Cold-Inducible RNA Binding Protein Impedes Breast Tumor Growth in the PyMT Murine Model for Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Transgenic Mice
2.2. Collection of Tissue
2.3. Whole Mounts
2.4. Histology and Immunostaining
2.5. Syngeneic Py2T Tumor Cell Grafts
2.6. CIRP Knockdown
2.7. Tissue Supernatants and Luminex Arrays
2.8. CIRP ELISA
2.9. Murine Breast Tumor Digestion
2.10. Cell Staining and Flow Cytometry
2.11. Statistics
3. Results
3.1. Human CIRP Expression Impeded Early Tumorigenesis in MMTV-PyMT Mice
3.2. CIRP Impeded Late Tumorigenesis and Pulmonary Metatasis
3.3. CIRP Knockdown Increased Tumor Growth in Py2T Tumor Cell Grafts
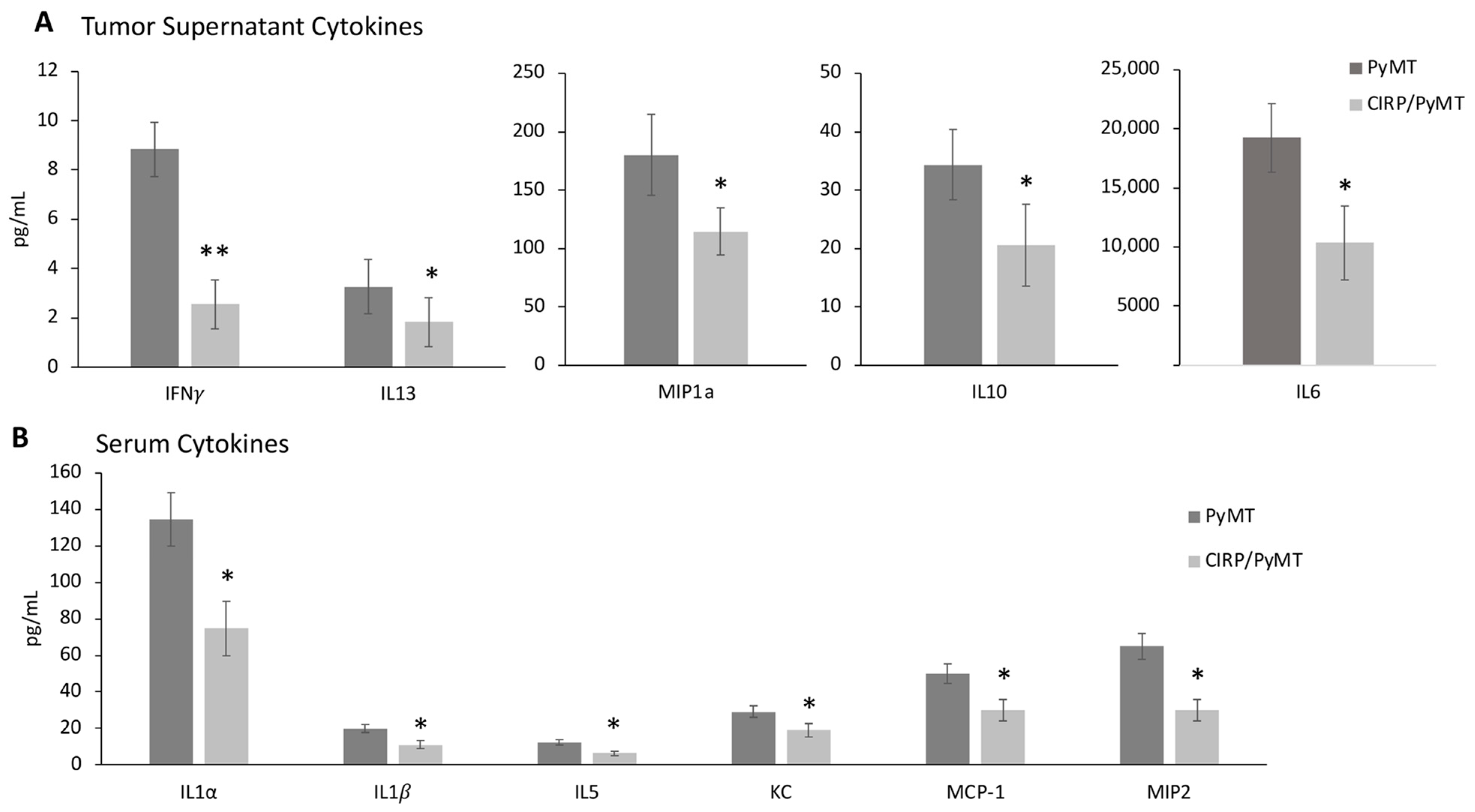
3.4. CIRP Decreases Pro-Tumorigenic Cytokines in Mammary Tumors
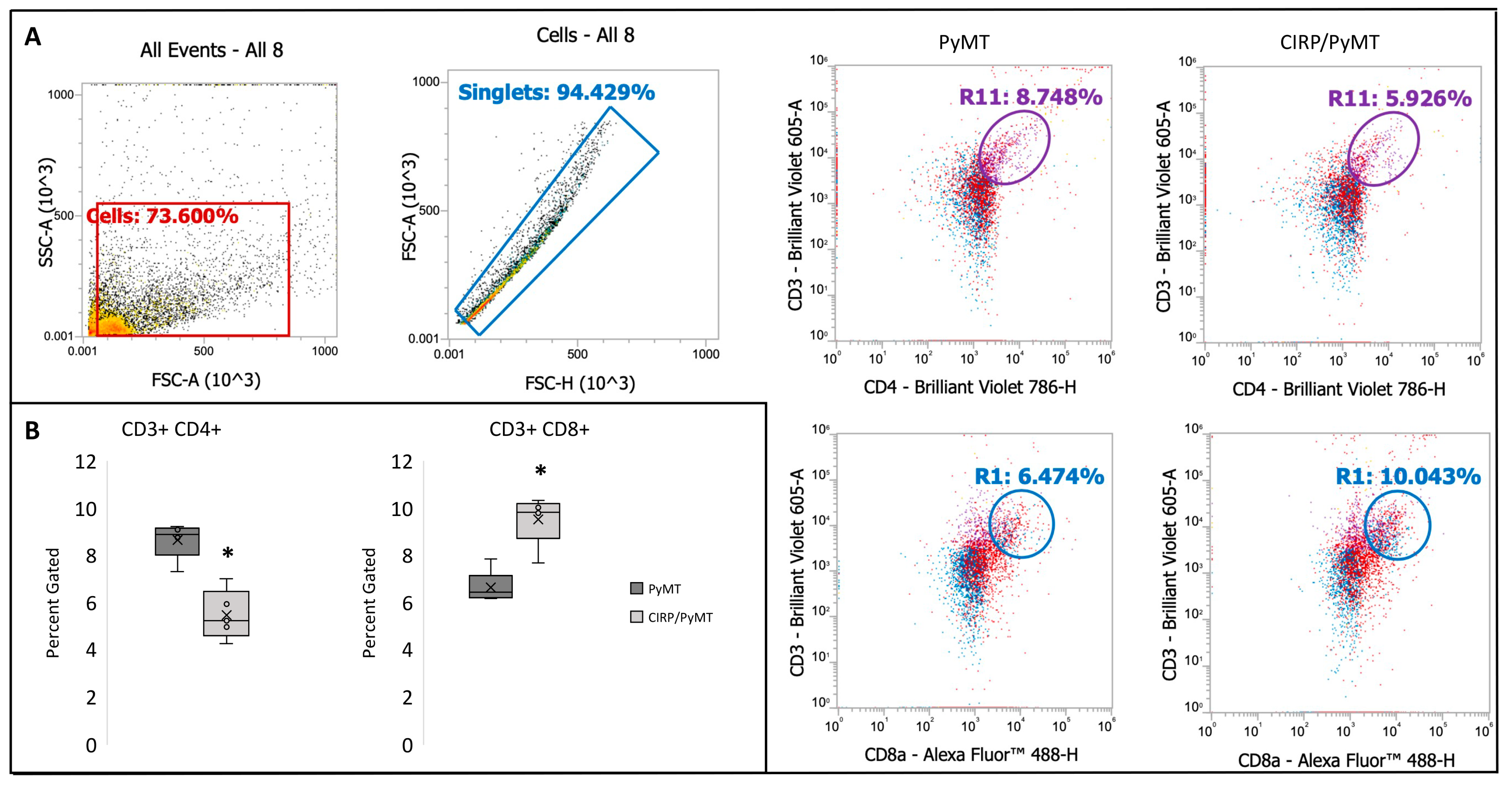
3.5. CIRP Decreases CD4+ Helper T Cells and Increases CD8+ Cytotoxic T Cells in Mammary Tumors during Late Stage Tumorigenesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hasan, A.; Cotobal, C.; Duncan, C.D.S.; Mata, J. Systematic analysis of the role of RNA-binding proteins in the regulation of RNA stability. PLoS Genet. 2014, 10, e1004684. [Google Scholar] [CrossRef]
- He, S.; Valkov, E.; Cheloufi, S.; Murn, J. The nexus between RNA-binding proteins and their effectors. Nat. Rev. Genet. 2023, 24, 276–294. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Cruz, A.; Baños-Jaime, B.; Díaz-Quintana, A.; De la Rosa, M.A.; Díaz-Moreno, I. Post-translational Control of RNA-Binding Proteins and Disease-Related Dysregulation. Front. Mol. Biosci. 2021, 8, 658852. [Google Scholar] [CrossRef] [PubMed]
- Sachse, M.; Tual-Chalot, S.; Ciliberti, G.; Amponsah-Offeh, M.; Stamatelopoulos, K.; Gatsiou, A.; Stellos, K. RNA-binding proteins in vascular inflammation and atherosclerosis. Atherosclerosis 2023, 374, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.-L. RNA-Binding Proteins as Critical Post-Transcriptional Regulators of Cardiac Regeneration. Int. J. Mol. Sci. 2023, 24, 12004. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Lee, Y.; Lee, J.-S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef]
