | 85 (42.1%) |
| Yes | 117 (57.9%) |
| Variable | Mean ± SEM | Range | Quartiles | ||
|---|---|---|---|---|---|
| 1st | 2nd (Median) | 3rd | |||
| circ-CCT3 expression (RQU) | |||||
| In cancerous tissues (n = 216) | 5.13 ± 0.123 | 2.07–7.97 | 3.46 | 5.12 | 6.88 |
| In normal tissues (n = 86) | 2.25 ± 0.099 | 0.814–3.76 | 1.54 | 2.17 | 3.07 |
| Covariate | HR | 95% CI | p Value 1 | BCa 95% CI | Bootstrap p Value 1 | |
|---|---|---|---|---|---|---|
| Univariate analysis | circ-CCT3 expression (continuous variable) | 1.25 | 1.08–1.43 | 0.002 | 1.09–1.44 | 0.003 |
| circ-CCT3 expression status | ||||||
| Low | 1.00 | |||||
| High | 2.35 | 1.40–3.95 | 0.001 | 1.42–4.13 | 0.001 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.64 | 0.98–2.74 | 0.059 | 0.98–2.71 | 0.052 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 1.73 | 0.62–4.81 | 0.29 | 0.74–9.10 | 0.24 | |
| III | 3.06 | 0.97–9.61 | 0.056 | 1.04–19.73 | 0.035 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 4.37 | 1.03–18.49 | 0.045 | 1.47–2.65 × 104 | 0.029 | |
| III | 9.95 | 2.39–41.47 | 0.002 | 3.30–6.85 × 104 | 0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.78 | 0.99–3.19 | 0.052 | 0.97–3.10 | 0.044 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 2.32 | 1.32–4.06 | 0.003 | 1.31–4.31 | 0.004 | |
| Multivariate analysis | circ-CCT3 expression status | |||||
| Low | 1.00 | |||||
| High | 1.75 | 1.03–2.98 | 0.039 | 1.04–3.27 | 0.044 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 2.47 | 1.22–5.00 | 0.012 | 1.12–5.91 | 0.012 | |
| TNM stage | 0.005 | |||||
| I | 1.00 | |||||
| II | 4.69 | 1.04–21.19 | 0.045 | 1.45–3.03 × 104 | 0.026 | |
| III | 9.32 | 1.94–44.67 | 0.005 | 2.60–7.67 × 104 | 0.002 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 0.55 | 0.25–1.24 | 0.15 | 0.20–1.28 | 0.19 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.30 | 0.69–2.43 | 0.42 | 0.63–2.58 | 0.44 |
| Covariate | HR | 95% CI | p Value 1 | BCa 95% CI | Bootstrap p Value 1 | |
|---|---|---|---|---|---|---|
| Univariate analysis | circ-CCT3 expression (continuous variable) | 1.27 | 1.13–1.43 | <0.001 | 1.14–1.44 | 0.001 |
| circ-CCT3 expression status | ||||||
| Low | 1.00 | |||||
| High | 2.12 | 1.39–3.25 | <0.001 | 1.39–3.30 | 0.002 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.53 | 1.01–2.33 | 0.047 | 0.98–2.39 | 0.056 | |
| Histological grade | ||||||
| I | 1.00 | |||||
| II | 1.32 | 0.61–2.88 | 0.48 | 0.61–3.61 | 0.51 | |
| III | 2.72 | 1.14–6.48 | 0.024 | 1.17–7.56 | 0.023 | |
| TNM stage | ||||||
| I | 1.00 | |||||
| II | 2.60 | 0.91–7.48 | 0.075 | 1.03–13.44 | 0.062 | |
| III | 5.76 | 2.03–16.29 | 0.001 | 2.30–28.71 | 0.005 | |
| IV | 34.85 | 11.71–103.72 | <0.001 | 13.78–203.79 | 0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.67 | 1.02–2.75 | 0.042 | 0.97–2.84 | 0.057 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.78 | 1.15–2.77 | 0.010 | 1.14–2.87 | 0.013 | |
| Multivariate analysis | circ-CCT3 expression status | |||||
| Low | 1.00 | |||||
| High | 1.26 | 0.80–1.98 | 0.33 | 0.80–2.12 | 0.35 | |
| Tumor location | ||||||
| Colon | 1.00 | |||||
| Rectum | 1.38 | 0.80–2.38 | 0.25 | 0.72–2.74 | 0.30 | |
| TNM stage | <0.001 | |||||
| I | 1.00 | |||||
| II | 3.04 | 1.02–9.04 | 0.046 | 1.08–19.36 | 0.040 | |
| III | 7.02 | 2.27–21.74 | <0.001 | 2.39–47.43 | 0.003 | |
| IV | 40.12 | 12.15–132.49 | <0.001 | 12.67–307.01 | 0.001 | |
| Radiotherapy | ||||||
| No | 1.00 | |||||
| Yes | 1.16 | 0.60–2.24 | 0.65 | 0.49–2.52 | 0.68 | |
| Chemotherapy | ||||||
| No | 1.00 | |||||
| Yes | 0.71 | 0.42–1.18 | 0.19 | 0.41–1.27 | 0.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokoropoulos, P.; Christodoulou, S.; Tsiakanikas, P.; Poulios, E.; Vassiliu, P.; Kontos, C.K.; Arkadopoulos, N. A Retrospective Study in Colorectal Adenocarcinoma Uncovers the Potential of Circ-CCT3 as a Predictor of Tumor Recurrence. Biomedicines 2025, 13, 2432. https://doi.org/10.3390/biomedicines13102432
Kokoropoulos P, Christodoulou S, Tsiakanikas P, Poulios E, Vassiliu P, Kontos CK, Arkadopoulos N. A Retrospective Study in Colorectal Adenocarcinoma Uncovers the Potential of Circ-CCT3 as a Predictor of Tumor Recurrence. Biomedicines. 2025; 13(10):2432. https://doi.org/10.3390/biomedicines13102432
Chicago/Turabian StyleKokoropoulos, Panagiotis, Spyridon Christodoulou, Panagiotis Tsiakanikas, Efthimios Poulios, Panteleimon Vassiliu, Christos K. Kontos, and Nikolaos Arkadopoulos. 2025. "A Retrospective Study in Colorectal Adenocarcinoma Uncovers the Potential of Circ-CCT3 as a Predictor of Tumor Recurrence" Biomedicines 13, no. 10: 2432. https://doi.org/10.3390/biomedicines13102432
APA StyleKokoropoulos, P., Christodoulou, S., Tsiakanikas, P., Poulios, E., Vassiliu, P., Kontos, C. K., & Arkadopoulos, N. (2025). A Retrospective Study in Colorectal Adenocarcinoma Uncovers the Potential of Circ-CCT3 as a Predictor of Tumor Recurrence. Biomedicines, 13(10), 2432. https://doi.org/10.3390/biomedicines13102432






