Delivery of a Hepatitis C Virus Vaccine Encoding NS3 Linked to the MHC Class II Chaperone Protein Invariant Chain Using Bacterial Ghosts
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Ethics Declaration
2.2. Preparation of E. coli Bacterial Ghosts
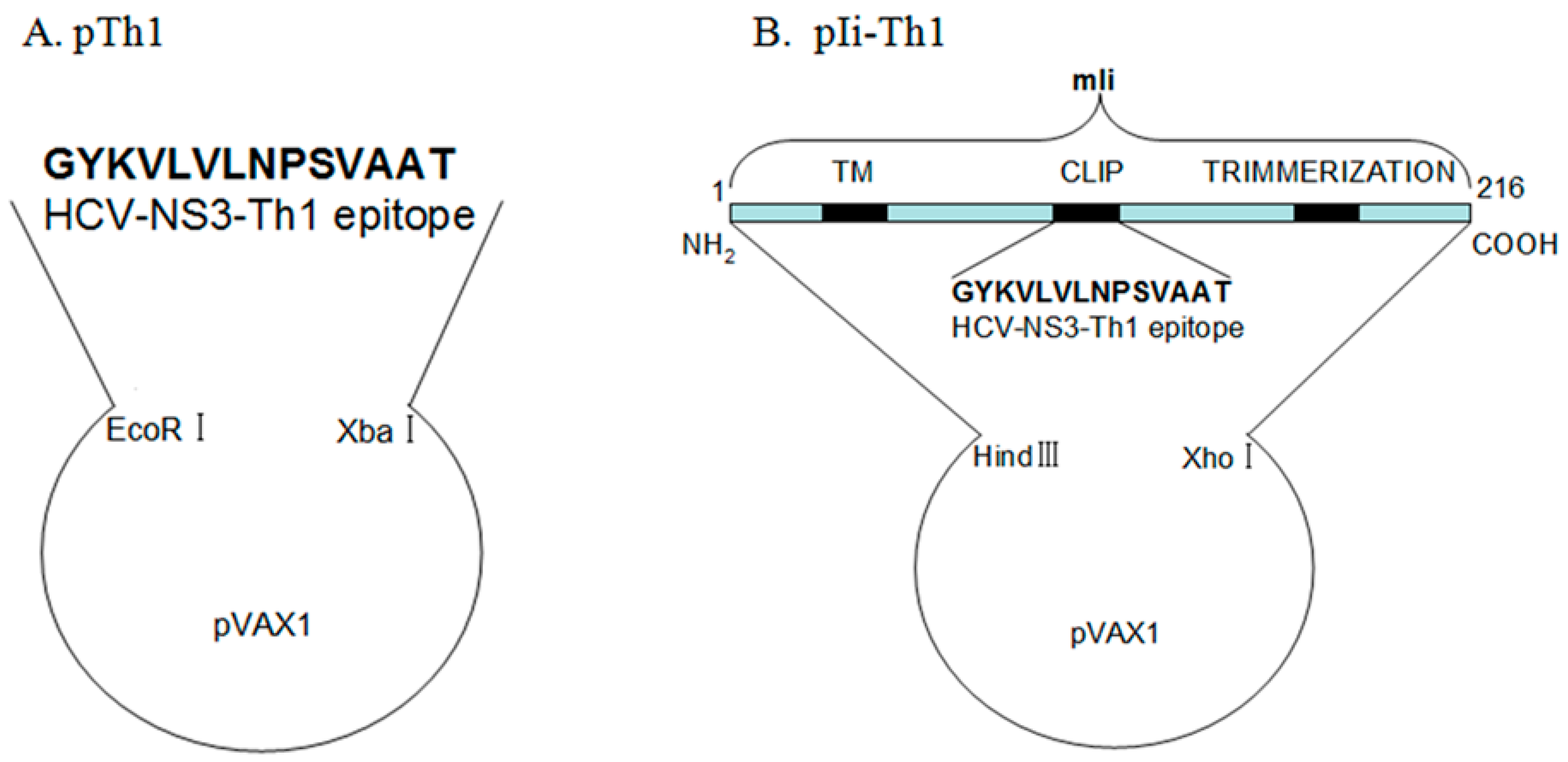
2.3. Plasmid Construction
2.4. Loading the BGs with the pVAX1-Ii-NS3 Th1 Plasmid
2.5. Transfection Experiments
2.6. Mouse Immunization
2.7. Anti-NS3 ELISA
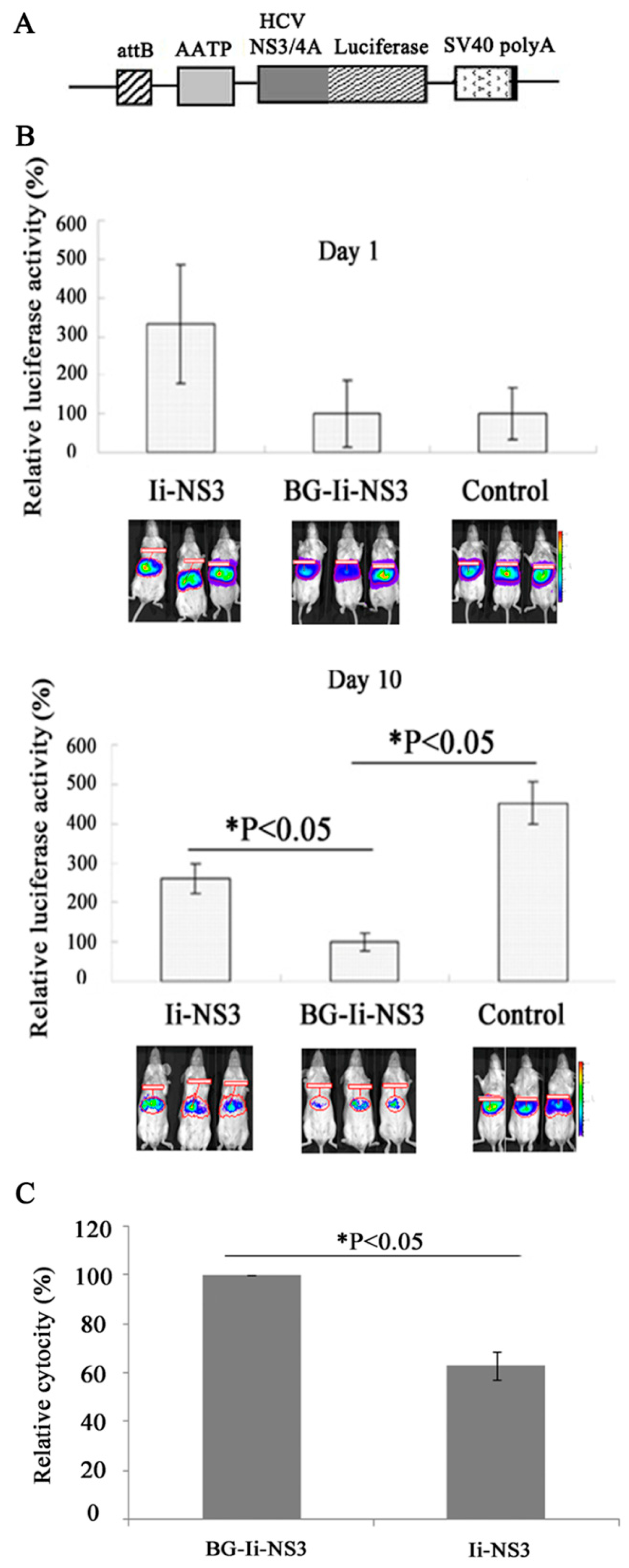
2.8. A Reporter Mouse Model That Allows Noninvasive Detection of HCV NS3/4A Expression in the Liver
2.9. Luciferase Activity Monitoring in Living Mice Using an IVIS Camera [28]
2.10. Assessment of Specific Cell Lysis [29,30]
(Maximal Target value − Spontaneous Target value).
2.11. Statistical Analysis
3. Results
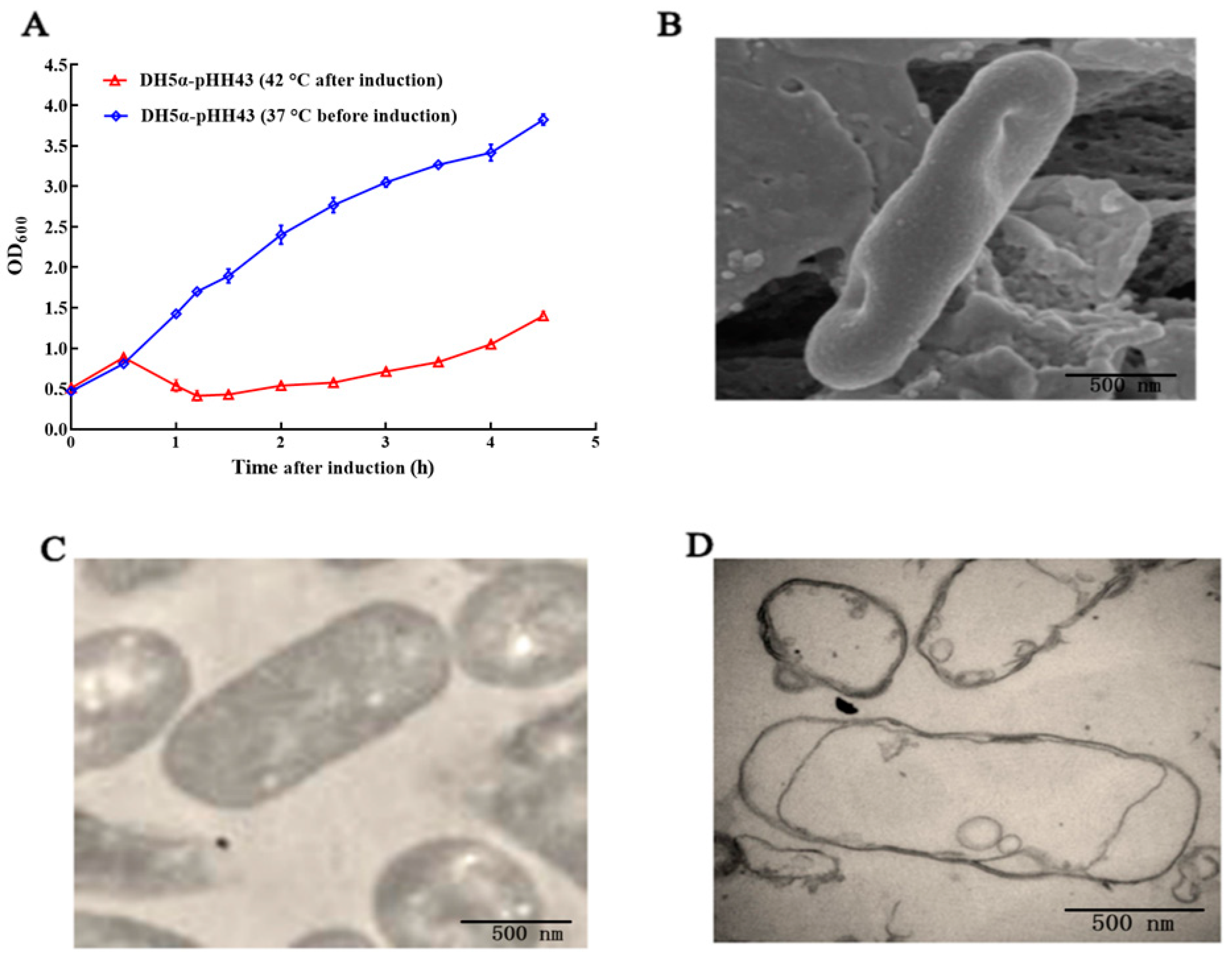
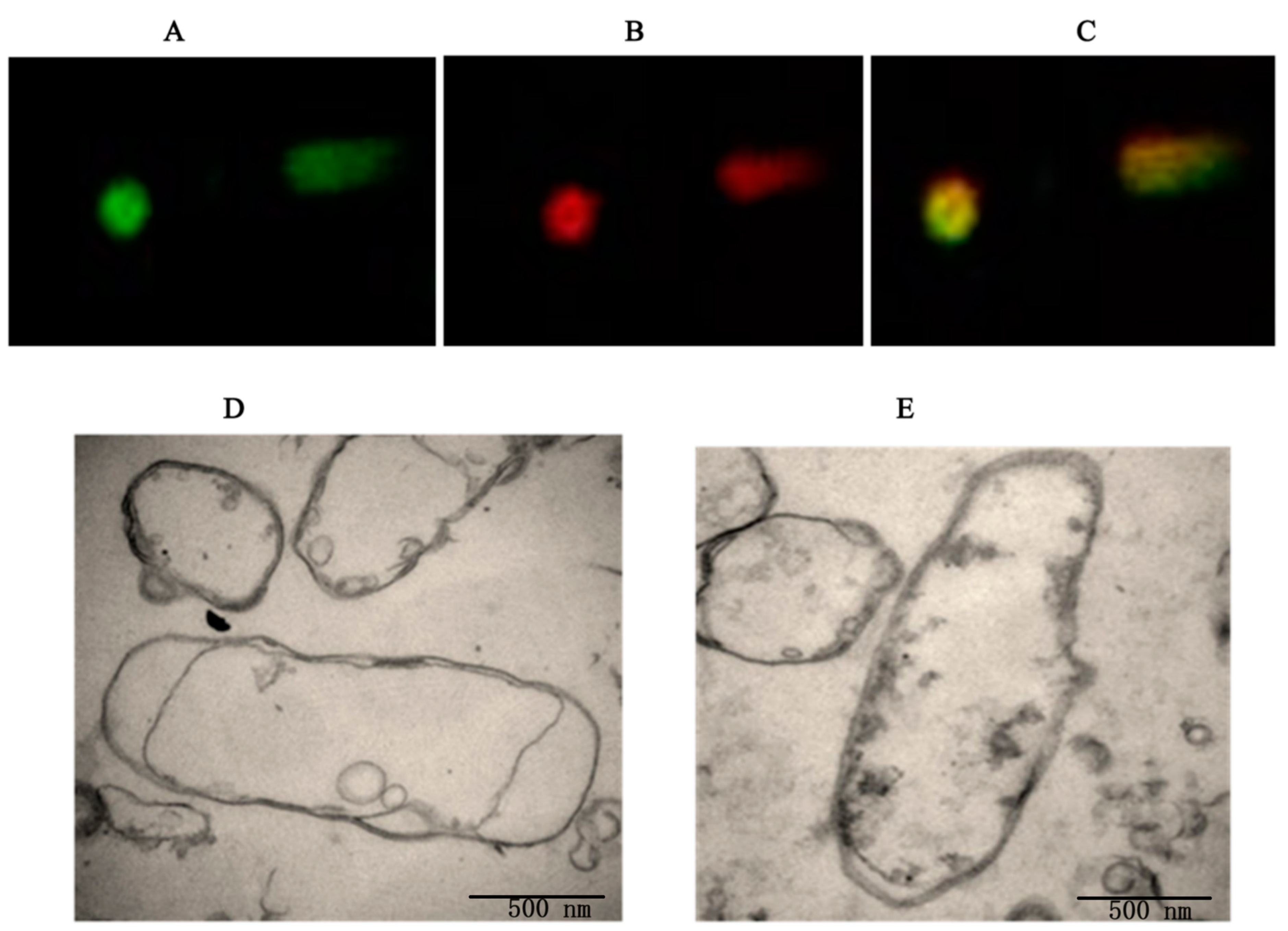
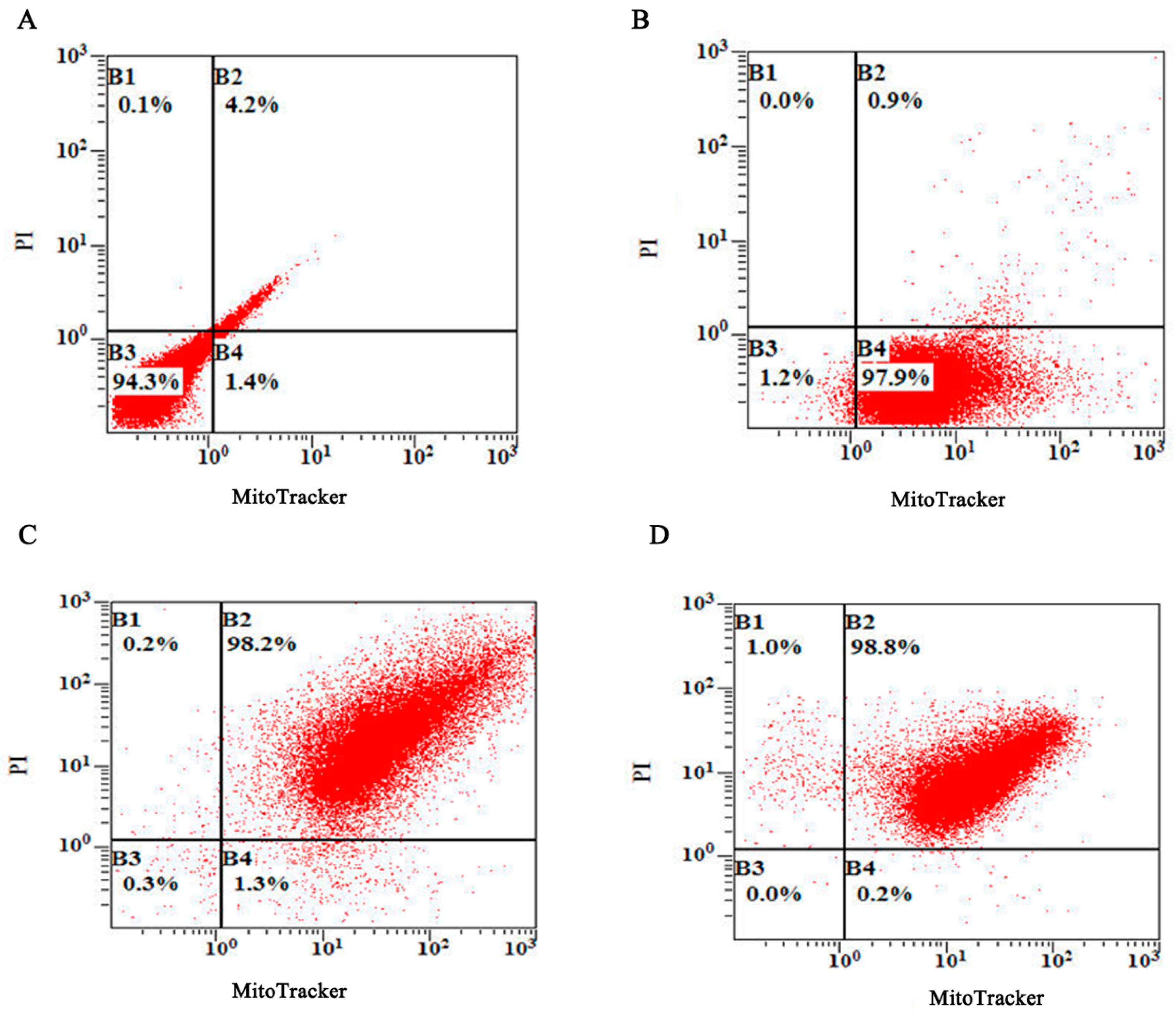
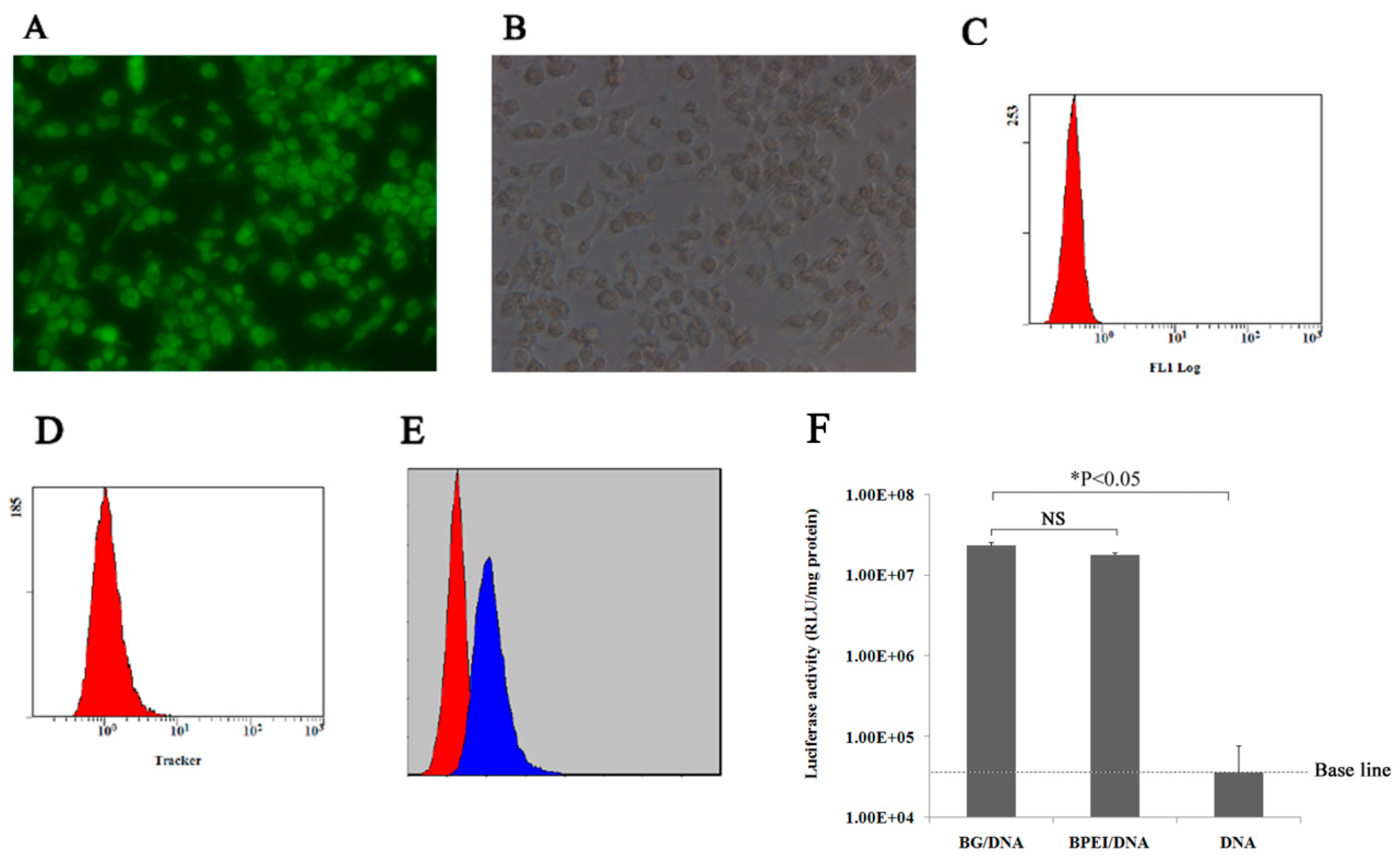
3.1. Preparation and Characterization of the DH5α BGs
3.2. Determination of the Plasmid DNA Loading Efficiency of the BGs
3.3. Evaluating the Efficacy of BG-Mediated Transfection In Vitro
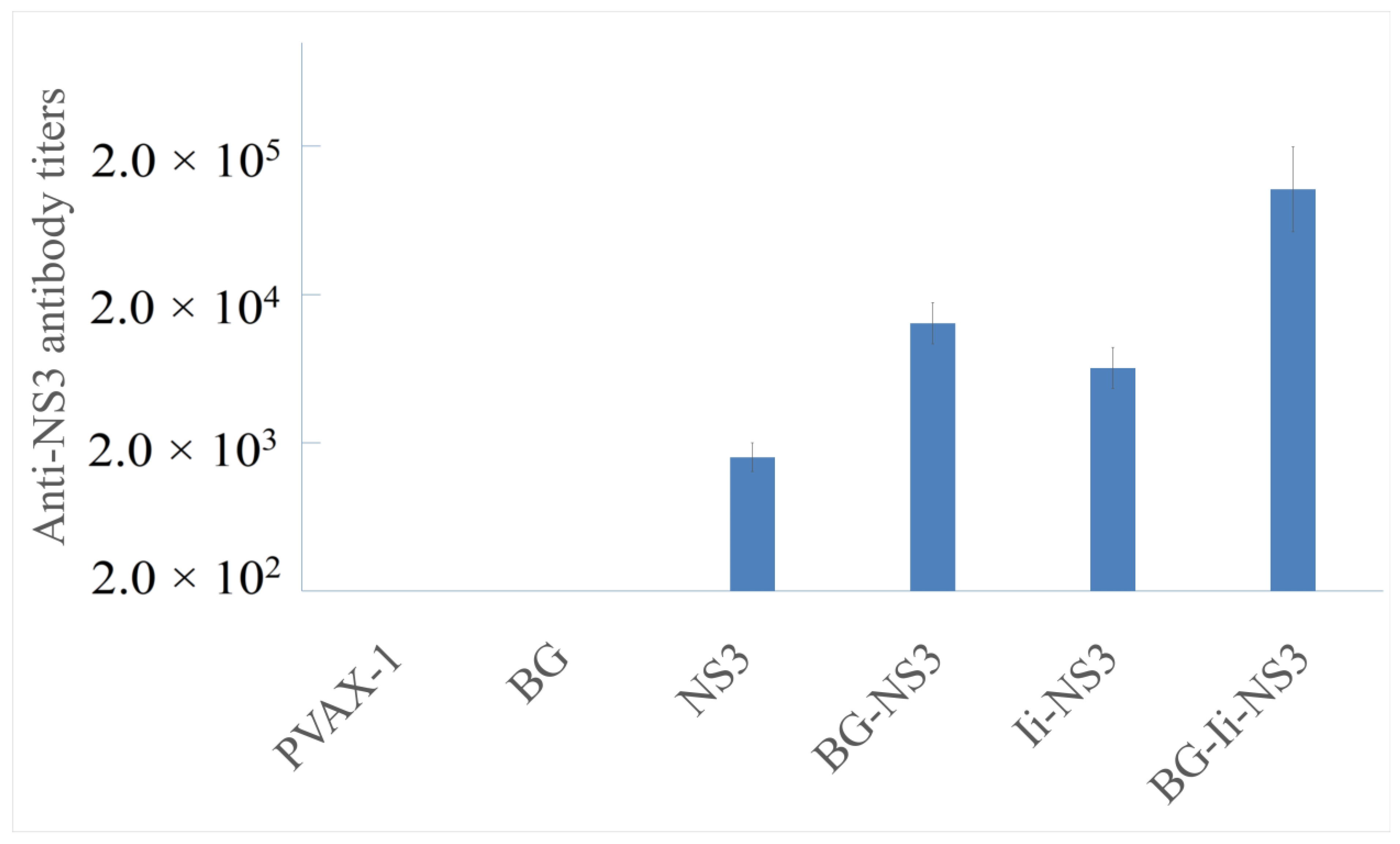
3.4. Humoral Immune Responses following Immunization with Plasmid Vaccines Loaded in Bacterial Ghosts or Free Plasmids
3.5. Evaluation of the Immunogenicity of the DNA Vaccine in the Mouse Model
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roudot-Thoraval, F. Epidemiology of hepatitis C virus infection. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101596. [Google Scholar] [CrossRef] [PubMed]
- Diepolder, H.M.; Zachoval, R.; Hoffmann, R.M.; Wierenga, E.A.; Santantonio, T.; Jung, M.C.; Eichenlaub, D.; Pape, G.R. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 1995, 346, 1006–1007. [Google Scholar] [CrossRef]
- Lang Kuhs, K.A.; Ginsberg, A.A.; Yan, J.; Wiseman, R.W.; Khan, A.S.; Sardesai, N.Y.; O’Connor, D.H.; Weiner, D.B. Hepatitis C virus NS3/NS4A DNA vaccine induces multiepitope T cell responses in rhesus macaques mimicking human immune responses. Mol. Ther. 2012, 20, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Basirnejad, M.; Bolhassani, A.; Sadat, S.M. The Distinct Role of Small Heat Shock Protein 20 on HCV NS3 Expression in HEK-293T Cell Line. Avicenna J. Med. Biotechnol. 2018, 10, 152–157. [Google Scholar] [PubMed]