- Li, W.; Deng, X.; Chen, J. RNA-binding proteins in regulating mRNA stability and translation: Roles and mechanisms in cancer. Semin. Cancer Biol. 2022, 86, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, L.; Zhao, S.; Chen, L.; Wei, Y.; Chen, W.; Ge, F. Research progress on RNA-binding proteins in breast cancer. Oncol. Lett. 2022, 23, 121. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qin, H.; Zheng, L. Research progress on RNA–binding proteins in breast cancer. Front. Oncol. 2022, 12, 974523. [Google Scholar] [CrossRef]
- Fornace, A.J.; Alamo, I.; Hollander, M.C. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. USA 1988, 85, 8800–8804. [Google Scholar] [CrossRef]
- Nishiyama, H.; Itoh, K.; Kaneko, Y.; Kishishita, M.; Yoshida, O.; Fujita, J. A Glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J. Cell Biol. 1997, 137, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, M.S.; Carrier, F.; Papathanasiou, M.A.; Hollander, M.C.; Zhan, Q.; Yu, K.; Fornace, A.J. Identification of several human homologs of hamster DNA damage-inducible transcripts cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J. Biol. Chem. 1997, 272, 26720–26726. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Carrier, F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J. Biol. Chem. 2001, 276, 47277–47284. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, F.; Zhang, T.; Wauquier, C.; Huez, G.; Kruys, V.; Gueydan, C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp. Cell Res. 2007, 313, 4130–4144. [Google Scholar] [CrossRef]
- Brochu, C.; Cabrita, M.A.; Melanson, B.D.; Hamill, J.D.; Lau, R.; Pratt, M.C.; McKay, B.C. NF-κB-dependent role for cold-inducible RNA binding protein in regulating interleukin 1β. PLoS ONE 2013, 8, e57426. [Google Scholar] [CrossRef]
- Qiang, X.; Yang, W.-L.; Wu, R.; Zhou, M.; Jacob, A.; Dong, W.; Kuncewitch, M.; Ji, Y.; Yang, H.; Wang, H.; et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat. Med. 2013, 19, 1489–1495. [Google Scholar] [CrossRef]
- Ren, W.H.; Zhang, L.M.; Liu, H.Q.; Gao, L.; Chen, C.; Qiang, C.; Wang, X.L.; Liu, C.Y.; Li, S.M.; Huang, C.; et al. Protein overexpression of CIRP and TLR4 in oral squamous cell carcinoma: An immunohistochemical and clinical correlation analysis. Med. Oncol. 2014, 31, 120. [Google Scholar] [CrossRef]
- Sakurai, T.; Kashida, H.; Watanabe, T.; Hagiwara, S.; Mizushima, T.; Iijima, H.; Nishida, N.; Higashitsuji, H.; Fujita, J.; Kudo, M. Stress response protein CIRP links inflammation and tumorigenesis in colitis-associated cancer. Cancer Res. 2014, 74, 6119–6128. [Google Scholar] [CrossRef]
