Activity of Epsilon-poly-L-lysine against Multidrug-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates of Urinary Tract Infections
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples and Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.3. Determination of Minimum Inhibitory Concentration
2.4. Biofilm Formation and Biomass Quantification
2.5. Antimicrobial Resistance and Virulence Genes
2.6. Multilocus Sequence Typing
3. Results and Discussion
3.1. Antimicrobial Susceptibility Testing
3.2. Antimicrobial Resistance Genes
3.3. Multilocus Sequence Typing
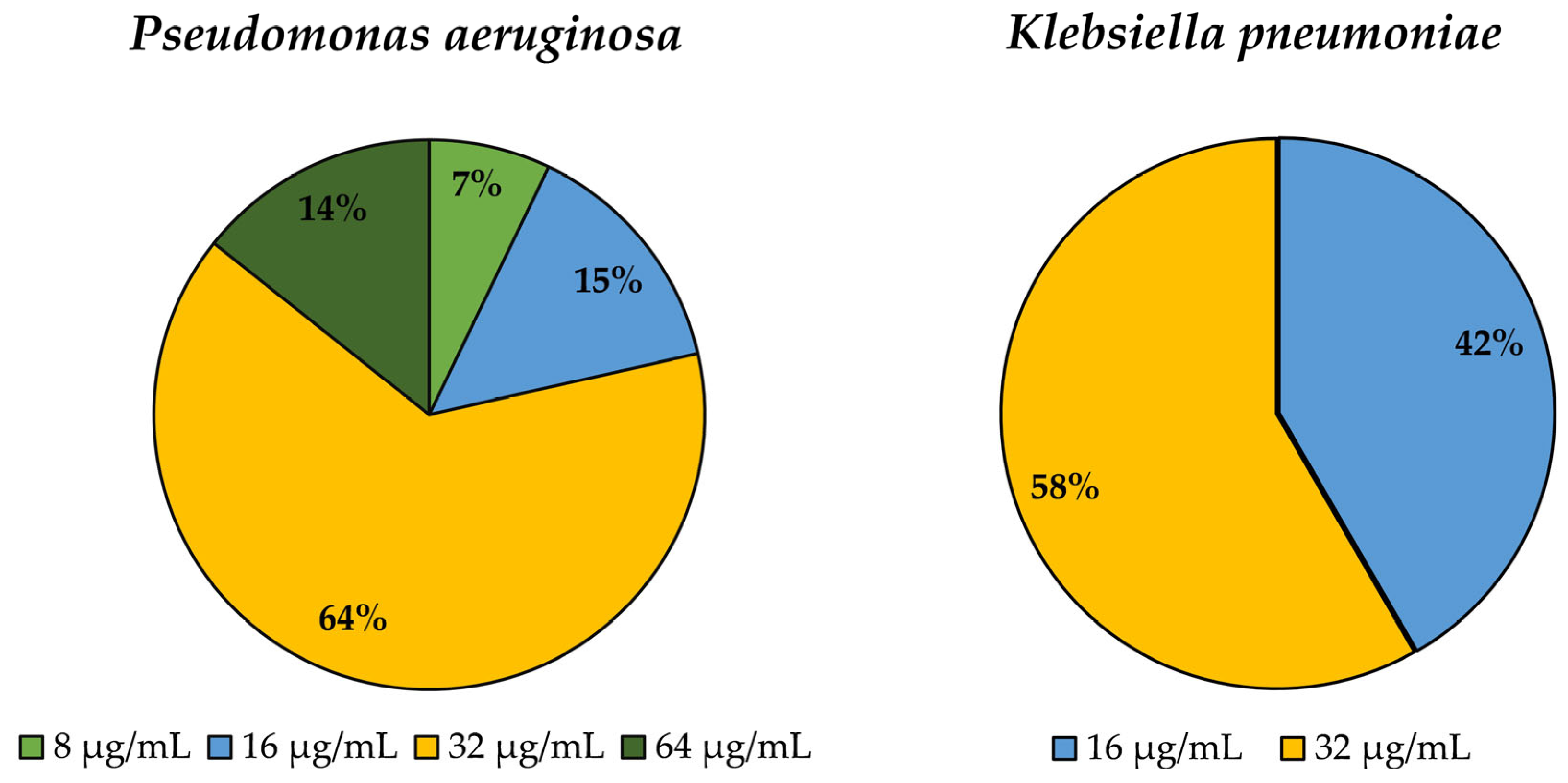
3.4. Determination of Minimum Inhibitory Concentration (MIC) for Epsilon-poly-L-lysine
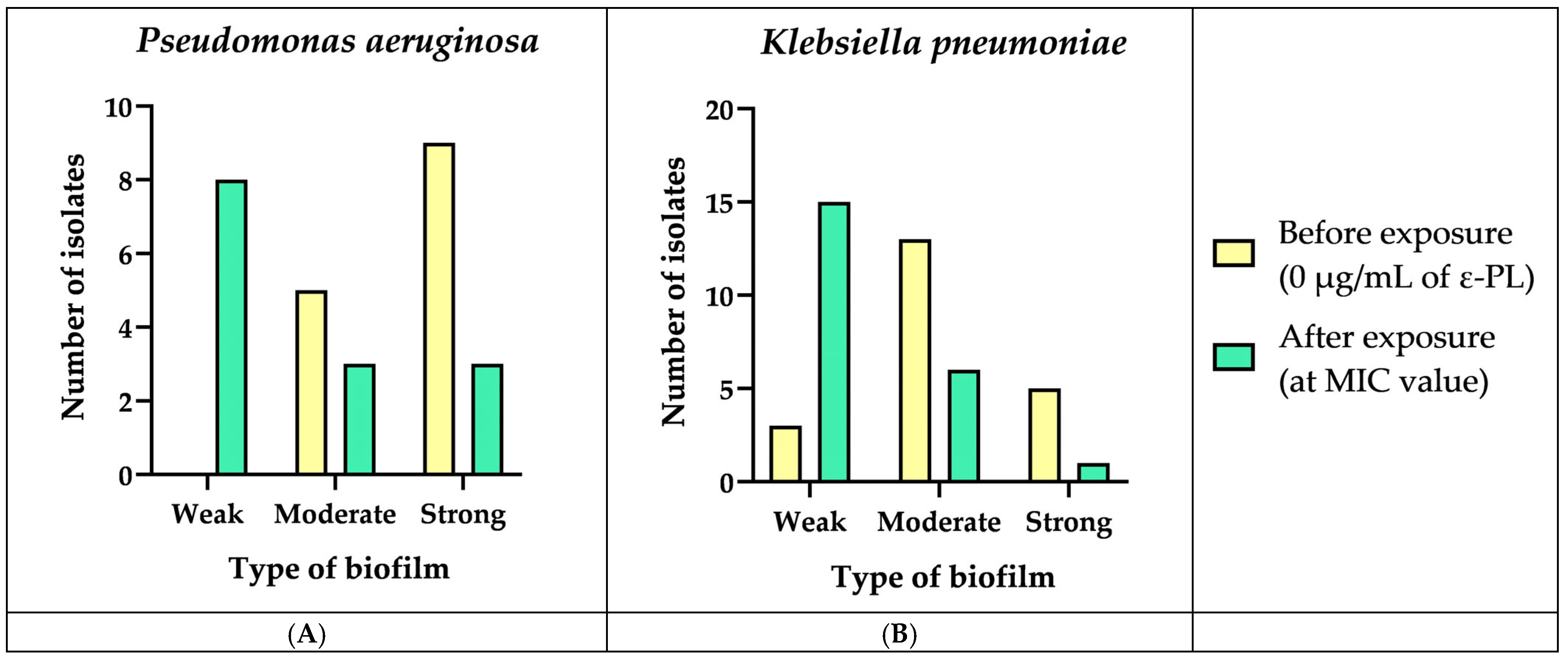
3.5. Biofilm Formation and Biomass Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riquelme, S.A.; Ahn, D.; Prince, A. Pseudomonas aeruginosa and Klebsiella pneumoniae Adaptation to Innate Immune Clearance Mechanisms in the Lung. J. Innate Immun. 2018, 10, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Yu, V.L.; Hansen, D.S.; Wen, C.K.; Sagnimeni, A.; Klugman, K.P.; Von Gottberg, A.; Goossens, H.; Wagener, M.M.; Benedi, V.J.; Casellas, J.M.; et al. Virulence Characteristics of Klebsiella and Clinical Manifestations of K. pneumoniae Bloodstream Infections. Emerg. Infect. Dis. 2007, 13, 986. [Google Scholar] [CrossRef] [PubMed]
- van Delden, C. Pseudomonas aeruginosa Bloodstream Infections: How Should We Treat Them? Int. J. Antimicrob. Agents 2007, 30, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Curran, C.S.; Bolig, T.; Torabi-Parizi, P. Mechanisms and Targeted Therapies for Pseudomonas aeruginosa Lung Infection. Am. J. Respir. Crit. Care Med. 2018, 197, 708–727. [Google Scholar] [CrossRef]
- Ferreiro, J.L.L.; Otero, J.Á.; González, L.G.; Lamazares, L.N.; Blanco, A.A.; Sanjurjo, J.R.B.; Conde, I.R.; Soneira, M.F.; De La Fuente Aguado, J. Pseudomonas aeruginosa Urinary Tract Infections in Hospitalized Patients: Mortality and Prognostic Factors. PLoS ONE 2017, 12, e0178178. [Google Scholar] [CrossRef]
- Mariana Cristea, O.; Silvia Avrămescu, C.; Bălășoiu, M.; Popescu, F.; Popescu, F.; Amzoiu, M. Original Paper Urinary Tract Infection with Klebsiella pneumoniae in Patients with Chronic Kidney Disease. Curr. Health Sci. J. 2017, 43, 137–148. [Google Scholar] [CrossRef]
- Kode, M.; Waso-Reyneke, M.; Reyneke, B.; Denissen, J.; Clements-Decker, T.; Havenga, B.; Khan, S.; Khan, W. Integration of Bdellovibrio spp. with SODIS and Moringa Oleifera Flocculation to Target Multi-Drug Resistant Klebsiella pneumoniae and Pseudomonas aeruginosa. J. Environ. Chem. Eng. 2022, 10, 107962. [Google Scholar] [CrossRef]
- Maraolo, A.E.; Cascella, M.; Corcione, S.; Cuomo, A.; Nappa, S.; Borgia, G.; De Rosa, F.G.; Gentile, I. Management of Multidrug-Resistant Pseudomonas aeruginosa in the Intensive Care Unit: State of the Art. Expert Rev. Anti-Infect. Ther. 2017, 15, 861–871. [Google Scholar] [CrossRef]
- Bassetti, M.; Righi, E.; Carnelutti, A.; Graziano, E.; Russo, A. Multidrug-Resistant Klebsiella pneumoniae: Challenges for Treatment, Prevention and Infection Control. Expert Rev. Anti-Infect. Ther. 2018, 16, 749–761. [Google Scholar] [CrossRef]
