Exploring the Impact of Skin Care Routines on the Skin Microbiome and Possible Skin Disease Risk—A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. Extraction of DNA
2.3. Next-Generation Sequencing
2.4. Analysis of the Microbiome
2.5. Statistical Analysis
3. Results
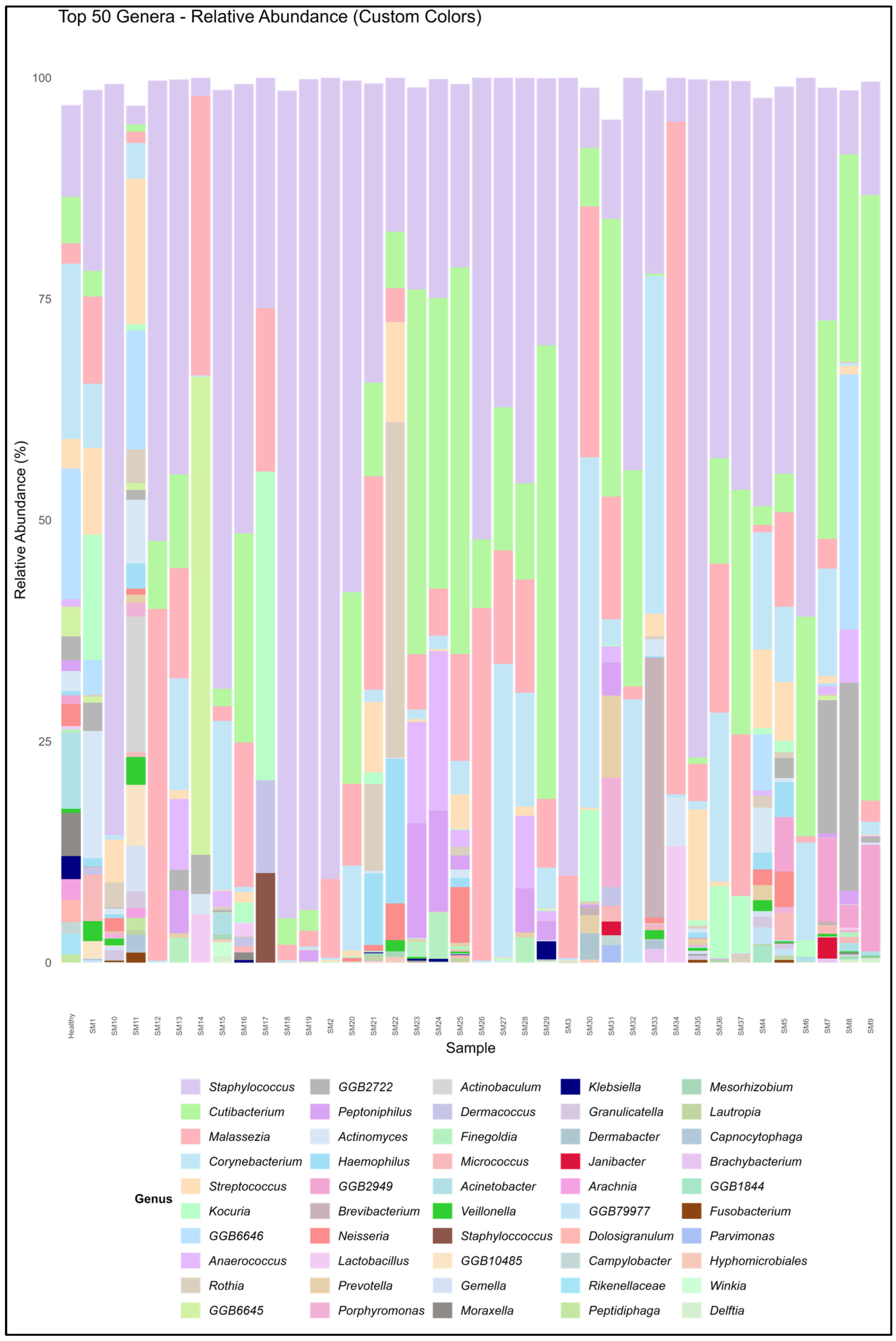
3.1. Microbiome Composition and Alpha Diversity
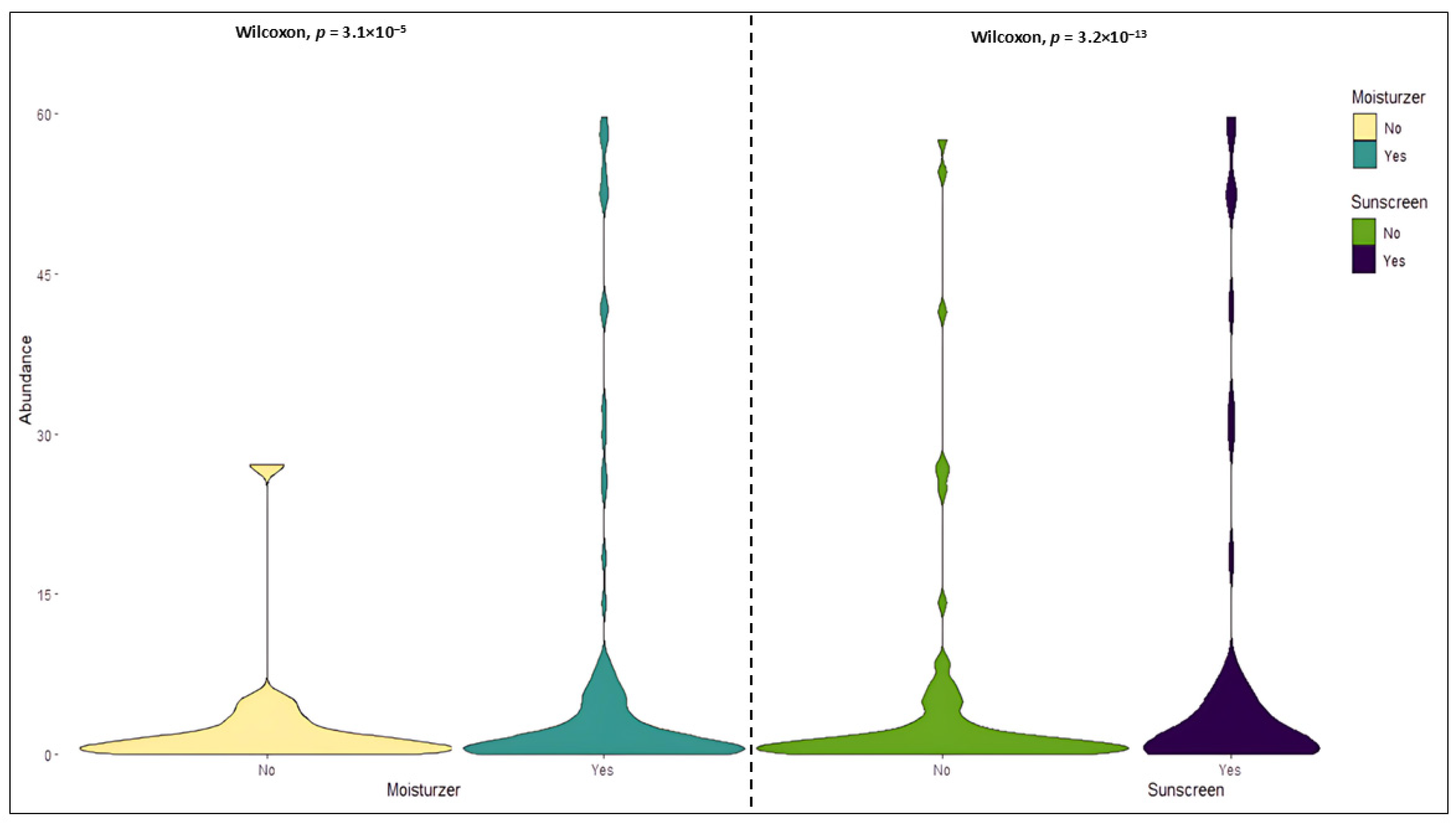
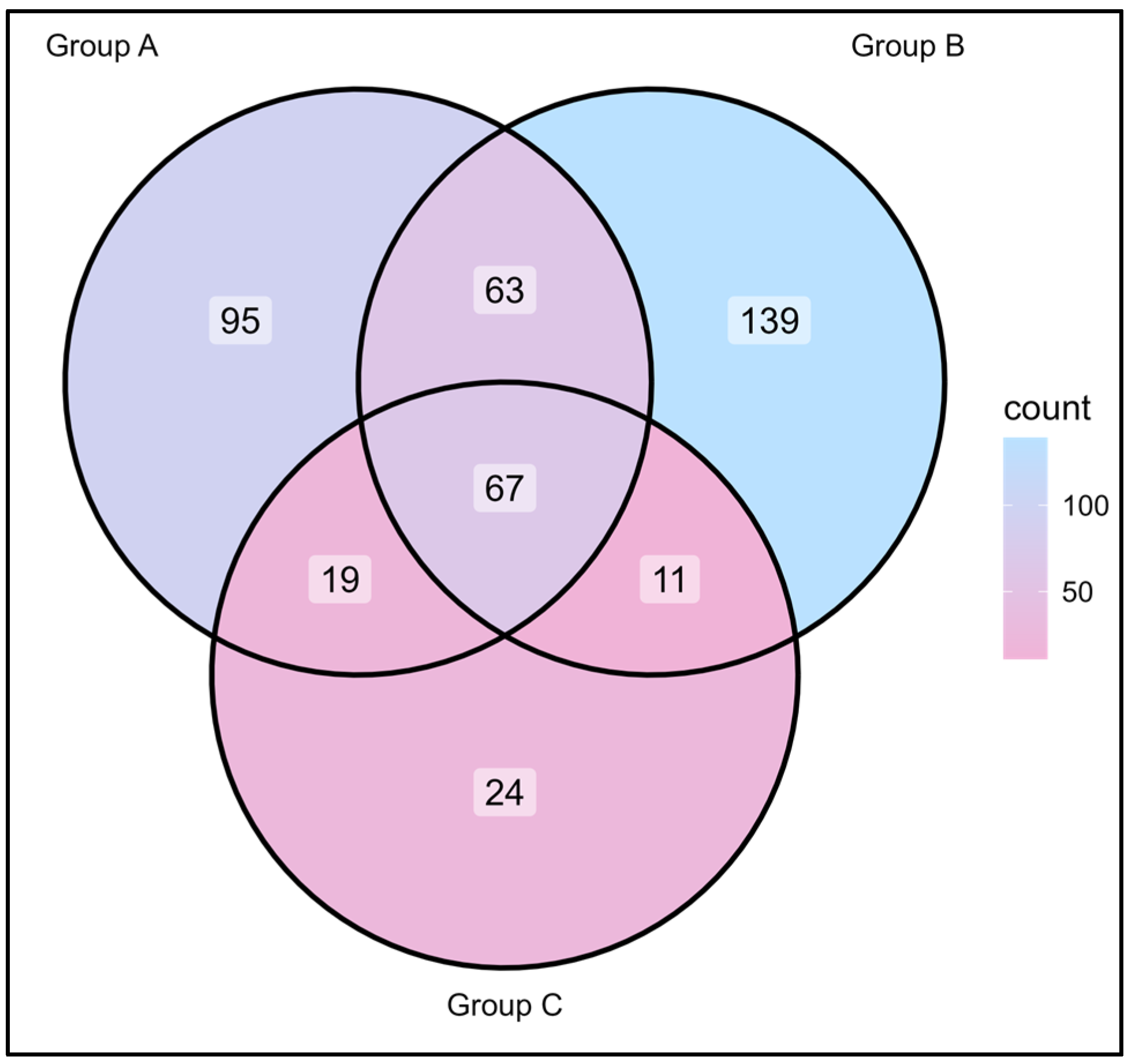
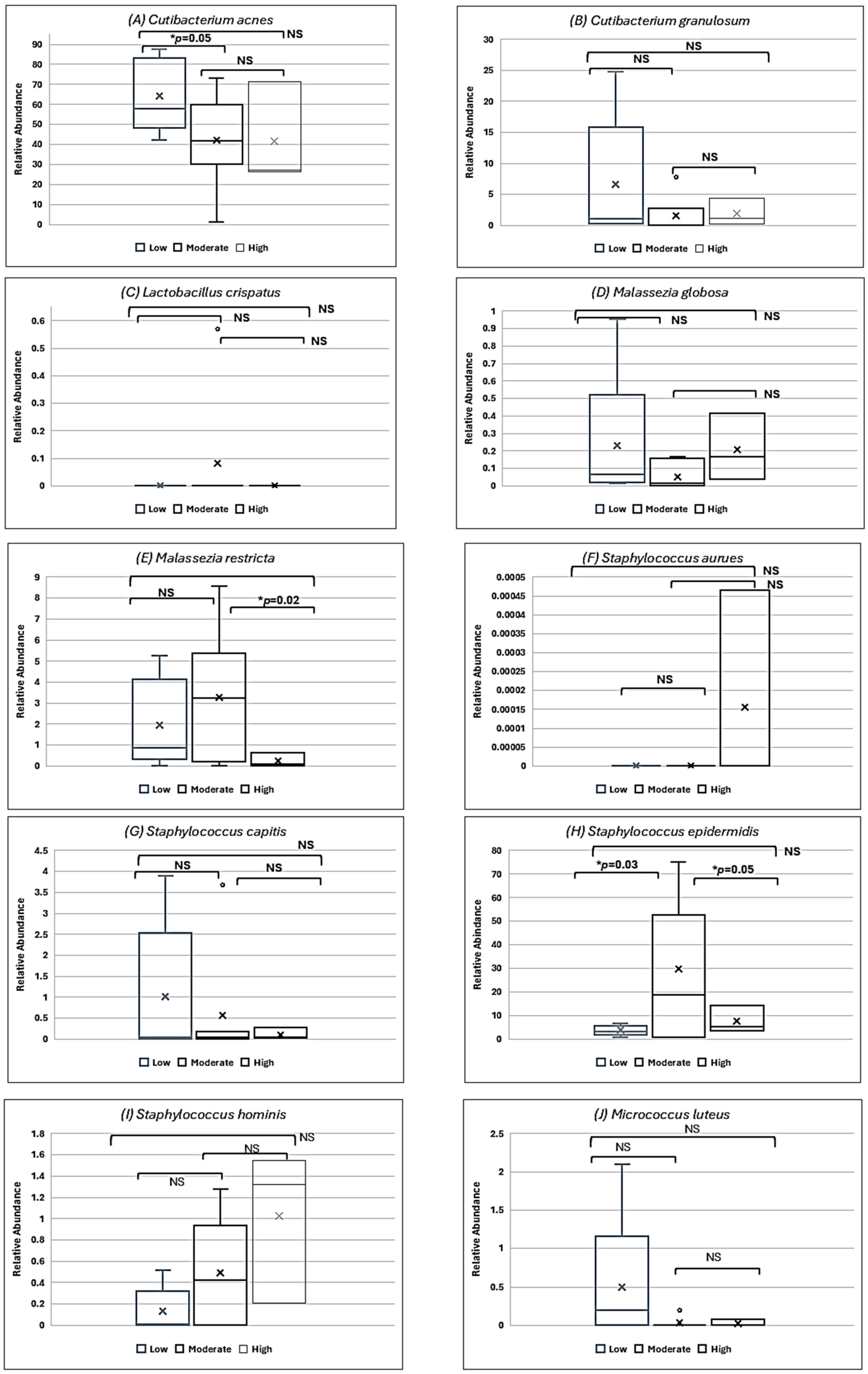
3.2. Relationship Between Skin Care Routine and Skin Microbiome
3.3. Skin Microbiome and Sun Exposure
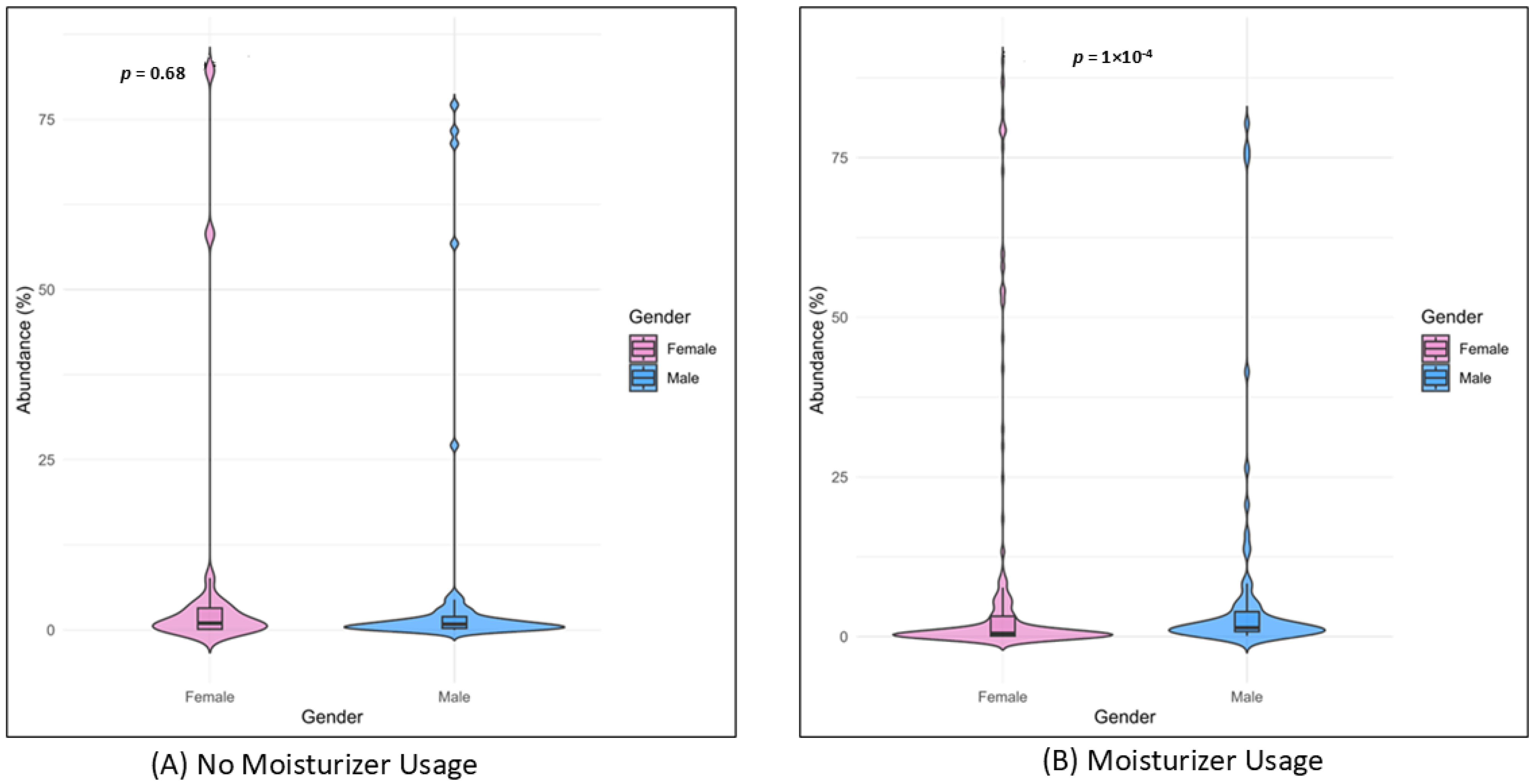
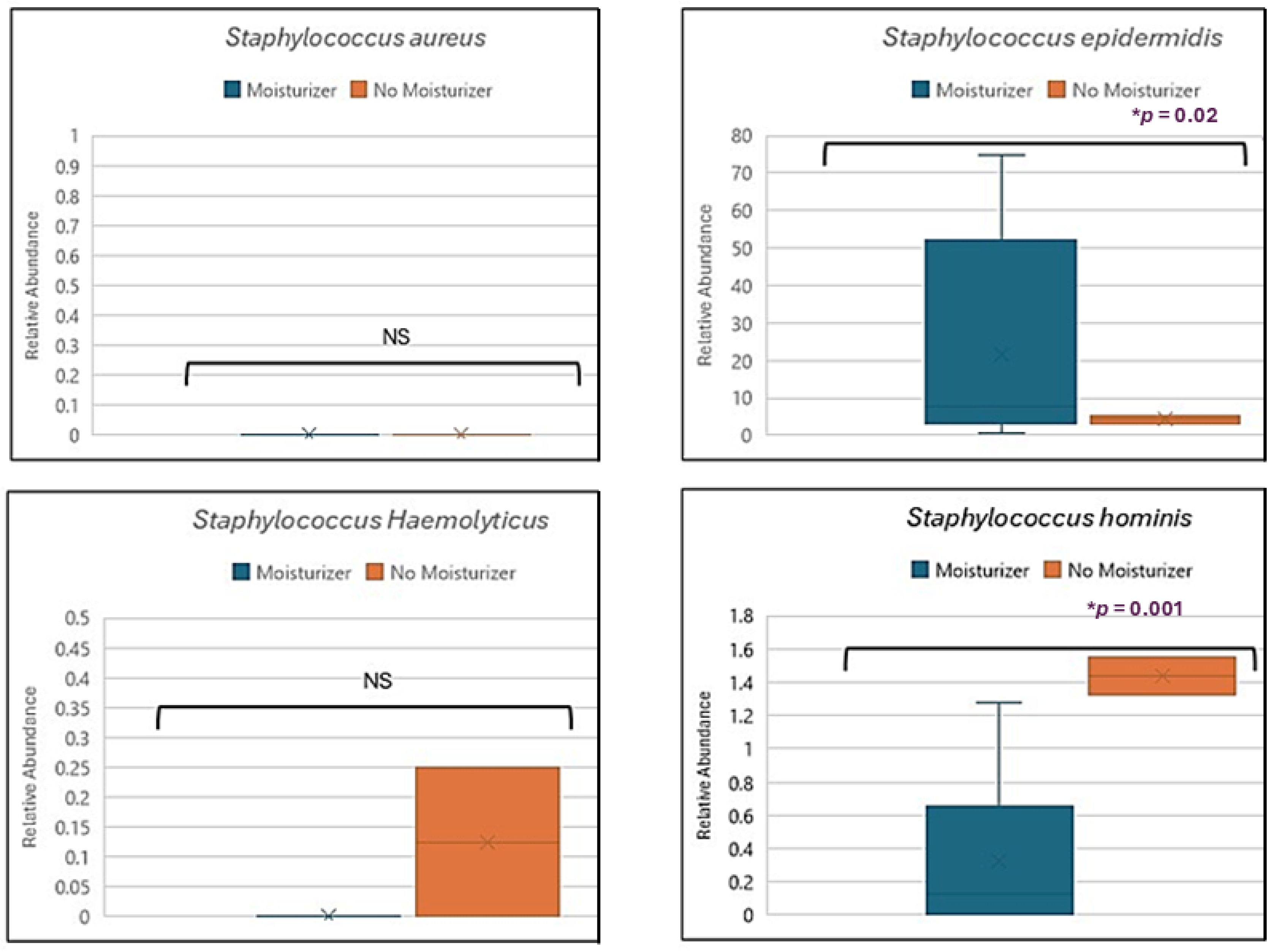
3.4. Skin Microbiome, Moisturizer and Atopic Dermatitis
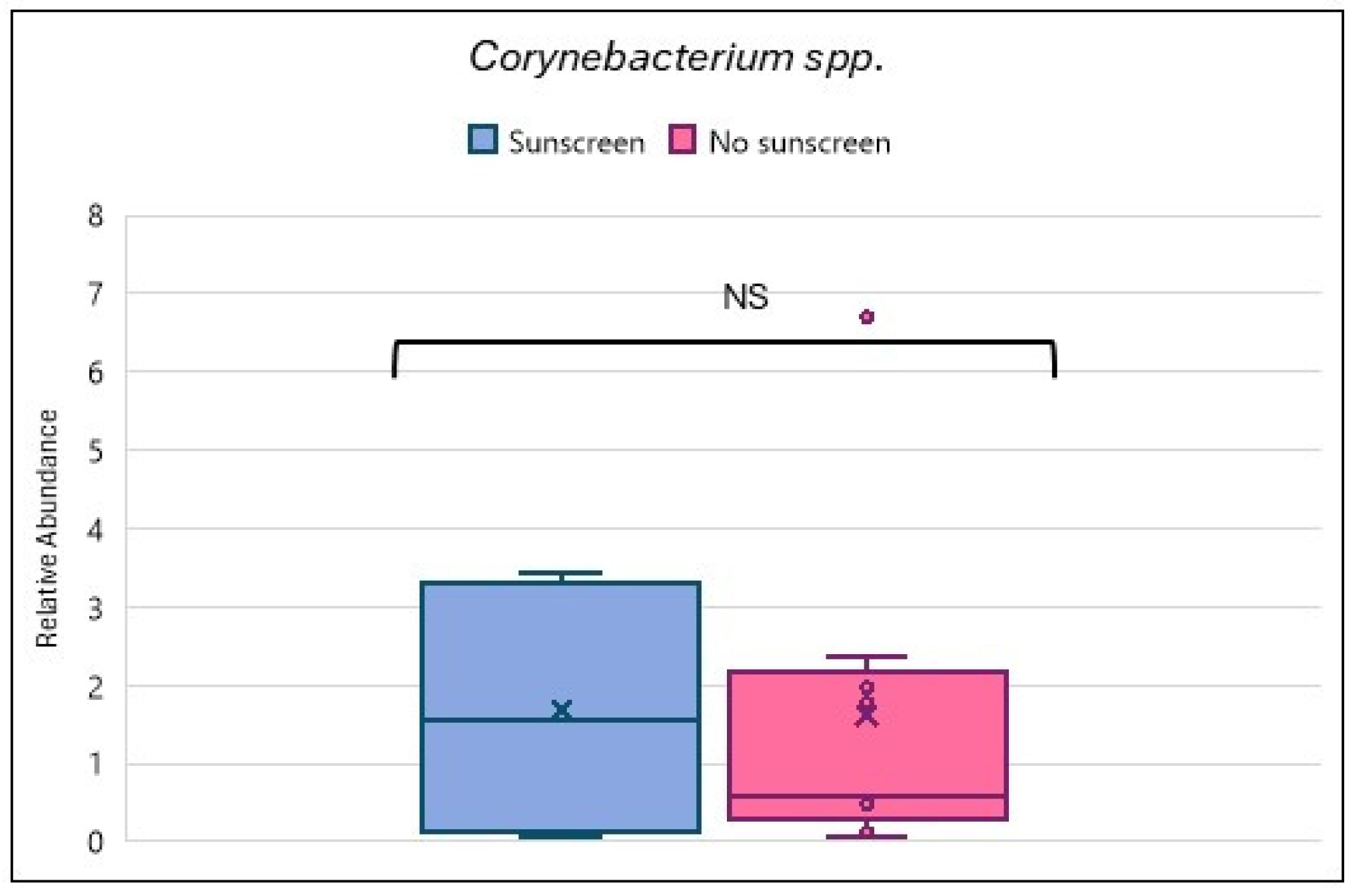
3.5. Skin Microbiome, Sunscreen, and Hyperpigmentation
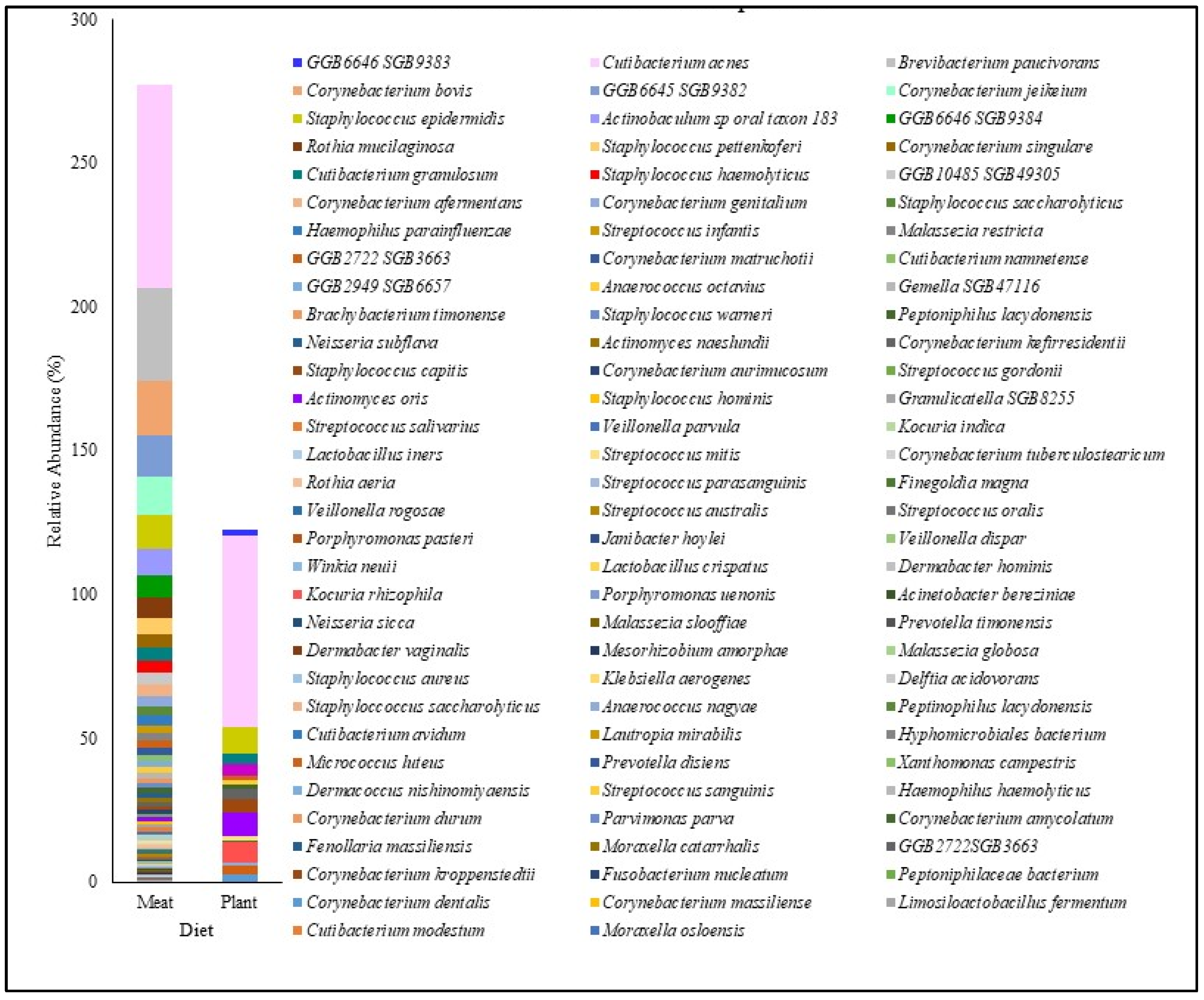
3.6. Skin Microbiome and Plant-Based Diet
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Murphy, B.; Grimshaw, S.; Hoptroff, M.; Paterson, S.; Arnold, D.; Cawley, A.; Adams, S.E.; Falciani, F.; Dadd, T.; Eccles, R.; et al. Alteration of barrier properties, stratum corneum ceramides and microbiome composition in response to lotion application on cosmetic dry skin. Sci. Rep. Nat. Res. 2022, 12, 5223. [Google Scholar] [CrossRef] [PubMed]
- Nagase, S.; Ogai, K.; Urai, T.; Shibata, K.; Matsubara, E.; Mukai, K.; Matsue, M.; Mori, Y.; Aoki, M.; Arisandi, D.; et al. Distinct Skin Microbiome and Skin Physiological Functions Between Bedridden Older Patients and Healthy People: A Single-Center Study in Japan. Front. Media 2020, 7, 101. [Google Scholar] [CrossRef]
- Dréno, B.; Araviiskaia, E.; Berardesca, E.; Gontijo, G.; Sanchez Viera, M.; Xiang, L.F.; Martin, R.; Bieber, T. Microbiome in healthy skin, update for dermatologists. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 2038–2047. [Google Scholar] [CrossRef] [PubMed]
- Sanford, J.A.; Gallo, R.L. Functions of the skin microbiota in health and disease. Semin. Immunol. 2013, 25, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.F.; Oddos, T.; Stamatas, G. Deciphering the Role of Skin Surface Microbiome in Skin Health: An Integrative Multiomics Approach Reveals Three Distinct Metabolite–Microbe Clusters. J. Investig. Dermatol. 2022, 142, 469–479.e5. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.K.; Lee, S.; Myoung, J.; Hwang, S.J.; Lim, J.M.; Jeong, E.T.; Park, S.G.; Youn, S.H. Effect of the skincare product on facial skin microbial structure and biophysical parameters: A pilot study. Microbiologyopen 2021, 10, e1236. [Google Scholar] [CrossRef] [PubMed]