- Hilligan, K.L.; Ronchese, F. Antigen presentation by dendritic cells and their instruction of CD4+ T helper cell responses. Cell Mol. Immunol. 2020, 17, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wu, S.; Blackwell, C.E.; Humphreys, R.E.; von Hofe, E.; Xu, M. Suppression of major histocompatibility complex class II-associated invariant chain enhances the potency of an HIV gp120 DNA vaccine. Immunology 2007, 120, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Senju, S.; Chen, Y.Z.; Ando, M.; Matsushita, S.; Nishimura, Y. The CLIP-substituted invariant chain efficiently targets an antigenic peptide to HLA class II pathway in L cells. Hum. Immunol. 1998, 59, 607–614. [Google Scholar] [CrossRef] [PubMed]
- Wälchli, S.; Kumari, S.; Fallang, L.E.; Sand, K.M.; Yang, W.; Landsverk, O.J.; Bakke, O.; Olweus, J.; Gregers, T.F. Invariant chain as a vehicle to load antigenic peptides on human MHC class I for cytotoxic T-cell activation. Eur. J. Immunol. 2014, 44, 774–784. [Google Scholar] [CrossRef]
- Mensali, N.; Grenov, A.; Pati, N.B.; Dillard, P.; Myhre, M.R.; Gaudernack, G.; Kvalheim, G.; Inderberg, E.M.; Bakke, O.; Wälchli, S. Antigen-delivery through invariant chain (CD74) boosts CD8 and CD4 T cell immunity. Oncoimmunology 2019, 8, 1558663. [Google Scholar] [CrossRef]
- Gao, M.; Wang, H.P.; Wang, Y.N.; Zhou, Y.; Wang, Q.L. Target HCV NS3 CD4+ Th1 epitope to major histocompatibility complex class II pathway. Biotechnol. Lett. 2006, 28, 3–8. [Google Scholar] [CrossRef]
- Gao, M.; Wang, H.P.; Wang, Y.N.; Zhou, Y.; Wang, Q.L. HCV-NS3 Th1 minigene vaccine based on invariant chain CLIP genetic substitution enhances CD4(+) Th1 cell responses in vivo. Vaccine 2006, 24, 5491–5497. [Google Scholar] [CrossRef] [PubMed]
- Pecora, A.; Malacari, D.A.; Perez Aguirreburualde, M.S.; Bellido, D.; Nuñez, M.C.; Dus Santos, M.J.; Escribano, J.M.; Wigdorovitz, A. Development of an APC-targeted multivalent E2-based vaccine against Bovine Viral Diarrhea Virus types 1 and 2. Vaccine 2015, 33, 5163–5171. [Google Scholar] [CrossRef][Green Version]