- Thi, M.T.T.; Wibowo, D.; Rehm, B.H.A. Pseudomonas aeruginosa Biofilms. Int. J. Mol. Sci. 2020, 21, 8671. [Google Scholar] [CrossRef]
- Wang, G.; Zhao, G.; Chao, X.; Xie, L.; Wang, H. The Characteristic of Virulence, Biofilm and Antibiotic Resistance of Klebsiella pneumoniae. Int. J. Environ. Res. Public Health 2020, 17, 6278. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, T.; Hébraud, M.; Enes Dapkevicius, M.L.N.; Maltez, L.; Pereira, J.E.; Capita, R.; Alonso-Calleja, C.; Igrejas, G.; Poeta, P. Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2021, 22, 12892. [Google Scholar] [CrossRef]
- Warszynski, P.; Li, Y.; Zhou, C.; Chen, S.; Huang, S. Recent Advances in Epsilon-Poly-L-Lysine and L-Lysine-Based Dendrimer Synthesis, Modification, and Biomedical Applications. Front. Chem. 2021, 1, 659304. [Google Scholar] [CrossRef]
- Shukla, S.C.; Singh, A.; Pandey, A.K.; Mishra, A. Review on Production and Medical Applications of ε-Polylysine. Biochem. Eng. J. 2012, 65, 70–81. [Google Scholar] [CrossRef]
- Takehara, M.; Hibino, A.; Saimura, M.; Hirohara, H. High-Yield Production of Short Chain Length Poly(ε-L-Lysine) Consisting of 5–20 Residues by Streptomyces aureofaciens, and Its Antimicrobial Activity. Biotechnol. Lett. 2010, 32, 1299–1303. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wu, R.; Wang, C.; Wu, Z. Effects of ε-Polylysine on Pseudomonas aeruginosa and Aspergillus fumigatus Biofilm in Vitro. Med. Sci. Monit. 2017, 23, 4225–4229. [Google Scholar] [CrossRef] [PubMed][Green Version]
- The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters, version 12.0; The European Committee on Antimicrobial Susceptibility Testing: Växjö, Sweden, 2022. [Google Scholar]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2021; ISBN 9781684401048. [Google Scholar]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011, 47, e2437. [Google Scholar] [CrossRef]
- Dashti, A.A.; Jadaon, M.M.; Abdulsamad, A.M.; Dashti, H.M. Heat Treatment of Bacteria: A Simple Method of DNA Extraction for Molecular Techniques. Kuwait Med. J. 2009, 41, 117–122. [Google Scholar]
- Perreten, V.; Boerlin, P. A New Sulfonamide Resistance Gene (Sul3) in Escherichia coli Is Widespread in the Pig Population of Switzerland. Antimicrob. Agents Chemother. 2003, 47, 1169–1172. [Google Scholar] [CrossRef]
- Schnellmann, C.; Gerber, V.; Rossano, A.; Jaquier, V.; Panchaud, Y.; Doherr, M.G.; Thomann, A.; Straub, R.; Perreten, V. Presence of New mecA and Mph(C) Variants Conferring Antibiotic Resistance in Staphylococcus spp. Isolated from the Skin of Horses before and after Clinic Admission. J. Clin. Microbiol. 2006, 44, 4444–4454. [Google Scholar] [CrossRef]
- Maynard, C.; Fairbrother, J.M.; Bekal, S.; Sanschagrin, F.; Levesque, R.C.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Antimicrobial Resistance Genes in Enterotoxigenic Escherichia coli O149:K91 Isolates Obtained over a 23-Year Period from Pigs. Antimicrob. Agents Chemother. 2003, 47, 3214–3221. [Google Scholar] [CrossRef] [PubMed]