- Wallen-Russell, C. The Role of Every-Day Cosmetics in Altering the Skin Microbiome: A Study Using Biodiversity. Cosmetics 2019, 6, 2. [Google Scholar] [CrossRef]
- Garcella, P.; Wijaya, T.H.; Kurniawan, D.W. Narrative Review: Herbal Nanocosmetics for Anti Aging. JPSCR J. Pharm. Sci. Clin. Res. 2023, 8, 63. [Google Scholar] [CrossRef]
- Hekmatpou, D.; Mehrabi, F.; Rahzani, K.; Aminiyan, A. The Effect of Aloe Vera Clinical Trials on Prevention and Healing of Skin Wound: A Systematic Review. Iran J Med Sci. 2019, 44, 1–9. [Google Scholar] [PubMed] [PubMed Central]
- Lodén, M. The clinical benefit of moisturizers. J. Eur. Acad. Dermatol. Venereol. 2005, 19, 672–688. [Google Scholar] [CrossRef] [PubMed]
- Purnamawati, S.; Indrastuti, N.; Danarti, R.; Saefudin, T. The Role of Moisturizers in Addressing Various Kinds of Der-matitis: A Review. Clin. Med. Res. 2017, 15, 75–87. [Google Scholar] [CrossRef]
- Lindh, J.D.; Bradley, M. Clinical Effectiveness of Moisturizers in Atopic Dermatitis and Related Disorders: A Systematic Review. Am. J. Clin. Dermatol. 2015, 16, 341–359. [Google Scholar] [CrossRef] [PubMed]
- Harel, N.; Reshef, L.; Biran, D.; Brenner, S.; Ron, E.Z.; Gophna, U. Effect of Solar Radiation on Skin Microbiome: Study of Two Populations. Microorganisms 2022, 10, 1523. [Google Scholar] [CrossRef]
- Mansuri, R.; Diwan, A.; Kumar, H.; Dangwal, K.; Yadav, D. Potential of Natural Compounds as Sunscreen Agents. Pharmacogn. Rev. EManuscript Technol. 2021, 15, 47–56. [Google Scholar] [CrossRef]
- Chavda, V.P.; Acharya, D.; Hala, V.; Daware, S.; Vora, L.K. Sunscreens: A comprehensive review with the application of nanotechnology. J. Drug Deliv. Sci. Technol. 2023, 86, 104720. [Google Scholar] [CrossRef]
- Muthumani, T.; Sudhahar, V.; Mukhopadhyay, T. Review on Sunscreens and Sun Protection factor. Res. J. Top. Cosmet. Sci. 2015, 6, 55–65. [Google Scholar] [CrossRef]
- Fatima, S.; Braunberger, T.; Mohammad, T.F.; Kohli, I.; Hamzavi, I.H. The Role of Sunscreen in Melasma and Postinflam-matory Hyperpigmentation. Indian J. Dermatol. 2020, 65, 5–10. [Google Scholar] [PubMed]