- McCann, N.; O’Connor, D.; Lambe, T.; Pollard, A.J. Viral vector vaccines. Curr. Opin. Immunol. 2022, 77, 102210. [Google Scholar] [CrossRef]
- Holst, P.J.; Sorensen, M.R.; Mandrup Jensen, C.M.; Orskov, C.; Thomsen, A.R.; Christensen, J.P. MHC class II-associated invariant chain linkage of antigen dramatically improves cell-mediated immunity induced by adenovirus vaccines. J. Immunol. 2008, 180, 3339–3346. [Google Scholar] [CrossRef] [PubMed]
- Shirley, J.L.; de Jong, Y.P.; Terhorst, C.; Herzog, R.W. Immune Responses to Viral Gene Therapy Vectors. Mol. Ther. 2020, 28, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Butt, M.H.; Zaman, M.; Ahmad, A.; Khan, R.; Mallhi, T.H.; Hasan, M.M.; Khan, Y.H.; Hafeez, S.; Massoud, E.E.S.; Rahman, M.H.; et al. Appraisal for the Potential of Viral and Nonviral Vectors in Gene Therapy: A Review. Genes 2022, 13, 1370. [Google Scholar] [CrossRef] [PubMed]
- Zu, H.; Gao, D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, and Prospects. AAPS J. 2021, 23, 78. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Ma, J.; Dong, Q.; Liu, Q. Live bacterial vaccine vector and delivery strategies of heterologous antigen: A review. Immunol Lett. 2018, 197, 70–77. [Google Scholar] [CrossRef]
- Huter, V.; Szostak, M.P.; Gampfer, J.; Prethaler, S.; Wanner, G.; Gabor, F.; Lubitz, W. Bacterial ghosts as drug carrier and targeting vehicles. J. Control. Release 1999, 61, 51–63. [Google Scholar] [CrossRef]
- Mayr, U.B.; Walcher, P.; Azimpour, C.; Riedmann, E.; Haller, C.; Lubitz, W. Bacterial ghosts as antigen delivery vehicles. Adv. Drug Deliv. Rev. 2005, 57, 1381–1391. [Google Scholar] [CrossRef]
- Muhammad, A.; Champeimont, J.; Mayr, U.B.; Lubitz, W.; Kudela, P. Bacterial ghosts as carriers of protein subunit and DNA-encoded antigens for vaccine applications. Expert. Rev. Vaccines 2012, 11, 97–116. [Google Scholar] [CrossRef] [PubMed]
- Hajam, I.A.; Dar, P.A.; Won, G.; Lee, J.H. Bacterial ghosts as adjuvants: Mechanisms and potential. Vet. Res. 2017, 48, 37. [Google Scholar] [CrossRef] [PubMed]