- Mazel, D.; Dychinco, B.; Webb, V.A.; Davies, J. Antibiotic Resistance in the ECOR Collection: Integrons and Identification of a Novel Aad Gene. Antimicrob. Agents Chemother. 2000, 44, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Sáenz, Y.; Briñas, L.; Domínguez, E.; Ruiz, J.; Zarazaga, M.; Vila, J.; Torres, C. Mechanisms of Resistance in Multiple-Antibiotic-Resistant Escherichia coli Strains of Human, Animal, and Food Origins. Antimicrob. Agents Chemother. 2004, 48, 3959–3967. [Google Scholar] [CrossRef] [PubMed]
- Guardabassi, L.; Dijkshoorn, L.; Collard, J.M.; Olsen, J.E.; Dalsgaard, A. Distribution and In-Vitro Transfer of Tetracycline Resistance Determinants in Clinical and Aquatic Acinetobacter Strains. J. Med. Microbiol. 2000, 49, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Yang, J.; Ju, Z.; Chang, W.; Sun, S. Molecular Characterization of Antimicrobial Resistance in Escherichia coli from Rabbit Farms in Tai’an, China. BioMed Res. Int. 2018, 2018, 8607647. [Google Scholar] [CrossRef] [PubMed]
- Madsen, L.; Aarestrup, F.M.; Olsen, J.E. Characterisation of Streptomycin Resistance Determinants in Danish Isolates of Salmonella typhimurium. Vet. Microbiol. 2000, 75, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hinz, A.J.; Nadeau, J.P.; Mah, T.F. Pseudomonas aeruginosa tssC1 Links Type VI Secretion and Biofilm-Specific Antibiotic Resistance. J. Bacteriol. 2011, 193, 5510–5513. [Google Scholar] [CrossRef]
- Van de Klundert, J.A.M.; Vliegenthart, J.S.; Rather, P.N.; Shaw, K.J.; Hare, R.S.; Miller, G.H. Nomenclature of Aminoglycoside Resistance Genes: A Comment. Antimicrob. Agents Chemother. 1993, 37, 927–928. [Google Scholar] [CrossRef][Green Version]
- Vila, J.; Ruiz, J.; Marco, F.; Barcelo, A.; Goni, P.; Giralt, E.; De Anta, T.J. Association between Double Mutation in gyrA Gene of Ciprofloxacin- Resistant Clinical Isolates of Escherichia coli and MICs. Antimicrob. Agents Chemother. 1994, 38, 2477–2479. [Google Scholar] [CrossRef]
- Schaber, J.A.; Carty, N.L.; McDonald, N.A.; Graham, E.D.; Cheluvappa, R.; Griswold, J.A.; Hamood, A.N. Analysis of Quorum Sensing-Deficient Clinical Isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 2004, 53, 841–853. [Google Scholar] [CrossRef]
- Neyestanaki, D.K.; Mirsalehian, A.; Rezagholizadeh, F.; Jabalameli, F.; Taherikalani, M.; Emaneini, M. Determination of Extended Spectrum Beta-Lactamases, Metallo-Beta-Lactamases and AmpC-Beta-Lactamases among Carbapenem Resistant Pseudomonas aeruginosa Isolated from Burn Patients. Burns 2014, 40, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Mona, S.; Nour, M.A.K.; ElSheshtawy, N.M. Genetic Identification of Pseudomonas aeruginosa Virulence Genes among Different Isolates. J. Microb. Biochem. Technol. 2015, 7, 274–277. [Google Scholar] [CrossRef]
- Hong, S.S.; Kim, K.; Huh, J.Y.; Jung, B.; Kang, M.S.; Hong, S.G. Multiplex PCR for Rapid Detection of Genes Encoding Class A Carbapenemases. Ann. Lab. Med. 2012, 32, 359–361. [Google Scholar] [CrossRef] [PubMed]