- Skowron, K.; Bauza-kaszewska, J.; Kraszewska, Z.; Wiktorczyk-kapischke, N.; Grudlewska-buda, K.; Kwiecińska-piróg, J.; Wałecka-zacharska, E.; Radtke, L.; Gospodarek-komkowska, E. Human skin microbiome: Impact of intrinsic and extrinsic factors on skin microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef]
- De Pessemier, B.; Grine, L.; Debaere, M.; Maes, A.; Paetzold, B.; Callewaert, C. Gut–Skin Axis: Current Knowledge of the Interrelationship between Microbial Dysbiosis and Skin Conditions. Microorganisms 2021, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Flores-Balderas, X.; Peña-Peña, M.; Rada, K.M.; Alvarez-Alvarez, Y.Q.; Guzmán-Martín, C.A.; Sánchez-Gloria, J.L.; Huang, F.; Ruiz-Ojeda, D.; Morán-Ramos, S.; Springall, R.; et al. Beneficial Effects of Plant-Based Diets on Skin Health and Inflammatory Skin Diseases. Nutrients 2023, 13, 2842. [Google Scholar] [CrossRef] [PubMed]
- Ying, S.; Zeng, D.N.; Chi, L.; Tan, Y.; Galzote, C.; Cardona, C.; Lax, S.; Gilbert, J.; Quan, Z.X. The influence of age and gender on skin-associated microbial communities in urban and rural human populations. PLoS ONE 2015, 10, e0141842. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. Babraham Bioinformatics. Babraham Institute. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 27 May 2025).
- Huttenhower Lab. KneadData: Quality control tool for metagenomic data. Harvard T.H. Chan School of Public Health. 2023. Available online: https://huttenhower.sph.harvard.edu/kneaddata/ (accessed on 27 May 2025).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with MetaPhlAn 4. Nat. Biotechnol. 2021, 39, 1174–1181. [Google Scholar]
- Blanco-Míguez, A.; Beghini, F.; Cumbo, F.; McIver, L.J.; Thompson, K.N.; Zolfo, M.; Manghi, P.; Dubois, L.; Huang, K.D.; Thomas, A.M. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat. Biotechnol. 2023, 41, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Luna, P.C. Skin Microbiome as Years Go By. Am. J. Clin. Dermatol. 2020, 21 (Suppl. S1), 12–17. [Google Scholar] [CrossRef] [PubMed]
- Isler, M.F.; Coates, S.J.; Boos, M.D. Climate change, the cutaneous microbiome and skin disease: Implications for a warming world. Int. J. Dermatol. 2023, 62, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Khmaladze, I.; Leonardi, M.; Fabre, S.; Messaraa, C.; Mavon, A. The skin interactome: A holistic “ge-nome-microbiome-exposome” approach to understand and modulate skin health and aging. Clin. Cosmet. Investig. Dermatol. 