- Sakurai, T.; Yada, N.; Watanabe, T.; Arizumi, T.; Hagiwara, S.; Ueshima, K.; Nishida, N.; Fujita, J.; Kudo, M. Cold-inducible RNA-binding protein promotes the development of liver cancer. Cancer Sci. 2015, 106, 352–358. [Google Scholar] [CrossRef]
- Juan, Y.; Haiqiao, W.; Xie, W.; Huaping, H.; Zhong, H.; Xiangdong, Z.; Kolosov, V.P.; Perelman, J.M. Cold-inducible RNA-binding protein mediates airway inflammation and mucus hypersecretion through a post-transcriptional regulatory mechanism under cold stress. Int. J. Biochem. Cell Biol. 2016, 78, 335–348. [Google Scholar] [CrossRef]
- Lujan, D.A.; Ochoa, J.L.; Hartley, R.S. Cold-inducible RNA binding protein in cancer and inflammation. Wiley Interdiscip. Rev. RNA 2018, 9, e1462. [Google Scholar] [CrossRef] [PubMed]
- Yoo, I.S.; Lee, S.Y.; Park, C.K.; Lee, J.C.; Kim, Y.; Yoo, S.J.; Shim, S.C.; Choi, Y.S.; Lee, Y.; Kang, S.W. Serum and synovial fluid concentrations of cold-inducible RNA-binding protein in patients with rheumatoid arthritis. Int. J. Rheum. Dis. 2018, 21, 148–154. [Google Scholar] [CrossRef]
- Bolourani, S.; Sari, E.; Brenner, M.; Wang, P. Extracellular CIRP Induces an Inflammatory Phenotype in Pulmonary Fibroblasts via TLR4. Front. Immunol. 2021, 12, 721970. [Google Scholar] [CrossRef]
- He, Q.; Gao, H.; Chang, Y.-L.; Wu, X.; Lin, R.; Li, G.; Lin, J.; Lu, H.; Chen, H.; Li, Z.; et al. ETS-1 facilitates Th1 cell-mediated mucosal inflammation in inflammatory bowel diseases through upregulating CIRBP. J. Autoimmun. 2022, 132, 102872. [Google Scholar] [CrossRef]
- Godwin, A.; Yang, W.-L.; Sharma, A.; Khader, A.; Wang, Z.; Zhang, F.; Nicastro, J.; Coppa, G.F.; Wang, P. Blocking Cold-Inducible RNA-Binding Protein (CIRP) Protects Liver from Ischemia/Reperfusion Injury. Shock 2015, 43, 24. [Google Scholar] [CrossRef]
- Biade, S.; Marinucci, M.; Schick, J.; Roberts, D.; Workman, G.; Sage, E.H.; O’Dwyer, P.J.; LiVolsi, V.A.; Johnson, S.W. Gene expression profiling of human ovarian tumours. Br. J. Cancer 2006, 95, 1092–1100. [Google Scholar] [CrossRef]
- Hamid, A.A.M.; Mandai, M.M.; Fujita, J.M.; Nanbu, K.M.; Kariya, M.M.; Kusakari, T.M.; Fukuhara, K.M.; Fujii, S.M. Expression of cold-inducible RNA-binding protein in the normal endometrium, endometrial hyperplasia, and endometrial carcinoma. Int. J. Gynecol. Pathol. 2003, 22, 240–247. [Google Scholar] [CrossRef]