- Galdino, A.C.M.; Viganor, L.; Ziccardi, M.; Nunes, A.P.F.; dos Santos, K.R.N.; Branquinha, M.H.; Santos, A.L.S. Heterogeneous Production of Proteases from Brazilian Clinical Isolates of Pseudomonas aeruginosa. Enferm. Infecc. Microbiol. Clin. 2017, 35, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Ellington, M.J.; Kistler, J.; Livermore, D.M.; Woodford, N. Multiplex PCR for Rapid Detection of Genes Encoding Acquired Metallo-β-Lactamases. J. Antimicrob. Chemother. 2007, 59, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Shanthi Amudhan, M.; Sekar, U.; Kamalanathan, A.; Balaraman, S. blaIMP and blaVIM Mediated Carbapenem Resistance in Pseudomonas and Acinetobacter Species in India. J. Infect. Dev. Ctries. 2012, 6, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Alm, R.A.; Mattick, J.S. Identification of a Gene, pilV, Required for Type 4 Fimbrial Biogenesis in Pseudomonas aeruginosa, Whose Product Possesses a Pre-pilin-like Leader Sequence. Mol. Microbiol. 1995, 16, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Ali, Z.; Riaz, M.; Zeshan, B.; Wattoo, J.I.; Aslam, M.N. Evaluation of Antibiotic Resistance and Virulence Genes among Clinical Isolates of Pseudomonas aeruginosa from Cancer Patients. Asian Pac. J. Cancer Prev. 2020, 21, 1333–1338. [Google Scholar] [CrossRef]
- Papa-Ezdra, R.; Bado, I.; Caiata, L.; Vignoli, R.; Seija, V. First Report of Pseudomonas aeruginosa Co-Harbouring blaVIM-2 and blaPER-1 in Latin America. J. Glob. Antimicrob. Resist. 2018, 15, 121–122. [Google Scholar] [CrossRef]
- Holbrook, S.Y.L.; Garneau-Tsodikova, S. Evaluation of Aminoglycoside and Carbapenem Resistance in a Collection of Drug-Resistant Pseudomonas aeruginosa Clinical Isolates. Microb. Drug Resist. 2018, 24, 1020–1030. [Google Scholar] [CrossRef]
- Lomonaco, S.; Crawford, M.A.; Lascols, C.; Timme, R.E.; Anderson, K.; Hodge, D.R.; Fisher, D.J.; Pillai, S.P.; Morse, S.A.; Khan, E.; et al. Resistome of Carbapenem- and Colistin-Resistant Klebsiella pneumoniae Clinical Isolates. PLoS ONE 2018, 13, e0198526. [Google Scholar] [CrossRef] [PubMed]
- Founou, R.C.; Founou, L.L.; Allam, M.; Ismail, A.; Essack, S.Y. Whole Genome Sequencing of Extended Spectrum β-Lactamase (ESBL)-Producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa. Sci. Rep. 2019, 9, 6266. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, M.; Castillo-Polo, J.A.; Cordero, D.G.; Pérez-Viso, B.; García-Castillo, M.; de la Fuente, J.S.; Morosini, M.I.; Cantón, R.; Ruiz-Garbajosa, P. Impact of Ceftazidime-Avibactam Treatment in the Emergence of Novel KPC Variants in the ST307-Klebsiella pneumoniae High-Risk Clone and Consequences for Their Routine Detection. J. Clin. Microbiol. 2022, 60, e02245-21. [Google Scholar] [CrossRef] [PubMed]
- Tekeli, A.; Dolapci, I.; Evren, E.; Oguzman, E.; Karahan, Z.C. Characterization of Klebsiella pneumoniae Coproducing KPC and NDM-1 Carbapenemases from Turkey. Microb. Drug Resist. 2020, 26, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Mendes, G.; Ramalho, J.F.; Bruschy-Fonseca, A.; Lito, L.; Duarte, A.; Melo-Cristino, J.; Caneiras, C. First Description of Ceftazidime/Avibactam Resistance in a ST13 KPC-70-Producing Klebsiella pneumoniae Strain from Portugal. Antibiotics 2022, 11, 167. [Google Scholar] [CrossRef]