2020, 13, 1021–1040. [Google Scholar] [CrossRef]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; Green, E.D.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Budding, A.E.; van der Lugt-Degen, M.; Du-Thumm, L.; Vandeven, M.; Fan, A. The influence of age, gender and race/ethnicity on the composition of the human axillary microbiome. Int. J. Cosmet. Sci. 2019, 41, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.L.; O’Neill, C.A.; Dickinson, M.R.; Chavan, B.; McBain, A.J. Exploring associations between skin, the dermal mi-crobiome, and ultraviolet radiation: Advancing possibilities for next-generation sunscreens. Front. Microbiomes 2023, 2, 1102315. [Google Scholar] [CrossRef]
- Grant, G.J.; Kohli, I.; Mohammad, T.F. A narrative review of the impact of ultraviolet radiation and sunscreen on the skin microbiome. Photodermatol. Photoimmunol. Photomed. 2024, 40, e12943. [Google Scholar] [CrossRef] [PubMed]
- Willmott, T.; Campbell, P.M.; Griffiths, C.E.M.; O’Connor, C.; Bell, M.; Watson, R.E.B.; McBain, A.J.; Langton, A.K. Behaviour and sun exposure in holidaymakers alters skin microbiota composition and diversity. Front. Aging 2023, 4, 1217635. [Google Scholar] [CrossRef] [PubMed]
- Patra, V.K.; Byrne, S.N.; Wolf, P. The skin microbiome: Is it affected by UV-induced immune suppression? Front. Microbiol. 2016, 7, 1235. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, R.; Claypool, J.; Sfriso, R.; Volhardt, J. Sunscreens can preserve human skin microbiome upon erythemal UV exposure. Int. J. Cosmet. Sci. 2023, 46, 71–84. [Google Scholar] [CrossRef]
- Keshari, S.; Balasubramaniam, A.; Myagmardoloonjin, B.; Herr, D.R.; Negari, I.P.; Huang, C.M. Butyric acid from probiotic staphylococcus epidermidis in the skin microbiome down-regulates the ultraviolet-induced pro-inflammatory IL-6 cytokine via short-chain fatty acid receptor. Int. J. Mol. Sci. 2019, 20, 4477. [Google Scholar] [CrossRef] [PubMed]
- Schalka, S. New data on hyperpigmentation disorders. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Zanchetta, C.; Vilanova, D.; Jarrin, C.; Scandolera, A.; Chapuis, E.; Auriol, D.; Robe, P.; Dupont, J.; Lapierre, L.; Reynaud, R. Bacterial taxa predictive of hyperpigmented skins. Health Sci. Rep. 2022, 5, e609. [Google Scholar] [CrossRef]
- Dimitriu, P.A.; Iker, B.; Malik, K.; Leung, H.; Mohn, W.W.; Hillebrand, G.G. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. MBio 2019, 10, 10–128. [Google Scholar] [CrossRef]
