Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Patient Selection
2.2. Clinical Characteristics Evaluation
2.3. Genetic and Epigenetic Analyses
2.4. miRNA Signature and Target Genes
2.5. Survival Analyses
3. Results
3.1. Clinical Characteristics of the Enrolled Subjects
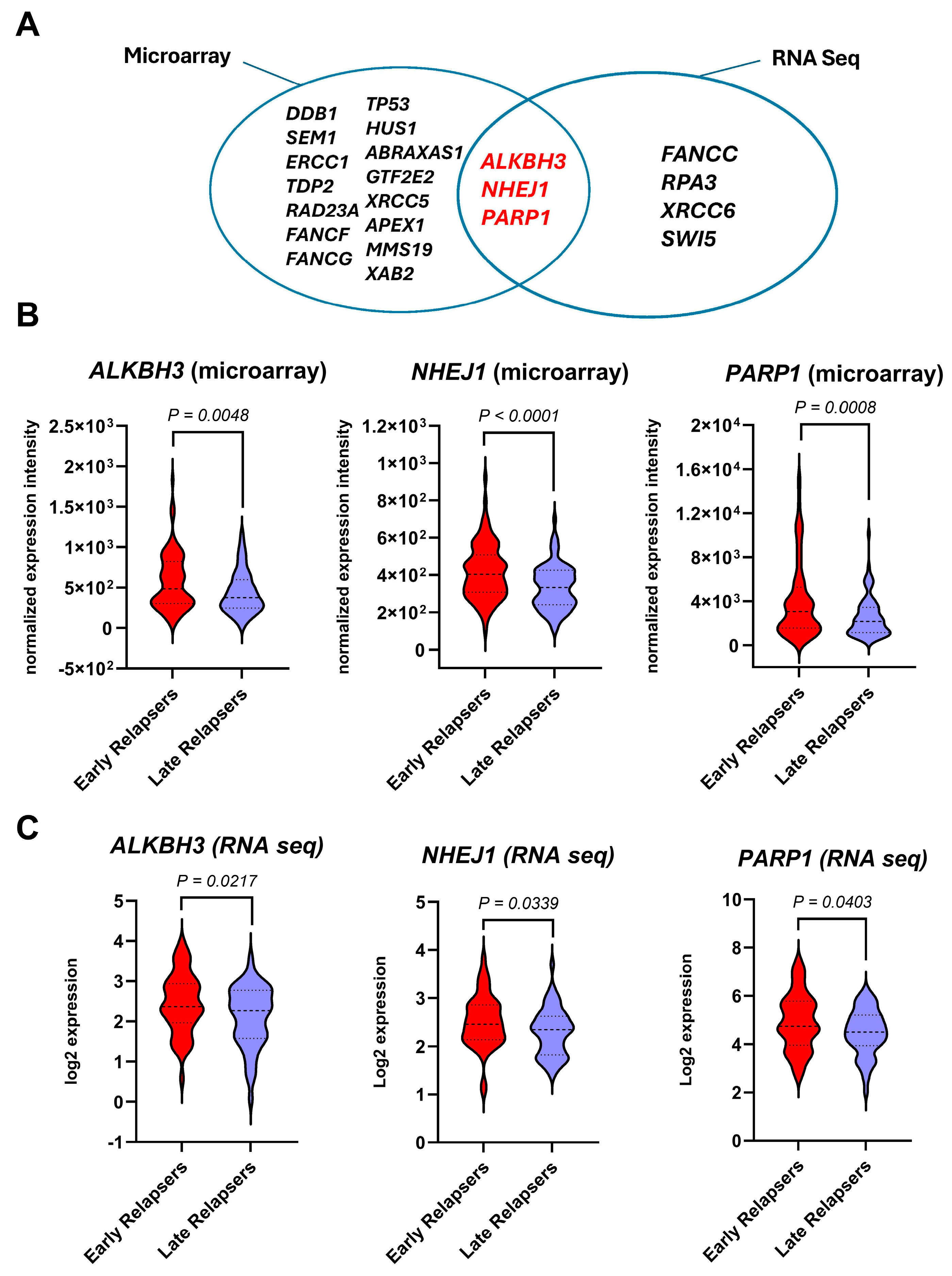
3.2. Differentially Expressed DNA-Repair Genes of Early-Relapsed Patients Relative to Late-Relapsed Patients
3.3. Genetic Alternations and Methylation Status of the Identified Candidate Genes
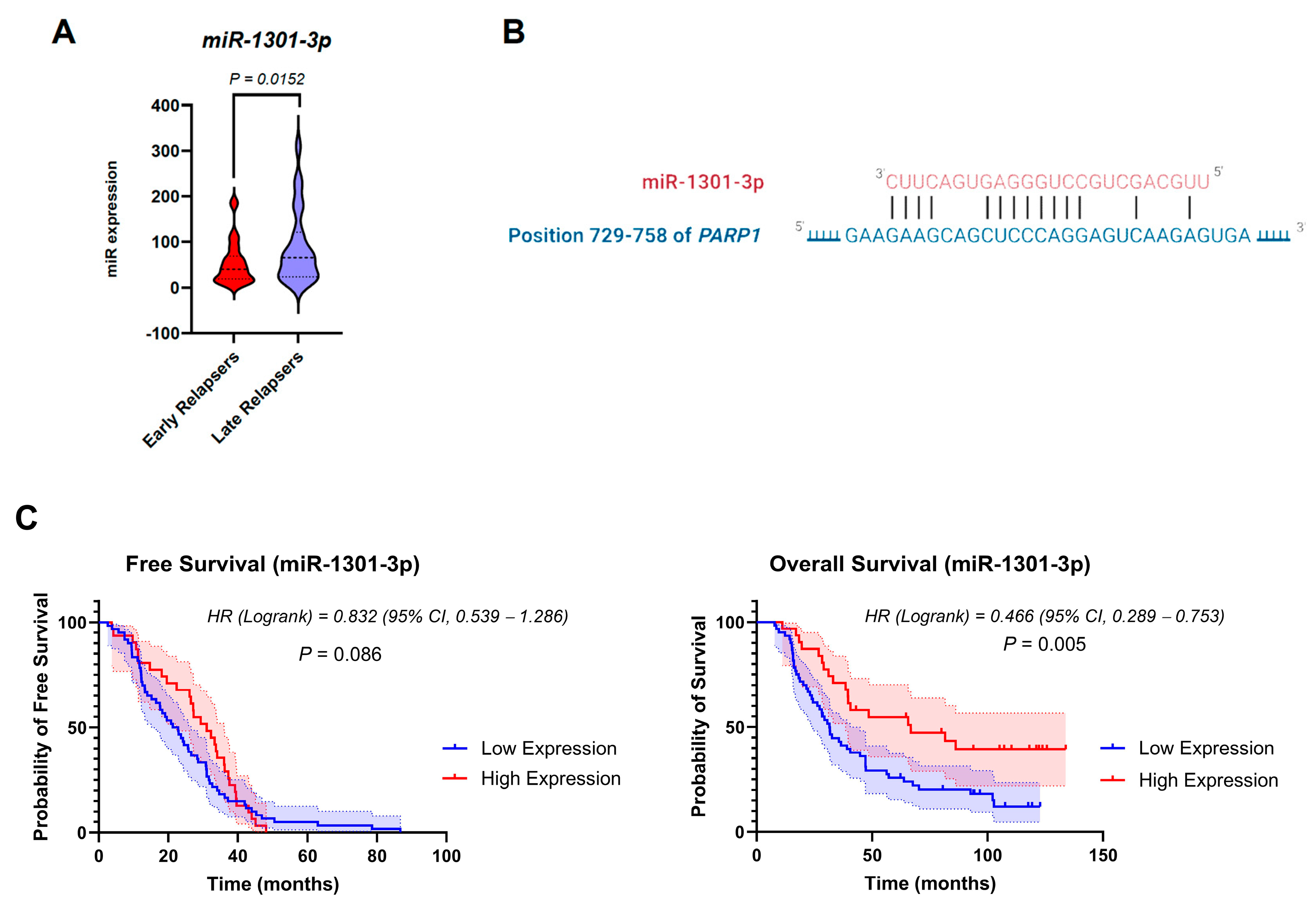
3.4. miRNA Dysregulation in the Cohort Groups
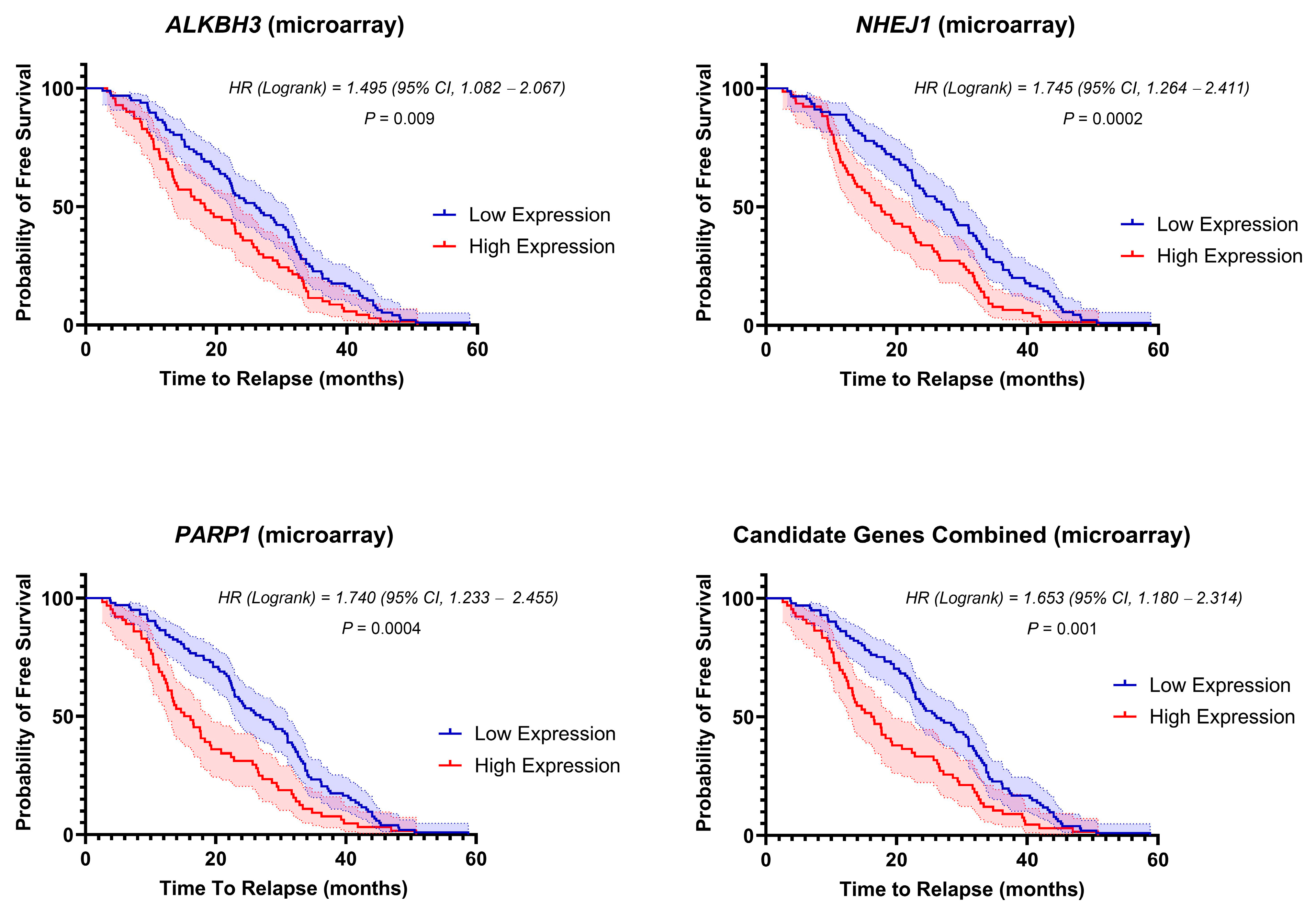
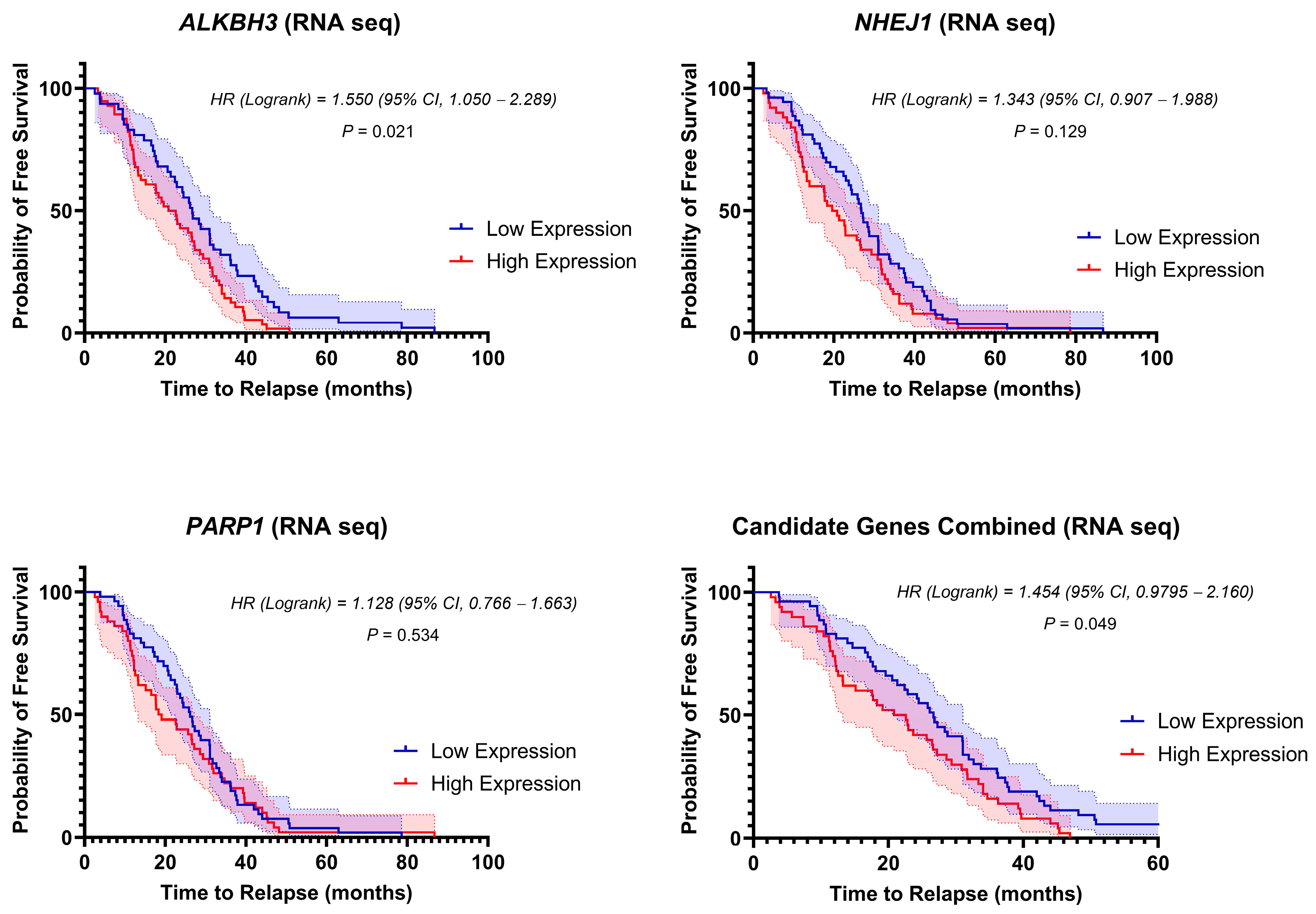
3.5. Stratification of Clinical Outcome Based on the DNA-Repair Gene Expressions at Diagnosis
3.6. Expression Levels of miR-1301-3p Correlate with Poorer Patient Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Inaba, H.; Mullighan, C.G. Pediatric Acute Lymphoblastic Leukemia. Haematologica 2020, 105, 2524–2539. [Google Scholar] [CrossRef] [PubMed]
- Nabizadeh, F.; Momtaz, S.; Ghanbari-Movahed, M.; Qalekhani, F.; Mohsenpour, H.; Aneva, I.Y.; Bishayee, A.; Farzaei, M.H.; Bishayee, A. Pediatric Acute Lymphoblastic Leukemia Management Using Multitargeting Bioactive Natural Compounds: A Systematic and Critical Review. Pharmacol. Res. 2022, 177, 106116. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mohty, M. Acute Lymphoblastic Leukaemia. Lancet 2020, 395, 1146–1162. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Alecsa, M.S.; Popescu, R.; Starcea, M.I.; Mocanu, A.M.; Rusu, C.; Miron, I.C. Pediatric Acute Lymphoblastic Leukemia Emerging Therapies-From Pathway to Target. Int. J. Mol. Sci. 2023, 24, 4661. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, J.; Gogoi, M.P.; Krishnan, S.; Saha, V. Relapsed Acute Lymphoblastic Leukemia. Indian J. Pediatr. 2024, 91, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Makimoto, A.; Yuza, Y. Treatment of Pediatric Acute Lymphoblastic Leukemia: A Historical Perspective. Cancers 2024, 16, 723. [Google Scholar] [CrossRef]
- Pieters, R.; Huismans, D.R.; Loonen, A.H.; Hählen, K.; van der Does-van den Berg, A.; van Wering, E.R.; Veerman, A.J. Relation of Cellular Drug Resistance to Long-Term Clinical Outcome in Childhood Acute Lymphoblastic Leukaemia. Lancet 1991, 338, 399–403. [Google Scholar] [CrossRef]
- Johnstone, R.W.; Ruefli, A.A.; Lowe, S.W. Apoptosis: A Link between Cancer Genetics and Chemotherapy. Cell 2002, 108, 153–164. [Google Scholar] [CrossRef]
- Kelley, M.R.; Logsdon, D.; Fishel, M.L. Targeting DNA Repair Pathways for Cancer Treatment: What’s New? Future Oncol. Lond. Engl. 2014, 10, 1215–1237. [Google Scholar] [CrossRef]