- Biedrzycka, M.; Urbanowicz, P.; Guzek, A.; Brisse, S.; Gniadkowski, M.; Izdebski, R. Dissemination of Klebsiella pneumoniae ST147 NDM-1 in Poland, 2015–2019. J. Antimicrob. Chemother. 2021, 76, 2538–2545. [Google Scholar] [CrossRef] [PubMed]
- Novais, Â.; Ferraz, R.V.; Viana, M.; da Costa, P.M.; Peixe, L. NDM-1 Introduction in Portugal through a ST11 KL105 Klebsiella pneumoniae Widespread in Europe. Antibiotics 2022, 11, 92. [Google Scholar] [CrossRef] [PubMed]
- Messaoudi, A.; Haenni, M.; Bouallègue, O.; Saras, E.; Chatre, P.; Chaouch, C.; Boujâafar, N.; Mansour, W.; Madec, J.Y. Dynamics and Molecular Features of OXA-48-like-Producing Klebsiella pneumoniae Lineages in a Tunisian Hospital. J. Glob. Antimicrob. Resist. 2020, 20, 87–93. [Google Scholar] [CrossRef]
- Gurung, S.; Kafle, S.; Dhungel, B.; Adhikari, N.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Detection of Oxa-48 Gene in Carbapenem-Resistant Escherichia coli and Klebsiella pneumoniae from Urine Samples. Infect. Drug Resist. 2020, 13, 2311–2321. [Google Scholar] [CrossRef]
- Protonotariou, E.; Poulou, A.; Politi, L.; Sgouropoulos, I.; Metallidis, S.; Kachrimanidou, M.; Pournaras, S.; Tsakris, A.; Skoura, L. Hospital Outbreak Due to a Klebsiella pneumoniae ST147 Clonal Strain Co-Producing KPC-2 and VIM-1 Carbapenemases in a Tertiary Teaching Hospital in Northern Greece. Int. J. Antimicrob. Agents 2018, 52, 331–337. [Google Scholar] [CrossRef]
- Dziri, R.; Ayari, I.; Barguellil, F.; Ouzari, H.I.; El Asli, M.S.; Klibi, N. First Report of NDM and VIM Coproducing Klebsiella pneumoniae in Tunisia and Emergence of Novel Clones. Microb. Drug Resist. 2019, 25, 1282–1286. [Google Scholar] [CrossRef] [PubMed]
- Khodadadian, R.; Rahdar, H.A.; Javadi, A.; Safari, M.; Khorshidi, A. Detection of VIM-1 and IMP-1 Genes in Klebsiella pneumoniae and Relationship with Biofilm Formation. Microb. Pathog. 2018, 115, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, X.; Wang, Q.; Chen, H.; Li, H.; Wang, S.; Wang, R.; Wang, H. Emergence of Tigecycline Nonsusceptible and IMP-4 Carbapenemase-Producing K2-ST65 Hypervirulent Klebsiella pneumoniae in China. Microbiol. Spectr. 2021, 9, e01305-21. [Google Scholar] [CrossRef] [PubMed]
- Literacka, E.; Izdebski, R.; Urbanowicz, P.; Zabicka, D.; Klepacka, J.; Sowa-Sierant, I.; Zak, I.; Garus-Jakubowska, A.; Hryniewicz, W.; Gniadkowski, M. Spread of Klebsiella pneumoniae ST45 Producing GES-5 Carbapenemase or GES-1 Extended-Spectrum β-Lactamase in Newborns and Infants. Antimicrob. Agents Chemother. 2020, 64, e00595-20. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Suh, H.S. Klebsiella pneumoniae ST11 Producing GES-5 Carbapenemase Isolated from Tertiary-Care Hospital. Clin. Lab. 2021, 67, 2189–2194. [Google Scholar] [CrossRef] [PubMed]
- Mbelle, N.M.; Feldman, C.; Sekyere, J.O.; Maningi, N.E.; Modipane, L.; Essack, S.Y. Pathogenomics and Evolutionary Epidemiology of Multi-Drug Resistant Clinical Klebsiella pneumoniae Isolated from Pretoria, South Africa. Sci. Rep. 2020, 10, 1232. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, I.; Chenouf, N.S.; Carvalho, J.A.; Castro, A.P.; Silva, V.; Capita, R.; Alonso-Calleja, C.; de Lurdes Nunes Enes Dapkevicius, M.; Igrejas, G.; Torres, C.; et al. Multidrug-Resistant Klebsiella pneumoniae Harboring Extended Spectrum β-Lactamase Encoding Genes Isolated from Human Septicemias. PLoS ONE 2021, 16, e0250525. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhao, Z.; Ye, C.; Zhou, S.; Wu, S.; Han, L.; Han, Z.; Ye, H. Molecular Characterization of Carbapenem-Resistant Klebsiella pneumoniae Isolates with Focus on Antimicrobial Resistance. BMC Genom. 2019, 20, 822. [Google Scholar] [CrossRef]