- Langemann, T.; Koller, V.J.; Muhammad, A.; Kudela, P.; Mayr, U.B.; Lubitz, W. The Bacterial Ghost platform system: Production and applications. Bioeng. Bugs. 2010, 1, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.; Reis e Sousa, C.; Germain, R.N. Production, specificity, and functionality of monoclonal antibodies to specific peptide–major histocompatibility complex class II complexes formed by processing of exogenousprotein. Proc. Natl. Acad. Sci. USA 1997, 94, 13856–13861. [Google Scholar] [CrossRef] [PubMed]
- Yamato, I.; Anraku, Y.; Hirosawa, K. Cytoplasmic membrane vesicles of Escherichia coli. A simple method for preparing the cytoplasmic and outer membranes. J. Biochem. 1975, 77, 705–718S. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.K.; Gong, L.; Zhang, X.; Yun, Z.M.; Li, S.B.; Gao, H.W.; Dai, C.J.; Yuan, J.J.; Chen, J.M.; Gong, F.; et al. Antimicrobial peptide AR-23 derivatives with high endosomal disrupting ability enhance poly(l-lysine)-mediated gene transfer. J. Gene Med. 2020, 22, e3259. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Wang, Y.; Fu, Q.; Zhou, Y.; Jia, S.; Du, J.; Peng, J.; Wang, Y.; Yang, S.; Zhan, L. Long-term hepatitis C internal ribosome entry site-dependent gene expression mediated by phage phiC31 integrase in mouse model. Antivir. Ther. 2009, 14, 393–400. [Google Scholar] [CrossRef]
- Wang, L.; Fu, Q.; Dong, Y.; Zhou, Y.; Jia, S.; Du, J.; Zhao, F.; Wang, Y.; Wang, X.; Peng, J.; et al. Bioluminescence imaging of Hepatitis C virus NS3/4A serine protease activity in cells and living animals. Antivir. Res. 2010, 87, 50–56. [Google Scholar] [CrossRef]
- Hermans, I.F.; Silk, J.D.; Yang, J.; Palmowski, M.J.; Gileadi, U.; McCarthy, C.; Salio, M.; Ronchese, F.; Cerundolo, V. The VITAL assay: A versatile fluorometric technique for assessing CTL- and NKT-mediated cytotoxicity against multiple targets in vitro and in vivo. J. Immunol. Methods 2004, 285, 25–40. [Google Scholar] [CrossRef]
- Gordon, E.J.; Bhat, R.; Liu, Q.; Wang, Y.F.; Tackney, C.; Prince, A.M. Immune responses to hepatitis C virus structural and nonstructural proteins induced by plasmid DNA immunizations. J. Infect. Dis. 2000, 181, 42–50. [Google Scholar] [CrossRef]