- Martin, L.P.; Hamilton, T.C.; Schilder, R.J. Platinum Resistance: The Role of DNA Repair Pathways. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 1291–1295. [Google Scholar] [CrossRef]
- Li, Y.; Bai, O.; Cui, J.; Li, W. Genetic Polymorphisms in the DNA Repair Gene, XRCC1 Associate with Non-Hodgkin Lymphoma Susceptibility: A Systematic Review and Meta-Analysis. Eur. J. Med. Genet. 2016, 59, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zhou, P.-K. DNA Damage Repair: Historical Perspectives, Mechanistic Pathways and Clinical Translation for Targeted Cancer Therapy. Signal Transduct. Target. Ther. 2021, 6, 254. [Google Scholar] [CrossRef] [PubMed]
- Li, L.-Y.; Guan, Y.; Chen, X.-S.; Yang, J.-M.; Cheng, Y. DNA Repair Pathways in Cancer Therapy and Resistance. Front. Pharmacol. 2020, 11, 629266. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. Causes and Consequences of DNA Single-Strand Breaks. Trends Biochem. Sci. 2024, 49, 68–78. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA Double-Strand Breaks: Signaling, Repair and the Cancer Connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. DNA Interstrand Crosslink Repair and Cancer. Nat. Rev. Cancer 2011, 11, 467–480. [Google Scholar] [CrossRef]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing Repair and Tolerance of DNA Damage Caused by Alkylating Agents. Nat. Rev. Cancer 2012, 12, 104–120. [Google Scholar] [CrossRef]
- Malu, S.; Malshetty, V.; Francis, D.; Cortes, P. Role of Non-Homologous End Joining in V(D)J Recombination. Immunol. Res. 2012, 54, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Marston, E.; Weston, V.; Jesson, J.; Maina, E.; McConville, C.; Agathanggelou, A.; Skowronska, A.; Mapp, K.; Sameith, K.; Powell, J.E.; et al. Stratification of Pediatric ALL by in Vitro Cellular Responses to DNA Double-Strand Breaks Provides Insight into the Molecular Mechanisms Underlying Clinical Response. Blood 2009, 113, 117–126. [Google Scholar] [CrossRef][Green Version]
- Novara, F.; Beri, S.; Bernardo, M.E.; Bellazzi, R.; Malovini, A.; Ciccone, R.; Cometa, A.M.; Locatelli, F.; Giorda, R.; Zuffardi, O. Different Molecular Mechanisms Causing 9p21 Deletions in Acute Lymphoblastic Leukemia of Childhood. Hum. Genet. 2009, 126, 511–520. [Google Scholar] [CrossRef]
- Wiemels, J.L.; Alexander, F.E.; Cazzaniga, G.; Biondi, A.; Mayer, S.P.; Greaves, M. Microclustering of TEL-AML1 Translocation Breakpoints in Childhood Acute Lymphoblastic Leukemia. Genes Chromosomes Cancer 2000, 29, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Castaño, J.; Herrero, A.B.; Bursen, A.; González, F.; Marschalek, R.; Gutiérrez, N.C.; Menendez, P. Expression of MLL-AF4 or AF4-MLL Fusions Does Not Impact the Efficiency of DNA Damage Repair. Oncotarget 2016, 7, 30440–30452. [Google Scholar] [CrossRef] [PubMed]