- Skočková, A.; Cupáková, Š.; Karpíšková, R.; Janštová, B.; Skočková, A.; Cupáková, Š.; Karpíšková, R.; Janštová, B. Detection of Tetracycline Resistance Genes in Escherichia coli from Raw Cow’s Milk. J. Microbiol. Biotechnol. Food Sci. 2012, 1, 777–784. [Google Scholar]
- Grossman, T.H. Tetracycline Antibiotics and Resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025387. [Google Scholar] [CrossRef]
- Vubil, D.; Figueiredo, R.; Reis, T.; Canha, C.; Boaventura, L.; Da Silva, G.J. Outbreak of KPC-3-Producing ST15 and ST348 Klebsiella pneumoniae in a Portuguese Hospital. Epidemiol. Infect. 2017, 145, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Aires-De-Sousa, M.; De La Rosa, J.M.O.; Gonçalves, M.L.; Pereira, A.L.; Nordmann, P.; Poirel, L. Epidemiology of Carbapenemase-Producing Klebsiella pneumoniae in a Hospital, Portugal. Emerg. Infect. Dis. 2019, 25, 1632. [Google Scholar] [CrossRef] [PubMed]
- Perdigão, J.; Modesto, A.; Pereira, A.L.; Neto, O.; Matos, V.; Godinho, A.; Phelan, J.; Charleston, J.; Spadar, A.; de Sessions, P.F.; et al. Whole-Genome Sequencing Resolves a Polyclonal Outbreak by Extended-Spectrum Beta-Lactam and Carbapenem-Resistant Klebsiella pneumoniae in a Portuguese Tertiary-Care Hospital. Microb. Genom. 2020, 7, 349. [Google Scholar] [CrossRef] [PubMed]
- Bachiri, T.; Bakour, S.; Ladjouzi, R.; Thongpan, L.; Rolain, J.M.; Touati, A. High Rates of CTX-M-15-Producing Escherichia coli and Klebsiella pneumoniae in Wild Boars and Barbary Macaques in Algeria. J. Glob. Antimicrob. Resist. 2017, 8, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Koreň, J.; Andrezál, M.; Drahovská, H.; Hubenáková, Z.; Liptáková, A.; Maliar, T. Next-Generation Sequencing of Carbapenem-Resistant Klebsiella pneumoniae Strains Isolated from Patients Hospitalized in the University Hospital Facilities. Antibiotics 2022, 11, 1538. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, D.; Ji, B.; Zhang, X.; Anbo, M.; Jelsbak, L. Whole-Genome Sequencing Reveals High-Risk Clones of Pseudomonas aeruginosa in Guangdong, China. Front. Microbiol. 2023, 14, 1134. [Google Scholar] [CrossRef] [PubMed]
- Gomila, M.; Del Carmen Gallegos, M.; Fernández-Baca, V.; Pareja, A.; Pascual, M.; Díaz-Antolín, P.; García-Valdés, E.; Lalucat, J. Genetic Diversity of Clinical Pseudomonas aeruginosa Isolates in a Public Hospital in Spain. BMC Microbiol. 2013, 13, 138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, C.; Liu, Z.; Pan, F.; Meng, R.; Bu, X.; Xing, H.; Deng, Y.; Guo, N.; Yu, L. Antibacterial Activity and Mode of Action of ε-Polylysine against Escherichia coli O157:H7. J. Med. Microbiol. 2018, 67, 838–845. [Google Scholar] [CrossRef]