- Raymond, C.S.; Soriano, P. High-efficiency FLP and φC31 site-specific recombination in mammalian cells. PLoS ONE 2007, 2, e162. [Google Scholar] [CrossRef] [PubMed]
- van Pijkeren, J.P.; Morrissey, D.; Monk, I.R.; Cronin, M.; Rajendran, S.; O’Sullivan, G.C.; Gahan, C.G.; Tangney, M. A novel Listeria monocytogenes-based DNA delivery system for cancer gene therapy. Hum. Gene Ther. 2010, 21, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Jawalagatti, V.; Kirthika, P.; Park, J.Y.; Hewawaduge, C.; Lee, J.H. Highly feasible immunoprotective multicistronic SARS-CoV-2 vaccine candidate blending novel eukaryotic expression and Salmonella bactofection. J. Adv Res. 2021, 36, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Báez, E.I.; Meza-Toledo, S.E.; Muñoz-López, P.; Flores-Martínez, L.F.; Fraga-Pérez, K.; Magaño-Bocanegra, K.J.; Juárez-Hernández, U.; Mateos-Chávez, A.A.; Luria-Pérez, R. Recombinant Attenuated Salmonella enterica as a Delivery System of Heterologous Molecules in Cancer Therapy. Cancers 2022, 14, 4224. [Google Scholar] [CrossRef]
- Yurina, V. Live Bacterial Vectors-A Promising DNA Vaccine Delivery System. Med. Sci. 2018, 6, 27. [Google Scholar] [CrossRef]
- Yoon, W.; Park, Y.; Kim, S.; Bang, I.S. Development of an Oral Salmonella-Based Vaccine Platform against SARS-CoV-2. Vaccines 2022, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Seow, Y.; Wood, M.J. Biological gene delivery vehicles: Beyond viral vectors. Mol. Ther. 2009, 17, 767–777. [Google Scholar] [CrossRef]
- Ebensen, T.; Paukner, S.; Link, C.; Kudela, P.; de Domenico, C.; Lubitz, W.; Guzmán, C.A. Bacterial ghosts are an efficient delivery system for DNA vaccines. J. Immunol. 2004, 172, 6858–6865. [Google Scholar] [CrossRef]
- Abtin, A.; Kudela, P.; Mayr, U.B.; Koller, V.J.; Mildner, M.; Tschachler, E.; Lubitz, W. Escherichia coli ghosts promote innate immune responses in human keratinocytes. Biochem. Biophys. Res. Commun. 2010, 400, 78–82. [Google Scholar] [CrossRef]