- Edslev, S.M.; Agner, T.; Andersen, P.S. Skin microbiome in atopic dermatitis. Acta Derm Venereol. Med. J. 2020, 100, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Fölster-Holst, R. The role of the skin microbiome in atopic dermatitis—Correlations and consequences. J. Ger. Soc. Dermatol. 2022, 20, 571–577. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, R.D.; Bandier, J.; Skov, L.; Engstrand, L.; Johansen, J.D. The role of the skin microbiome in atopic dermatitis: A sys-tematic review. Br. J. Dermatol. 2017, 177, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Khadka, V.D.; Key, F.M.; Romo-González, C.; Martínez-Gayosso, A.; Campos-Cabrera, B.L.; Gerónimo-Gallegos, A.; Lynn, T.C.; Durán-McKinster, C.; Coria-Jiménez, R.; Lieberman, T.D.; et al. The Skin Microbiome of Patients with Atopic Dermatitis Normalizes Gradually During Treatment. Front. Cell Infect. Microbiol. 2021, 11, 720674. [Google Scholar] [CrossRef] [PubMed]
- Carmona-Cruz, S.; Orozco-Covarrubias, L.; Sáez-de-Ocariz, M. The Human Skin Microbiome in Selected Cutaneous Diseases. Front. Cell. Infect. Microbiol. 2022, 12, 834135. [Google Scholar] [CrossRef] [PubMed]
- Nicol, N.H.; Rippke, F.; Weber, T.M.; Hebert, A.A. Daily Moisturization for Atopic Dermatitis: Importance, Recommen-dations, and Moisturizer Choices. J. Nurse Pract. 2021, 17, 920–925. [Google Scholar] [CrossRef]
- Pace, L.A.; Crowe, S.E. Complex Relationships Between Food, Diet, and the Microbiome. Gastroenterol. Clin. North Am. 2016, 45, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Akter, S.; Hasan, M.R.; Acharjee, M.; Hossain, M.S.; et al. Impact of gut microbiome on skin health: Gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes 2022, 14, 2096995. [Google Scholar] [CrossRef] [PubMed]
- Thye, A.Y.K.; Bah, Y.R.; Law, J.W.F.; Tan, L.T.H.; He, Y.W.; Wong, S.H.; Thurairajasingam, S.; Chan, K.G.; Lee, L.H.; Letchumanan, V. Gut–Skin Axis: Unravelling the Connection between the Gut Microbiome and Psoriasis. Biomedicines 2022, 10, 1037. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, E.; Park, Y.M.; Hong, S.J. Microbiome in the gut-skin axis in atopic dermatitis. Allergy, Asthma and Immunology Research. Korean Acad. Asthma Allergy Clin. Immunol. 2018, 10, 354–362. [Google Scholar]