- Cezard, A.; Fouquenet, D.; Vasseur, V.; Jeannot, K.; Launay, F.; Si-Tahar, M.; Hervé, V. Poly-L-Lysine to Fight Antibiotic Resistances of Pseudomonas aeruginosa. Int. J. Mol. Sci. 2023, 24, 2851. [Google Scholar] [CrossRef]
- Shen, C.; Islam, M.T.; Masuda, Y.; Honjoh, K.I.; Miyamoto, T. Transcriptional Changes Involved in Inhibition of Biofilm Formation by ε-Polylysine in Salmonella typhimurium. Appl. Microbiol. Biotechnol. 2020, 104, 5427–5436. [Google Scholar] [CrossRef]
- Shi, C.L.; Wang, X.W.; Li, A.Q.; Qian, S.H.; Wang, Z.; Zhao, S.G.; Liu, Y.; Xue, Z.L. Effect of ε-Polylysine on the Cell Structure and Biofilm Formation of Cronobacter sakazakii. Biotechnol. Bull. 2022, 38, 147. [Google Scholar] [CrossRef]
- Syed, A.K.; Ghosh, S.; Love, N.G.; Boles, B.R. Triclosan Promotes Staphylococcus aureus Nasal Colonization. mBio 2014, 5, e01015-13. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, C.; Zhang, J.; Rao, Z.; Xu, X.; Mao, Z.; Chen, X. Epsilon-Poly-L-Lysine: Recent Advances in Biomanufacturing and Applications. Front. Bioeng. Biotechnol. 2021, 9, 748976. [Google Scholar] [CrossRef]
- Li, S.; Mao, Y.; Zhang, L.; Wang, M.; Meng, J.; Liu, X.; Bai, Y.; Guo, Y. Recent Advances in Microbial ε-Poly-L-Lysine Fermentation and Its Diverse Applications. Biotechnol. Biofuels Bioprod. 2022, 15, 65. [Google Scholar] [CrossRef]
| Species | Isolate | MDR | Phenotype of Resistance | Resistance Genes |
|---|---|---|---|---|
| Pa | HU1 | − | IMI, MEM, LEV | blaKPC |
| HU2 | − | IMI | blaKPC | |
| HU4 | + | PTZ, CAZ, ATM, IMI, MEM, DOR | blaKPC | |
| HU5 | − | IMI, MEM, LEV | blaSPM | |
| HU6 | − | IMI | blaKPC, blaSPM | |
| HU7 | − | IMI | blaSPM | |
| HU8 | + | PTZ, TTC, CAZ, ATM, CIP, | blaCTX-M, blaVIM2, blaSPM | |
| HU9 | − | IMI | blaSHV | |
| HU10 | − | IMI | blaSHV, blaVIM | |
| HU11 | − | IMI | blaTEM, blaSPM | |
| HU12 | − | IMI | blaCTX-M, blaOXA | |
| HU13 | − | TOB, CN | aac(3)-IV | |
| HU14 | − | IMI, CN | blaPER, blaOXA, aac(3)-IV | |
| HU15 | − | IMI | blaKPC | |
| Kp | HS2 | + | TTC, CAZ, CTX, FEP, ATM, CN, TET, CIP, SXT | blaCTX-M, aac(3)-II, aadA1, sul2 |
| HS8 | + | TTC, FOX, CAZ, CTX, FEP, ATM, CN, TET, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, sul2 | |
| HS14 | + | TTC, CAZ, CTX, FEP, ATM, AK, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, sul2 | |
| HS17 | + | TTC, FOX, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, cmlA, sul2 | |
| HS29 | + | TTC, CAZ, CTX, FEP, ATM, TET, CIP, SXT | blaCTX-M, blaTEM, blaSHV, tetA, sul2 | |
| HS30 | + | TTC, FOX, CAZ, CTX, FEP, ATM, MEM, ERT, IMI, AK, CN, CIP, SXT | blaTEM, blaSHV, blaKPC, aac(3)-II, aadA1, sul2 | |
| HS34 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaSHV, aac(3)-II, aadA1, cmlA, sul2 | |
| HS36 | + | TTC, FOX, CAZ, CTX, FEP, ATM, MEM, ERT, AK, CN, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, blaKPC, aac(3)-II, aadA1, sul2 | |
| HS42 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, sul2, sul1 | |
| HS50 | + | TTC, CAZ, CTX, FEP, ATM, ERT, AK, CN, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, sul1, sul2 | |
| HS54 | + | TTC, CAZ, CTX, FEP, ATM, ERT, CIP, SXT | blaTEM, blaSHV, blaNDM, sul2, sul1 | |
| HS55 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, cmlA, sul2 | |