- Hähnel, P.S.; Enders, B.; Sasca, D.; Roos, W.P.; Kaina, B.; Bullinger, L.; Theobald, M.; Kindler, T. Targeting Components of the Alternative NHEJ Pathway Sensitizes KRAS Mutant Leukemic Cells to Chemotherapy. Blood 2014, 123, 2355–2366. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, E.; Avigad, S.; Stark, B.; Zilberstein, J.; Freedman, L.; Gorfine, M.; Gavriel, H.; Cohen, I.J.; Goshen, Y.; Yaniv, I.; et al. Germ-Line ATM Gene Alterations Are Associated with Susceptibility to Sporadic T-Cell Acute Lymphoblastic Leukemia in Children. Genes Chromosomes Cancer 2004, 39, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Stankovic, T.; Marston, E. Molecular Mechanisms Involved in Chemoresistance in Paediatric Acute Lymphoblastic Leukaemia. Srp. Arh. Celok. Lek. 2008, 136, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, O.M.; As Sobeai, H.M.; Grant, S.G.; Latimer, J.J. Nucleotide Excision Repair Is a Predictor of Early Relapse in Pediatric Acute Lymphoblastic Leukemia. BMC Med. Genom. 2018, 11, 95. [Google Scholar] [CrossRef] [PubMed]
- Christmann, M.; Kaina, B. Epigenetic Regulation of DNA Repair Genes and Implications for Tumor Therapy. Mutat. Res. Rev. Mutat. Res. 2019, 780, 15–28. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.-P.; et al. An Estimate of the Total Number of True Human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef]
- Alshamrani, A.A. Roles of microRNAs in Ovarian Cancer Tumorigenesis: Two Decades Later, What Have We Learned? Front. Oncol. 2020, 10, 1084. [Google Scholar] [CrossRef]
- Wan, G.; Mathur, R.; Hu, X.; Zhang, X.; Lu, X. miRNA Response to DNA Damage. Trends Biochem. Sci. 2011, 36, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tong, Y.; Liu, J.; Lou, J. The Role of MicroRNA in DNA Damage Response. Front. Genet. 2022, 13, 850038. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, V. Regulation of DNA Repair by Non-Coding miRNAs. Non-Coding RNA Res. 2016, 1, 64–68. [Google Scholar] [CrossRef]
- The Therapeutically Applicable Research to Generate Effective Treatments (TARGET) Initiative, Phs000218, Managed by the NCI. Phs000464.V18.P7. Available online: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000464.v18.p7#attribution-section (accessed on 7 March 2022).
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Wood, R.D.; Mitchell, M.; Sgouros, J.; Lindahl, T. Human DNA Repair Genes. Science 2001, 291, 1284–1289. [Google Scholar] [CrossRef]
- Wood, R.D.; Mitchell, M.; Lindahl, T. Human DNA Repair Genes, 2005. Mutat. Res. 2005, 577, 275–283. [Google Scholar] [CrossRef]