- Szántó, M.; Dózsa, A.; Antal, D.; Szabó, K.; Kemény, L.; Bai, P. Targeting the gut-skin axis—Probiotics as new tools for skin disorder management? Exp. Dermatol. 2019, 28, 1210–1218. [Google Scholar] [CrossRef] [PubMed]
- Prescott, S.L.; Larcombe, D.L.; Logan, A.C.; West, C.; Burks, W.; Caraballo, L.; Levin, M.; Van Etten, E.; Horwitz, P.; Kozyrskyj, A.; et al. The skin microbiome: Impact of modern environments on skin ecology, barrier integrity, and systemic immune programming. World Allergy Organ. J. 2017, 10, 29. [Google Scholar] [CrossRef]
- Bouslimani, A.; Da Silva, R.; Kosciolek, T.; Janssen, S.; Callewaert, C.; Amir, A.; Dorrestein, K.; Melnik, A.V.; Zaramela, L.S.; Kim, J.N.; et al. The impact of skin care products on skin chemistry and microbiome dynamics. BMC Biol. 2019, 17, 47. [Google Scholar] [CrossRef] [PubMed]

| Sample $ | Gender | Skin Type | Diet | Moisturizer Use | Sunscreen Use | Sun Exposure * |
|---|---|---|---|---|---|---|
| SM1 | Female | Oily | Plant | Yes | Yes | Low |
| SM2 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM3 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM4 | Male | Dry | Meat | Yes | No | High |
| SM5 | Female | Dry | Meat | Yes | No | Low |
| SM6 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM7 | Male | Dry | Meat | No | No | High |
| SM8 | Female | Oily | Meat | Yes | No | Low |
| SM9 | Female | Dry | Meat | Yes | No | Low |
| SM10 | Male | Dry | Meat | Yes | No | Moderate |
| SM11 | Male | Oily | Meat | Yes | No | Moderate |
| SM12 | Female | Oily | Meat | Yes | Yes | Low |
| SM13 | Female | Oily | Plant | Yes | Yes | Moderate |
| SM14 | Female | Dry | Meat | Yes | No | Moderate |
| SM15 | Male | Dry | Meat | No | No | High |
| SM16 | Female | Oily | Meat | Yes | Yes | Low |
| SM17 | Female | Oily | Meat | Yes | Yes | Low |
| SM18 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM19 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM20 | Female | Dry | Meat | No | No | Moderate |
| SM21 | Female | Dry | Meat | Yes | Yes | Moderate |
| SM22 | Female | Dry | Meat | Yes | Yes | Low |
| SM23 | Female | Oily | Meat | Yes | Yes | Moderate |
| SM24 | Female | Oily | Meat | Yes | Yes | Moderate |
| SM25 | Male | Dry | Meat | Yes | Yes | Low |
| SM26 | Female | Oily | Meat | Yes | Yes | Moderate |
| SM27 | Male | Dry | Meat | No | No | High |
| SM28 | Female | Oily | Plant | No | No | Moderate |
| SM29 | Male | Dry | Meat | Yes | Yes | Low |