| HS56 | + | TTC, FOX, CAZ, CTX, FEP, ATM, MEM, ERT, AK, CN, CIP, NA, CHL | blaCTX-M, blaSHV, blaKPC, blaNDM, aac(3)-II, aadA1, cmlA | |
| HS57 | + | TTC, CAZ, CTX, FEP, ATM, TET, CIP, SXT | blaCTX-M, blaTEM, blaSHV, tetA, sul2 | |
| HS58 | + | TTC, CAZ, CTX, FEP, ATM, CN, TET, CIP, NA, SXT, CHL | blaCTX-M, blaSHV, aac(3)-II, cmlA, sul1 | |
| HS67 | + | CAZ, CTX, FEP, ATM, CN, CIP, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, sul2 | |
| HS77 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, sul1, sul2, sul3 | |
| HS79 | + | TTC, FOX, CAZ, CTX, FEP, ATM, ERT, CIP | blaCTX-M, blaTEM, blaKPC, blaOXA, blaNDM | |
| HS81 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, cmlA, sul1, sul2, sul3 | |
| HS84 | + | TTC, CAZ, CTX, FEP, ATM, CN, CIP, NA, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, cmlA, sul1, sul2, dfrA | |
| HS87 | + | CTX, FEP, ATM, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, sul1, sul2, dfrA | |
| HS91 | + | TTC, CAZ, CTX, FEP, ATM, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, sul1, sul2 | |
| HS92 | + | TTC, CAZ, CTX, FEP, ATM, AK, CIP, NA, SXT | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, sul1, sul2 | |
| HS101 | + | CAZ, CTX, FEP, ATM, CN, TET, CIP, SXT, CHL | blaCTX-M, blaTEM, blaSHV, aac(3)-II, aadA1, tetA, cmlA, sul1, sul2 |
| P. aeruginosa | K. pneumoniae | ||
|---|---|---|---|
| Isolate | ST | Isolate | ST |
| HU2 | 699 | HS8 | 307 |
| HU5 | X * | HS29 | 348 |
| HU6 | X * | HS34 | 15 |
| HU7 | 1338 | HS36 | 307 |
| HU10 | 285 | HS50 | 584 |
| HU14 | 274 | HS55 | 15 |
| HU15 | 1404 | HS58 | 307 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Sousa, T.; Sabença, C.; Ribeiro, M.; Pino-Hurtado, M.; Torres, C.; Hébraud, M.; Alves, O.; Sousa, S.; Costa, E.; Igrejas, G.; et al. Activity of Epsilon-poly-L-lysine against Multidrug-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates of Urinary Tract Infections. Biomedicines 2024, 12, 638. https://doi.org/10.3390/biomedicines12030638
de Sousa T, Sabença C, Ribeiro M, Pino-Hurtado M, Torres C, Hébraud M, Alves O, Sousa S, Costa E, Igrejas G, et al. Activity of Epsilon-poly-L-lysine against Multidrug-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates of Urinary Tract Infections. Biomedicines. 2024; 12(3):638. https://doi.org/10.3390/biomedicines12030638
Chicago/Turabian Stylede Sousa, Telma, Carolina Sabença, Miguel Ribeiro, Mario Pino-Hurtado, Carmen Torres, Michel Hébraud, Olimpia Alves, Sara Sousa, Eliana Costa, Gilberto Igrejas, and et al. 2024. "Activity of Epsilon-poly-L-lysine against Multidrug-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates of Urinary Tract Infections" Biomedicines 12, no. 3: 638. https://doi.org/10.3390/biomedicines12030638
APA Stylede Sousa, T., Sabença, C., Ribeiro, M., Pino-Hurtado, M., Torres, C., Hébraud, M., Alves, O., Sousa, S., Costa, E., Igrejas, G., & Poeta, P. (2024). Activity of Epsilon-poly-L-lysine against Multidrug-Resistant Pseudomonas aeruginosa and Klebsiella pneumoniae Isolates of Urinary Tract Infections. Biomedicines, 12(3), 638. https://doi.org/10.3390/biomedicines12030638