- Lujan, D.A.; Garcia, S.; Vanderhoof, J.; Sifuentes, J.; Brandt, Y.; Wu, Y.; Guo, X.; Mitchell, T.; Howard, T.; Hathaway, H.J.; et al. Cold-inducible RNA binding protein in mouse mammary gland development. Tissue Cell 2016, 48, 577–587. [Google Scholar] [CrossRef]
- Peri, S.; de Cicco, R.L.; Santucci-Pereira, J.; Slifker, M.; Ross, E.A.; Russo, I.H.; Russo, P.A.; Arslan, A.A.; Belitskaya-Lévy, I.; Zeleniuch-Jacquotte, A.; et al. Defining the genomic signature of the parous breast. BMC Med. Genom. 2012, 5, 46. [Google Scholar] [CrossRef]
- Yang, R.; Weber, D.J.; Carrier, F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 2006, 34, 1224–1236. [Google Scholar] [CrossRef]
- Yang, R.; Zhan, M.; Nalabothula, N.R.; Yang, Q.; Indig, F.E.; Carrier, F. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J. Biol. Chem. 2010, 285, 8887–8893. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.-Y.; Chen, Y.; Jia, J.-S.; Zhou, C.; Lian, M.; Wen, Y.-T.; Li, X.-Y.; Chen, H.-W.; Lin, X.-L.; Zhang, X.-L.; et al. Loss of Cirbp expression is correlated with the malignant progression and poor prognosis in nasopharyngeal carcinoma. Cancer Manag. Res. 2019, 11, 6959. [Google Scholar] [CrossRef]
- Artero-Castro, A.; Callejas, F.B.; Castellvi, J.; Kondoh, H.; Carnero, A.; Fernández-Marcos, P.J.; Serrano, M.; Cajal, S.R.Y.; Lleonart, M.E. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol. Cell. Biol. 2009, 29, 1855–1868. [Google Scholar] [CrossRef]
- Jian, F.; Chen, Y.; Ning, G.; Fu, W.; Tang, H.; Chen, X.; Zhao, Y.; Zheng, L.; Pan, S.; Wang, W.; et al. Cold inducible RNA binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via Erk signaling pathway. Oncotarget 2016, 7, 9175–9187. [Google Scholar] [CrossRef]
- Lee, H.N.; Ahn, S.-M.; Jang, H.H. Cold-inducible RNA-binding protein, CIRP, inhibits DNA damage-induced apoptosis by regulating p53. Biochem. Biophys. Res. Commun. 2015, 464, 916–921. [Google Scholar] [CrossRef]
- Lee, H.N.; Ahn, S.-M.; Jang, H.H. Cold-inducible RNA-binding protein promotes epithelial-mesenchymal transition by activating ERK and p38 pathways. Biochem. Biophys. Res. Commun. 2016, 477, 1038–1044. [Google Scholar] [CrossRef]