| SM30 | Male | Dry | Meat | No | No | High |
| SM31 | Female | Dry | Meat | Yes | Yes | Low |
| SM32 | Male | Dry | Meat | Yes | Yes | Moderate |
| SM33 | Male | Dry | Meat | Yes | Yes | Moderate |
| SM34 | Female | Dry | Meat | Yes | Yes | Low |
| SM35 | Female | Oily | Meat | Yes | Yes | Moderate |
| SM36 | Male | Dry | Meat | No | No | High |
| SM37 | Female | Oily | Meat | Yes | Yes | Moderate |
| Sample | Shannon Diversity | Corynebacterium spp. | Cutibacterium spp. | Staphylococcus spp. | Malassezia spp. |
|---|---|---|---|---|---|
| SM1 | 2.12 | 2.90 ± 0.73 | 43.13 ± 19.52 | 8.15 ± 2.68 | 3.92 ± 1.01 |
| SM2 | 0.93 | 0.15 ± 0.00 | 29.86 ± 0.00 | 53.50 ± 22.57 | 5.25 ± 2.47 |
| SM3 | 0.94 | 0.16 ± 0.01 | 32.48 ± 0.01 | 53.25 ± 24.78 | 5.51 ± 2.59 |
| SM4 | 2.4 | 6.62 ± 2.34 | 27.46 ± 12.68 | 23.03 ± 5.96 | 0.41 ± 0.17 |
| SM5 | 0.43 | 0.57 ± 0.12 | 87.61 ± 41.10 | 2.93 ± 1.36 | 0.72 ± 0.27 |
| SM6 | 1.02 | 3.43 ± 1.63 | 67.36 ± 25.99 | 18.88 ± 8.58 | 0.21 ± 0.09 |
| SM7 | 1.04 | 2.35 ± 0.48 | 76.24 ± 26.32 | 5.09 ± 1.30 | 0.65 ± 0.29 |
| SM8 | 1.32 | 0.10 ± 0.05 | 64.29 ± 25.71 | 2.07 ± 0.48 | 0.02 ± 0.01 |
| SM9 | 1.14 | 0.54 ± 0.14 | 79.54 ± 21.52 | 4.69 ± 0.01 | 0.87 ± 0.41 |
| SM10 | 0.99 | 0.47 ± 0.05 | 1.00 ± 0.01 | 76.10 ± 34.95 | 0.00 ± 0.00 |
| SM11 | 2.73 | 1.49 ± 0.26 | 41.76 ± 19.43 | 0.78 ± 0.34 | 0.47 ± 0.23 |
| SM12 | 0.61 | 0.03 ± 0.00 | 80.05 ± 34.07 | 6.97 ± 0.40 | 5.32 ± 2.59 |
| SM13 | 1.27 | 3.24 ± 1.59 | 60.88 ± 26.80 | 11.47 ± 3.04 | 3.20 ± 0.00 |
| SM14 | 0.91 | 0.04 ± 0.00 | 72.93 ± 0.00 | 0.56 ± 0.00 | 8.55 ± 0.00 |
| SM15 | 1.09 | 1.96 ± 0.50 | 27.32 ± 11.71 | 6.95 ± 1.97 | 0.17 ± 0.00 |
| SM16 | 0.32 | 0.03 ± 0.01 | 83.32 ± 38.45 | 2.52 ± 0.64 | 0.81 ± 0.39 |
| SM17 | 0.27 | 0.00 ± 0.00 | 79.54 ± 0.00 | 0.54 ± 0.00 | 0.28 ± 0.00 |
| SM18 | 0.48 | 0.02 ± 0.00 | 80.70 ± 34.76 | 9.86 ± 0.00 | 0.18 ± 4.17 |
| SM19 | 0.46 | 0.04 ± 0.01 | 79.49 ± 34.26 | 10.98 ± 4.30 | 0.20 ± 0.10 |
| SM20 | 0.41 | 0.77 ± 0.24 | 83.94 ± 32.72 | 4.64 ± 0.00 | 0.74 ± 0.32 |
| SM21 | 1.33 | 0.37 ± 0.05 | 56.64 ± 25.50 | 9.03 ± 2.34 | 6.43 ± 3.79 |
| SM22 | 1.52 | 0.00 ± 0.00 | 48.98 ± 21.50 | 6.13 ± 3.06 | 1.35 ± 0.00 |
| SM23 | 1.34 | 0.35 ± 0.14 | 67.48 ± 23.09 | 7.33 ± 2.64 | 2.01 ± 0.75 |
| SM24 | 1.19 | 0.41 ± 0.10 | 69.18 ± 25.12 | 6.65 ± 2.45 | 1.43 ± 0.50 |
| SM25 | 0.57 | 0.36 ± 0.06 | 84.55 ± 29.67 | 1.99 ± 0.93 | 1.16 ± 0.50 |