- Oliveros, J.C. (2007–2015) Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 6 May 2022).
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. miRWalk: An Online Resource for Prediction of microRNA Binding Sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef]
- Huang, H.-Y.; Lin, Y.-C.-D.; Cui, S.; Huang, Y.; Tang, Y.; Xu, J.; Bao, J.; Li, Y.; Wen, J.; Zuo, H.; et al. miRTarBase Update 2022: An Informative Resource for Experimentally Validated miRNA-Target Interactions. Nucleic Acids Res. 2022, 50, D222–D230. [Google Scholar] [CrossRef]
- Jia, M.; Hu, B.-F.; Xu, X.-J.; Zhang, J.-Y.; Li, S.-S.; Tang, Y.-M. Clinical Features and Prognostic Impact of TCF3-PBX1 in Childhood Acute Lymphoblastic Leukemia: A Single-Center Retrospective Study of 837 Patients from China. Curr. Probl. Cancer 2021, 45, 100758. [Google Scholar] [CrossRef]
- Helwak, A.; Kudla, G.; Dudnakova, T.; Tollervey, D. Mapping the Human miRNA Interactome by CLASH Reveals Frequent Noncanonical Binding. Cell 2013, 153, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug Resistance in Cancer: An Overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Alhmoud, J.F.; Mustafa, A.G.; Malki, M.I. Targeting DNA Repair Pathways in Hematological Malignancies. Int. J. Mol. Sci. 2020, 21, 7365. [Google Scholar] [CrossRef] [PubMed]
- Liefke, R.; Windhof-Jaidhauser, I.M.; Gaedcke, J.; Salinas-Riester, G.; Wu, F.; Ghadimi, M.; Dango, S. The Oxidative Demethylase ALKBH3 Marks Hyperactive Gene Promoters in Human Cancer Cells. Genome Med. 2015, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhu, Q. The Role of Demethylase AlkB Homologs in Cancer. Front. Oncol. 2023, 13, 1153463. [Google Scholar] [CrossRef]
- Chen, T.; Tongpeng, S.; Lu, Z.; Topatana, W.; Juengpanich, S.; Li, S.; Hu, J.; Cao, J.; Lee, C.; Tian, Y.; et al. DNA Damage Response Inhibition-based Combination Therapies in Cancer Treatment: Recent Advances and Future Directions. Aging Cancer 2022, 3, 44–67. [Google Scholar] [CrossRef]
- Tasaki, M.; Shimada, K.; Kimura, H.; Tsujikawa, K.; Konishi, N. ALKBH3, a Human AlkB Homologue, Contributes to Cell Survival in Human Non-Small-Cell Lung Cancer. Br. J. Cancer 2011, 104, 700–706. [Google Scholar] [CrossRef]
- Yamato, I.; Sho, M.; Shimada, K.; Hotta, K.; Ueda, Y.; Yasuda, S.; Shigi, N.; Konishi, N.; Tsujikawa, K.; Nakajima, Y. PCA-1/ALKBH3 Contributes to Pancreatic Cancer by Supporting Apoptotic Resistance and Angiogenesis. Cancer Res. 2012, 72, 4829–4839. [Google Scholar] [CrossRef]