- Mangé, A.; Lacombe, J.; Bascoul-Mollevi, C.; Jarlier, M.; Lamy, P.-J.; Rouanet, P.; Maudelonde, T.; Solassol, J. Serum autoantibody signature of ductal carcinoma in situ progression to invasive breast cancer. Clin. Cancer Res. 2012, 18, 1992–2000. [Google Scholar] [CrossRef]
- Zeng, Y.; Kulkarni, P.; Inoue, T.; Getzenberg, R.H. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J. Cell. Biochem. 2009, 107, 179–188. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Mao, P.; Li, F.; Han, X.; Zhang, Y.; Jiang, S.; Chen, Y.; Huang, J.; Liu, D.; et al. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2015, 44, 761–775. [Google Scholar] [CrossRef]
- Chang, E.T.; Parekh, P.R.; Yang, Q.; Nguyen, D.M.; Carrier, F. Heterogenous ribonucleoprotein A18 (hnRNP A18) promotes tumor growth by increasing protein translation of selected transcripts in cancer cells. Oncotarget 2016, 7, 10578–10593. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, Y.; Hartley, R.S. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol. Carcinog. 2010, 49, 130–140. [Google Scholar] [CrossRef]
- Indacochea, A.; Guerrero, S.; Ureña, M.; Araujo, F.; Coll, O.; Lleonart, M.E.; Gebauer, F. Cold-inducible RNA binding protein promotes breast cancer cell malignancy by regulating Cystatin C levels. RNA 2021, 27, 190–201. [Google Scholar] [CrossRef]
- Lewis, M.T.; Porter, W.W. Methods in mammary gland biology and breast cancer research: An update. J. Mammary Gland. Biol. Neoplasia 2009, 14, 365. [Google Scholar] [CrossRef]
- Waldmeier, L.; Meyer-Schaller, N.; Diepenbruck, M.; Christofori, G. Py2T murine breast cancer cells, a versatile model of TGFβ-induced EMT in vitro and in vivo. PLoS ONE 2012, 7, e48651. [Google Scholar] [CrossRef]
- Franci, C.; Zhou, J.; Jiang, Z.; Modrusan, Z.; Good, Z.; Jackson, E.; Kouros-Mehr, H. Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS ONE 2013, 8, e58183. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Lin, E.Y.; Jones, J.G.; Li, P.; Zhu, L.; Whitney, K.D.; Muller, W.J.; Pollard, J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003, 163, 2113–2126. [Google Scholar] [CrossRef]
- Marjon, N.A.; Hu, C.; Hathaway, H.J.; Prossnitz, E.R. G Protein–coupled estrogen receptor regulates mammary tumorigenesis and metastasis. Mol. Cancer Res. 2014, 12, 1644–1654. [Google Scholar] [CrossRef]