- Stein, E.; Inic-Kanada, A.; Belij, S.; Montanaro, J.; Bintner, N.; Schlacher, S.; Mayr, U.B.; Lubitz, W.; Stojanovic, M.; Najdenski, H.; et al. In vitro and in vivo uptake study of Escherichia coli Nissle 1917 bacterial ghosts: Cell-based delivery system to target ocular surface diseases. Investig. Ophthalmol. Vis. Sci. 2013, 54, 6326–6333. [Google Scholar] [CrossRef]
- Adam, E.; Delbrassine, L.; Bouillot, C.; Reynders, V.; Mailleux, A.C.; Muraille, E.; Jacquet, A. Probiotic Escherichia coli Nissle 1917 activates DC and prevents house dust mite allergy through a TLR4-dependent pathway. Eur. J. Immunol. 2010, 40, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Quevedo-Diaz, M.A.; Song, C.; Xiong, Y.; Chen, H.; Wahl, L.M.; Radulovic, S.; Medvedev, A.E. Involvement of TLR2 and TLR4 in cell responses to Rickettsia akari. J. Leukoc. Biol. 2010, 88, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Hajam, I.A.; Dar, P.A.; Appavoo, E.; Kishore, S.; Bhanuprakash, V.; Ganesh, K. Bacterial Ghosts of Escherichia coli Drive Efficient Maturation of Bovine Monocyte-Derived Dendritic Cells. PLoS ONE 2015, 10, e0144397. [Google Scholar] [CrossRef] [PubMed]
- Hamza, T.; Barnett, J.B.; Li, B. Interleukin 12 a key immunoregulatory cytokine in infection applications. Int. J. Mol. Sci. 2010, 11, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, E.S.; Ebersold, M.; Garrett, W.; Pypaert, M.; Mellman, I. Activation of lysosomal function during dendritic cell maturation. Science 2003, 299, 1400–1403. [Google Scholar] [CrossRef] [PubMed]
- Trombetta, E.S.; Mellman, I. Cell biology of antigen processing in vitro and in vivo. Ann. Rev. Immunol. 2005, 23, 975–1028. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, C.A.; Walcher, P.; Mayr, U.B.; Stiedl, T.; Binder, M.; McGrath, J.; Lubitz, W. Bacterial ghosts-biological particles as delivery systems for antigens, nucleic acids and drugs. Curr. Opin. Biotechnol. 2004, 15, 530–537. [Google Scholar] [CrossRef]
- Felnerova, D.; Kudela, P.; Bizik, J.; Haslberger, A.; Hensel, A.; Saalmuller, A.; Lubitz, W. T cell-specific immune response induced by bacterial ghosts. Med. Sci. Monit. 2004, 10, BR362–BR370. [Google Scholar]