- Hotta, K.; Sho, M.; Fujimoto, K.; Shimada, K.; Yamato, I.; Anai, S.; Harada, H.; Tsujikawa, K.; Konishi, N.; Shinohara, N.; et al. Clinical Significance and Therapeutic Potential of Prostate Cancer Antigen-1/ALKBH3 in Human Renal Cell Carcinoma. Oncol. Rep. 2015, 34, 648–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, G.; Wang, Y.; Liu, C.; He, X. Association of AlkB Homolog 3 Expression with Tumor Recurrence and Unfavorable Prognosis in Hepatocellular Carcinoma. J. Gastroenterol. Hepatol. 2018, 33, 1617–1625. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. Comparison of Nonhomologous End Joining and Homologous Recombination in Human Cells. DNA Repair 2008, 7, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Sishc, B.J.; Davis, A.J. The Role of the Core Non-Homologous End Joining Factors in Carcinogenesis and Cancer. Cancers 2017, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Valikhani, M.; Rahimian, E.; Ahmadi, S.E.; Chegeni, R.; Safa, M. Involvement of Classic and Alternative Non-Homologous End Joining Pathways in Hematologic Malignancies: Targeting Strategies for Treatment. Exp. Hematol. Oncol. 2021, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Chiou, S.-S.; Huang, J.-L.; Tsai, Y.-S.; Chen, T.-F.; Lee, K.-W.; Juo, S.-H.H.; Jong, Y.-J.; Hung, C.-H.; Chang, T.-T.; Lin, C.-S. Elevated mRNA Transcripts of Non-Homologous End-Joining Genes in Pediatric Acute Lymphoblastic Leukemia. Leukemia 2007, 21, 2061–2064. [Google Scholar] [CrossRef][Green Version]
- Chen, T.; Chen, J.; Su, W.; Wu, M.; Tsao, C. Expression of DNA Repair Gene Ku80 in Lymphoid Neoplasm. Eur. J. Haematol. 2005, 74, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Gouttia, O.G.; Wang, L.; Peng, A. PARP1 Upregulation in Recurrent Oral Cancer and Treatment Resistance. Front. Cell Dev. Biol. 2021, 9, 804962. [Google Scholar] [CrossRef]
- Sessa, C. Update on PARP1 Inhibitors in Ovarian Cancer. Ann. Oncol. 2011, 22, viii72–viii76. [Google Scholar] [CrossRef]
- Mego, M.; Cierna, Z.; Svetlovska, D.; Macak, D.; Machalekova, K.; Miskovska, V.; Chovanec, M.; Usakova, V.; Obertova, J.; Babal, P.; et al. PARP Expression in Germ Cell Tumours. J. Clin. Pathol. 2013, 66, 607–612. [Google Scholar] [CrossRef] [PubMed]
- Newman, E.A.; Lu, F.; Bashllari, D.; Wang, L.; Opipari, A.W.; Castle, V.P. Alternative NHEJ Pathway Components Are Therapeutic Targets in High-Risk Neuroblastoma. Mol. Cancer Res. 2015, 13, 470–482. [Google Scholar] [CrossRef] [PubMed]
- Tomoda, T.; Kurashige, T.; Moriki, T.; Yamamoto, H.; Fujimoto, S.; Taniguchi, T. Enhanced Expression of Poly(ADP-ribose) Synthetase Gene in Malignant Lymphoma. Am. J. Hematol. 1991, 37, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F.; García-Parra, J.; Zazo, S.; Tusquets, I.; Ferrer-Lozano, J.; Menendez, S.; Eroles, P.; Chamizo, C.; Servitja, S.; Ramírez-Merino, N.; et al. Nuclear PARP-1 Protein Overexpression Is Associated with Poor Overall Survival in Early Breast Cancer. Ann. Oncol. 2012, 23, 1156–1164. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.; Garber, J.E. PARP Inhibition in Breast Cancer: Progress Made and Future Hopes. Npj Breast Cancer 2022, 8, 47. [Google Scholar] [CrossRef]