- Richert, M.M.; Schwertfeger, K.L.; Ryder, J.W.; Anderson, S.M. An atlas of mouse mammary gland development. J. Mammary Gland. Biol. Neoplasia 2000, 5, 227–241. [Google Scholar] [CrossRef]
- Nicolini, A.; Carpi, A.; Rossi, G. Cytokines in breast cancer. Cytokine Growth Factor Rev. 2006, 17, 325–337. [Google Scholar] [CrossRef]
- Ben-Baruch, A. Host microenvironment in breast cancer development: Inflammatory cells, cytokines and chemokines in breast cancer progression: Reciprocal tumor–microenvironment interactions. Breast Cancer Res. 2002, 5, 31. [Google Scholar] [CrossRef]
- DeNardo, D.G.; Coussens, L.M. Inflammation and breast cancer. Balancing immune response: Crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.T.; Castillo, E.F.; Ray, A.L.; Weston, L.L.; Nofchissey, R.A.; Hanson, J.A.; Samedi, V.G.; Pinchuk, I.V.; Hudson, L.G.; Beswick, E.J. Anti-G-CSF treatment induces protective tumor immunity in mouse colon cancer by promoting NK cell, macrophage and T cell responses. Oncotarget 2015, 6, 22338–22347. [Google Scholar] [CrossRef]
- Li, X.; Yang, Q.; Yu, H.; Wu, L.; Zhao, Y.; Zhang, C.; Yue, X.; Liu, Z.; Wu, H.; Haffty, B.G.; et al. LIF promotes tumorigenesis and metastasis of breast cancer through the AKT-mTOR pathway. Oncotarget 2014, 5, 788. [Google Scholar] [CrossRef]
- Yu, H.; Yue, X.; Zhao, Y.; Li, X.; Wu, L.; Zhang, C.; Liu, Z.; Lin, K.; Xu-Monette, Z.Y.; Young, K.H.; et al. LIF negatively regulates tumor suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat. Commun. 2014, 5, 5218. [Google Scholar] [CrossRef]
- Acharyya, S.; Oskarsson, T.; Vanharanta, S.; Malladi, S.; Kim, J.; Morris, P.G.; Manova-Todorova, K.; Leversha, M.; Hogg, N.; Seshan, V.E.; et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012, 150, 165–178. [Google Scholar] [CrossRef]
- Qian, B.-Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Sun, X.; Glynn, D.J.; Hodson, L.J.; Huo, C.; Britt, K.; Thompson, E.W.; Woolford, L.; Evdokiou, A.; Pollard, J.W.; Robertson, S.A.; et al. CCL2-driven inflammation increases mammary gland stromal density and cancer susceptibility in a transgenic mouse model. Breast Cancer Res. 2017, 19, 4. [Google Scholar] [CrossRef]
- García-Tuñón, I.; Ricote, M.; Ruiz, A.A.; Fraile, B.; Paniagua, R.; Royuela, M. Influence of IFN-gamma and its receptors in human breast cancer. BMC Cancer 2007, 7, 158. [Google Scholar] [CrossRef]