| SM26 | 0.6 | 0.03 ± 0.00 | 80.05 ± 34.07 | 6.97 ± 2.59 | 5.32 ± 0.40 |
| SM27 | 0.28 | 1.34 ± 0.67 | 77.79 ± 36.21 | 1.51 ± 0.44 | 0.52 ± 0.00 |
| SM28 | 1.23 | 3.21 ± 0.00 | 60.88 ± 26.80 | 11.47 ± 3.04 | 3.20 ± 0.00 |
| SM29 | 0.61 | 0.52 ± 0.26 | 82.19 ± 28.10 | 3.40 ± 0.26 | 0.88 ± 0.00 |
| SM30 | 1.05 | 6.31 ± 1.00 | 57.81 ± 27.85 | 1.08 ± 0.28 | 4.52 ± 2.19 |
| SM31 | 0.21 | 0.08 ± 0.01 | 91.04 ± 35.99 | 0.31 ± 0.09 | 0.38 ± 0.00 |
| SM32 | 2.17 | 10.37 ± 2.34 | 13.26 ± 3.32 | 15.47 ± 5.73 | 0.51 ± 0.00 |
| SM33 | 2.59 | 26.86 ± 4.50 | 1.43 ± 0.53 | 14.58 ± 1.53 | 0.00 ± 0.00 |
| SM34 | 0.926 | 0.04 ± 0.00 | 72.93 ± 0.00 | 0.56 ± 0.00 | 8.55 ± 0.00 |
| SM35 | 0.45 | 0.08 ± 0.02 | 76.72 ± 30.66 | 6.82 ± 3.16 | 0.37 ± 0.16 |
| SM36 | 0.69 | 2.33 ± 1.08 | 74.76 ± 34.22 | 5.22 ± 0.19 | 1.80 ± 0.68 |
| SM37 | 0.3 | 0.00 ± 0.00 | 87.64 ± 40.36 | 2.26 ± 0.27 | 0.89 ± 0.35 |
| Healthy | 34.95 ± 4.62 | 49.33 ± 14.58 | 18.35 ± 3.79 | 4.04 ± 1.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubli, K.; Balasundaram, P.; Chaudhari, R.; Vettrivelan, S.; Borawake, A.; Kapoor, R.; Kovalchuk, I.; Kapoor, A.; Singh, R.; Tripathi, M.B. Exploring the Impact of Skin Care Routines on the Skin Microbiome and Possible Skin Disease Risk—A Pilot Study. Biomedicines 2025, 13, 2371. https://doi.org/10.3390/biomedicines13102371
Dubli K, Balasundaram P, Chaudhari R, Vettrivelan S, Borawake A, Kapoor R, Kovalchuk I, Kapoor A, Singh R, Tripathi MB. Exploring the Impact of Skin Care Routines on the Skin Microbiome and Possible Skin Disease Risk—A Pilot Study. Biomedicines. 2025; 13(10):2371. https://doi.org/10.3390/biomedicines13102371
Chicago/Turabian StyleDubli, Kirti, Preethi Balasundaram, Rinku Chaudhari, Sarvesh Vettrivelan, Arman Borawake, Raman Kapoor, Igor Kovalchuk, Anmol Kapoor, Raja Singh, and Minal Borkar Tripathi. 2025. "Exploring the Impact of Skin Care Routines on the Skin Microbiome and Possible Skin Disease Risk—A Pilot Study" Biomedicines 13, no. 10: 2371. https://doi.org/10.3390/biomedicines13102371
APA StyleDubli, K., Balasundaram, P., Chaudhari, R., Vettrivelan, S., Borawake, A., Kapoor, R., Kovalchuk, I., Kapoor, A., Singh, R., & Tripathi, M. B. (2025). Exploring the Impact of Skin Care Routines on the Skin Microbiome and Possible Skin Disease Risk—A Pilot Study. Biomedicines, 13(10), 2371. https://doi.org/10.3390/biomedicines13102371