- Mayr, U.B.; Kudela, P.; Atrasheuskaya, A.; Bukin, E.; Ignatyev, G.; Lubitz, W. Rectal single dose immunization of mice with Escherichia coli O157:H7 bacterial ghosts induces efficient humoral and cellular immune responses and protects against the lethal heterologous challenge. Microb. Biotechnol. 2012, 5, 283–294. [Google Scholar] [CrossRef]
- Wen, J.; Yang, Y.; Zhao, G.; Tong, S.; Yu, H.; Jin, X.; Du, L.; Jiang, S.; Kou, Z.; Zhou, Y. Salmonella typhi Ty21a bacterial ghost vector augments HIV-1 gp140 DNA vaccine-induced peripheral and mucosal antibody responses via TLR4 pathway. Vaccine 2012, 30, 5733–5739. [Google Scholar] [CrossRef]
- Hensel, A.; van Leengoed, L.A.; Szostak, M.; Windt, H.; Weissenböck, H.; Stockhofe-Zurwieden, N.; Katinger, A.; Stadler, M.; Ganter, M.; Bunka, S.; et al. Induction of protective immunity by aerosol or oral application of candidatevaccines in a dose-controlled pig aerosol infection model. J. Biotechnol. 1996, 44, 171–181. [Google Scholar] [CrossRef]
- Mayr, U.B.; Haller, C.; Haidinger, W.; Atrasheuskaya, A.; Bukin, E.; Lubitz, W.; Ignatyev, G. Bacterial ghosts as an oral vaccine: A single dose of Escherichia coli O157:H7 bacterial ghosts protects mice against lethal challenge. Infect. Immun. 2005, 73, 4810–4817. [Google Scholar] [CrossRef]
- Mader, H.J.; Szostak, M.P.; Hensel, A.; Lubitz, W.; Haslberger, A.G. Endotoxicity does not limit the use of bacterial ghosts as candidate vaccines. Vaccine 1997, 15, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Meyer, R.; Padmanabhan, R.; Britto, J. Clinical safety of Lactobacillus casei shirota as a probiotic in critically ill children. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 171–173. [Google Scholar] [CrossRef] [PubMed]
- Frey, S.E.; Lottenbach, K.R.; Hill, H.; Blevins, T.P.; Yu, Y.; Zhang, Y.; Brenneman, K.E.; Kelly-Aehle, S.M.; McDonald, C.; Jansen, A.; et al. A Phase I, dose-escalation trial in adults of three recombinant attenuated Salmonella Typhi vaccine vectors producing Streptococcus pneumoniae surface protein antigen, PspA. Vaccine 2013, 31, 4874–4880. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Harro, C.; DeNearing, B.; Bream, J.; Bauers, N.; Dally, L.; Flores, J.; Van de Verg, L.; Sack, D.A.; Walker, R. Evaluation of the Safety, Tolerability, and Immunogenicity of an Oral, Inactivated Whole-Cell Shigella flexneri 2a Vaccine in Healthy Adult Subjects. Clin. Vaccine Immunol. 2016, 23, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Li, M.; Tang, T.; Wang, R.; Li, Y.; Xu, Y.; Tang, L.; Wang, L.; Liu, M.; Jiang, Y.; et al. Construction of Lactobacillus casei ghosts by Holin-mediated inactivation and the potential as a safe and effective vehicle for the delivery of DNA vaccines. BMC Microbiol. 2018, 18, 80. [Google Scholar] [CrossRef]
- Kim, E.; Won, G.; Lee, J.H. Construction of a novel tetravalent dengue vaccine with a Salmonella Typhimurium bacterial ghost and evaluation of its immunogenicity and protective efficacy using a murine model. Vaccine 2020, 38, 916–924. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Samir, R.; Amin, H.M. Production of highly immunogenic and safe Triton X-100 produced bacterial ghost vaccine against Shigella flexneri 2b serotype. Gut Pathog. 2023, 15, 41. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, Y.; Zhang, S.; Ji, S. Delivery of a Hepatitis C Virus Vaccine Encoding NS3 Linked to the MHC Class II Chaperone Protein Invariant Chain Using Bacterial Ghosts. Biomedicines 2024, 12, 525. https://doi.org/10.3390/biomedicines12030525
Chi Y, Zhang S, Ji S. Delivery of a Hepatitis C Virus Vaccine Encoding NS3 Linked to the MHC Class II Chaperone Protein Invariant Chain Using Bacterial Ghosts. Biomedicines. 2024; 12(3):525. https://doi.org/10.3390/biomedicines12030525
Chicago/Turabian StyleChi, Yulang, Shikun Zhang, and Shouping Ji. 2024. "Delivery of a Hepatitis C Virus Vaccine Encoding NS3 Linked to the MHC Class II Chaperone Protein Invariant Chain Using Bacterial Ghosts" Biomedicines 12, no. 3: 525. https://doi.org/10.3390/biomedicines12030525
APA StyleChi, Y., Zhang, S., & Ji, S. (2024). Delivery of a Hepatitis C Virus Vaccine Encoding NS3 Linked to the MHC Class II Chaperone Protein Invariant Chain Using Bacterial Ghosts. Biomedicines, 12(3), 525. https://doi.org/10.3390/biomedicines12030525