- Dziaman, T.; Ludwiczak, H.; Ciesla, J.M.; Banaszkiewicz, Z.; Winczura, A.; Chmielarczyk, M.; Wisniewska, E.; Marszalek, A.; Tudek, B.; Olinski, R. PARP-1 Expression Is Increased in Colon Adenoma and Carcinoma and Correlates with OGG1. PLoS ONE 2014, 9, e115558. [Google Scholar] [CrossRef]
- Bi, F.-F.; Li, D.; Yang, Q. Hypomethylation of ETS Transcription Factor Binding Sites and Upregulation of PARP1 Expression in Endometrial Cancer. BioMed Res. Int. 2013, 2013, 946268. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Bi, F.-F.; Cao, J.-M.; Cao, C.; Li, C.-Y.; Liu, B.; Yang, Q. Poly (ADP-Ribose) Polymerase 1 Transcriptional Regulation: A Novel Crosstalk between Histone Modification H3K9ac and ETS1 Motif Hypomethylation in BRCA1-Mutated Ovarian Cancer. Oncotarget 2014, 5, 291–297. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bi, F.-F.; Li, D.; Yang, Q. Promoter Hypomethylation, Especially around the E26 Transformation-Specific Motif, and Increased Expression of Poly (ADP-Ribose) Polymerase 1 in BRCA-Mutated Serous Ovarian Cancer. BMC Cancer 2013, 13, 90. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Jin, J.; Wang, J.; Huang, J.; Ma, Z.; Huang, X.; He, X.; Zhou, Y.; Xu, Y.; et al. High PARP-1 Expression Predicts Poor Survival in Acute Myeloid Leukemia and PARP-1 Inhibitor and SAHA-Bendamustine Hybrid Inhibitor Combination Treatment Synergistically Enhances Anti-Tumor Effects. eBioMedicine 2018, 38, 47–56. [Google Scholar] [CrossRef]
- Zhong, C.; Dong, Y.; Zhang, Q.; Yuan, C.; Duan, S. Aberrant Expression of miR-1301 in Human Cancer. Front. Oncol. 2021, 11, 789626. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Bagès, S.; Knobloch, G.; Ladurner, A.G.; Buschbeck, M. The Taming of PARP1 and Its Impact on NAD+ Metabolism. Mol. Metab. 2020, 38, 100950. [Google Scholar] [CrossRef]
- Masutani, M.; Nozaki, T.; Nishiyama, E.; Shimokawa, T.; Tachi, Y.; Suzuki, H.; Nakagama, H.; Wakabayashi, K.; Sugimura, T. Function of Poly(ADP-Ribose) Polymerase in Response to DNA Damage: Gene-Disruption Study in Mice. Mol. Cell. Biochem. 1999, 193, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Fu, K.; Hodgson, A.; Wier, E.M.; Wen, M.G.; Kamenyeva, O.; Xia, X.; Koo, L.Y.; Wan, F. Sam68 Is Required for DNA Damage Responses via Regulating Poly(ADP-Ribosyl)Ation. PLoS Biol. 2016, 14, e1002543. [Google Scholar] [CrossRef] [PubMed]