- Mandai, M.; Hamanishi, J.; Abiko, K.; Matsumura, N.; Baba, T.; Konishi, I. Dual faces of IFNγ in cancer progression: A role of PD-L1 induction in the determination of pro-and antitumor immunity. Clin. Cancer Res. 2016, 22, 2329–2334. [Google Scholar] [CrossRef]
- Feliciano, P. CXCL1 and CXCL2 link metastasis and chemoresistance. Nat. Genet. 2012, 44, 840. [Google Scholar] [CrossRef]
- Yang, C.; He, L.; He, P.; Liu, Y.; Wang, W.; He, Y.; Du, Y.; Gao, F. Increased drug resistance in breast cancer by tumor-associated macrophages through IL-10/STAT3/bcl-2 signaling pathway. Med. Oncol. 2015, 32, 14. [Google Scholar] [CrossRef] [PubMed]
- Llanes-Fernández, L.; Álvarez-Goyanes, R.I.; Arango-Prado, M.d.C.; Alcocer-González, J.M.; Mojarrieta, J.C.; Pérez, X.E.; López, M.O.; Odio, S.F.; Camacho-Rodríguez, R.; Guerra-Yi, M.E.; et al. Relationship between IL-10 and tumor markers in breast cancer patients. Breast 2006, 15, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Terai, M.; Tamura, Y.; Alexeev, V.; Mastrangelo, M.J.; Selvan, S.R. Interleukin 10 in the tumor microenvironment: A target for anticancer immunotherapy. Immunol. Res. 2011, 51, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Knüpfer, H.; Preiss, R. Significance of interleukin-6 (IL-6) in breast cancer. Breast Cancer Res Treat. 2007, 102, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, A.; Santini, D.; Bonafè, M.; Taffurelli, M.; Avenia, N. Interleukin-6 and pro inflammatory status in the breast tumor microenvironment. World J. Surg. Oncol. 2015, 13, 129. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Yao, Q.; Liu, Y.; Du, S.; Liu, A.; Guo, Z.; Sun, A.; Ruan, J.; Chen, L.; Ye, C.; et al. IL-6-induced epithelial-mesenchymal transition promotes the generation of breast cancer stem-like cells analogous to mammosphere cultures. Int. J. Oncol. 2012, 40, 1171–1179. [Google Scholar] [CrossRef]
- Srabovic, N.; Mujagic, Z.; Mujanovic-Mustedanagic, J.; Muminovic, Z.; Softic, A.; Begic, L. Interleukin 13 expression in the primary breast cancer tumor tissue. Biochem. Med. 2011, 21, 131–138. [Google Scholar] [CrossRef]
- Gil Del Alcazar, C.R.; Huh, S.J.; Ekram, M.B.; Trinh, A.; Liu, L.L.; Beca, F.; Zi, X.; Kwak, M.; Bergholtz, H.; Su, Y.; et al. Immune Escape in Breast Cancer During In Situ to Invasive Carcinoma Transition. Cancer Discov. 2017, 7, 1098–1115. [Google Scholar] [CrossRef]