- Gibbs-Seymour, I.; Fontana, P.; Rack, J.G.M.; Ahel, I. HPF1/C4orf27 Is a PARP-1-Interacting Protein That Regulates PARP-1 ADP-Ribosylation Activity. Mol. Cell 2016, 62, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Sisay, M.; Edessa, D. PARP Inhibitors as Potential Therapeutic Agents for Various Cancers: Focus on Niraparib and Its First Global Approval for Maintenance Therapy of Gynecologic Cancers. Gynecol. Oncol. Res. Pract. 2017, 4, 18. [Google Scholar] [CrossRef] [PubMed]
- Bhamidipati, D.; Haro-Silerio, J.I.; Yap, T.A.; Ngoi, N. PARP Inhibitors: Enhancing Efficacy through Rational Combinations. Br. J. Cancer 2023, 129, 904–916. [Google Scholar] [CrossRef] [PubMed]
- Kontandreopoulou, C.-N.; Diamantopoulos, P.T.; Tiblalexi, D.; Giannakopoulou, N.; Viniou, N.-A. PARP1 as a Therapeutic Target in Acute Myeloid Leukemia and Myelodysplastic Syndrome. Blood Adv. 2021, 5, 4794–4805. [Google Scholar] [CrossRef]
| Parameter | Early-Relapsed n = 109 (55.05%) | Late-Relapsed n = 89 (44.95%) | p-Value * |
|---|---|---|---|
| Age (years) | 9.55 ± 0.62 | 7.64 ± 0.53 | 0.023 |
| Vital status | Alive = 24 (22.02%) Deceased = 85 (77.98%) | Alive = 38 (42.70%) Deceased = 51 (53.30%) | 0.002 |
| Overall Survival (months) | 23.87 | 76.37 | <0.0001 |
| Sex | Male = 67 (61.47%) Female = 42 (38.53%) | Male = 45 (50.56%) Female = 44 (49.44%) | 0.124 |
| race | White = 73 (66.97%) African American = 14 (12.84%) Others = 3 (2.75%) Unknown = 19 (17.43%) | White = 69 (77.53%) African American = 5 (5.62%) Others = 2 (2.25%) Unknown = 13 (14.61%) | 0.294 |
| WBC | 103.47 ± 18.28 | 67.92 ± 12.48 | 0.127 |
| TCF3-PBX1 fusion status | Positive = 15 (13.76%) Negative = 60 (55.05%) Unknown = 34 (31.19%) | Positive = 3 (3.37%) Negative = 59 (66.29%) Unknown = 27 (30.34%) | 0.032 |
| Trisomy 4_10 | Positive = 5 (4.59%) Negative = 91 (83.49%) Unknown = 13 (11.93%) | Positive = 7 (7.86%) Negative = 67 (75.28%) Unknown = 15 (16.85%) | 0.356 |
| BCR-ABL1 fusion status | Positive = 2 (1.83%) Negative = 106 (97.25%) Unknown = 1 (0.92%) | Positive = 89 (100%) Negative = 0 (0%) Unknown = 0 (0%) | 0.288 |
| ETV6-RUNX1 fusion status | Positive = 5 (4.59%) Negative = 88 (80.73%) Unknown = 16 (14.68%) | Positive = 11 (12.36%) Negative = 64 (71.91%) Unknown = 14 (15.73%) | 0.123 |
| TP53 mutation status | Positive = 3 (2.75%) Negative = 62 (56.88%) Unknown = 44 (40.37%) | Positive = 0 (0%) Negative = 65 (62.92%) Unknown = 33 (37.08%) | 0.156 |
| ATM mutation status | Positive = 3 (2.75%) Negative = 62 (56.88%) Unknown = 44 (40.37%) | Positive = 0 (0%) Negative = 65 (62.92%) Unknown = 33 (37.08%) | 0.156 |
| Genes | Fold-Change | p-Value * | Pathway |
|---|---|---|---|
| PARP1 | 1.56 | 7.57 × 10−4 | Poly(ADP-ribose) polymerase (PARP) enzymes |
| TP53 | 1.49 | 8.67 × 10−4 | Regulation of the cell cycle |
| SEM1 | 1.31 | 8.28 × 10−4 | Homologous recombination |
| ERCC1 | 1.31 | 3.60 × 10−3 | Nucleotide excision repair |
| ALKBH3 | 1.31 | 4.78 × 10−3 | Direct reversal of damage |
| DDB1 | 1.29 | 6.80 × 10−5 | Nucleotide excision repair |
| FANCG | 1.29 | 0.011 | Fanconi Anemia |
| ABRAXAS1 | 1.26 | 0.0185 | Homologous recombination |
| NHEJ1 | 1.25 | 6.88 × 10−5 | Non-homologous end-joining |
| RAD23A | 1.24 | 7.29 × 10−3 | Nucleotide excision repair |
| TDP2 | 1.23 | 4.62 × 10−3 | Repair of DNA-protein crosslinks |
| FANCF | 1.23 | 9.49 × 10−3 | Fanconi Anemia |
| HUS1 | 1.18 | 7.04 × 10−3 | Subunits of PCNA-like sensor of damaged DNA |
| GTF2E2 | 1.18 | 0.022 | Nucleotide excision repair |
| XAB2 | 1.18 | 0.0493 | Nucleotide excision repair |
| APEX1 | 1.17 | 0.0476 | Base excision repair |
| XRCC5 | 1.14 | 0.0246 | Non-homologous end-joining |
| MMS19 | 1.13 | 0.0487 | Nucleotide excision repair |
| Genes | Log2 Ratio | p-Value * | Pathway |
|---|---|---|---|
| PARP1 | 0.43 | 0.0403 | Poly(ADP-ribose) polymerase (PARP) enzymes |
| RPA3 | 0.39 | 0.0217 | Nucleotide excision repair |
| ALKBH3 | 0.34 | 0.0217 | Direct reversal of damage |
| SWI5 | 0.31 | 0.048 | Homologous recombination |
| FANCC | 0.27 | 2.28 × 10−3 | Fanconi Anemia |
| XRCC6 | 0.25 | 0.023 | Non-homologous end-joining |
| NHEJ1 | 0.22 | 0.0339 | Non-homologous end-joining |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albaqami, W.F.; Alshamrani, A.A.; Almubarak, A.A.; Alotaibi, F.E.; Alotaibi, B.J.; Alanazi, A.M.; Alotaibi, M.R.; Alhoshani, A.; As Sobeai, H.M. Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes. Biomedicines 2024, 12, 1766. https://doi.org/10.3390/biomedicines12081766
Albaqami WF, Alshamrani AA, Almubarak AA, Alotaibi FE, Alotaibi BJ, Alanazi AM, Alotaibi MR, Alhoshani A, As Sobeai HM. Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes. Biomedicines. 2024; 12(8):1766. https://doi.org/10.3390/biomedicines12081766
Chicago/Turabian StyleAlbaqami, Walaa F., Ali A. Alshamrani, Ali A. Almubarak, Faris E. Alotaibi, Basil Jamal Alotaibi, Abdulrahman M. Alanazi, Moureq R. Alotaibi, Ali Alhoshani, and Homood M. As Sobeai. 2024. "Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes" Biomedicines 12, no. 8: 1766. https://doi.org/10.3390/biomedicines12081766
APA StyleAlbaqami, W. F., Alshamrani, A. A., Almubarak, A. A., Alotaibi, F. E., Alotaibi, B. J., Alanazi, A. M., Alotaibi, M. R., Alhoshani, A., & As Sobeai, H. M. (2024). Genetic and Epigenetic Biomarkers Associated with Early Relapse in Pediatric Acute Lymphoblastic Leukemia: A Focused Bioinformatics Study on DNA-Repair Genes. Biomedicines, 12(8), 1766. https://doi.org/10.3390/biomedicines12081766