- Idrovo, J.P.; Jacob, A.; Yang, W.L.; Wang, Z.; Yen, H.T.; Nicastro, J.; Coppa, G.F.; Wang, P. A deficiency in cold-inducible RNA-binding protein accelerates the inflammation phase and improves wound healing. Int. J. Mol. Med. 2016, 37, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Wu, C.; Ma, C.; Luo, T.; Yuan, J.; Zhou, P.; Wei, Z. Identification of an endoplasmic reticulum stress-related gene signature to predict prognosis and potential drugs of uterine corpus endometrial cancer. Math. Biosci. Eng. 2023, 20, 4018–4039. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Sun, X.; Wang, L. Prognostic Values from Integrated Analysis of the Nomogram Based on RNA-Binding Proteins and Clinical Factors in Endometrial Cancer. Clin. Med. Insights Oncol. 2022, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Feng, R.; Guo, J.; Pan, L.; Yao, Y.; Gao, J. Integrated single-cell and bulk RNA sequencing analysis identifies a neoadjuvant chemotherapy-related gene signature for predicting survival and therapy in breast cancer. BMC Med. Genom. 2023, 16, 300. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, Y.; Ge, P.; Dakal, T.C.; Wen, H.; Tang, S.; Luo, Y.; Yang, Q.; Hua, B.; Zhang, G.; et al. Exosome-derived CIRP: An amplifier of inflammatory diseases. Front. Immunol. 2023, 14, 1066721. [Google Scholar] [CrossRef]
- Brogowska, K.K.; Zajkowska, M.; Mroczko, B. Vascular Endothelial Growth Factor Ligands and Receptors in Breast Cancer. J. Clin. Med. 2023, 12, 2412. [Google Scholar] [CrossRef] [PubMed]
- Al Kawas, H.; Saaid, I.; Jank, P.; Westhoff, C.C.; Denkert, C.; Pross, T.; Weiler, K.B.S.; Karsten, M.M. How VEGF-A and its splice variants affect breast cancer development—Clinical implications. Cell. Oncol. 2022, 45, 227–239. [Google Scholar] [CrossRef]
- Goossens, E.A.C.; Zhang, L.; de Vries, M.R.; Jukema, J.W.; Quax, P.H.A.; Nossent, A.Y. Cold-Inducible RNA-Binding Protein but Not Its Antisense lncRNA Is a Direct Negative Regulator of Angiogenesis In Vitro and In Vivo via Regulation of the 14q32 angiomiRs-microRNA-329-3p and microRNA-495-3p. Int. J. Mol. Sci. 2021, 22, 12678. [Google Scholar] [CrossRef]
- Tulotta, C.; Ottewell, P.D. The role of IL-1B in breast cancer bone metastasis. Endocr.-Relat. Cancer 2018, 25, R421–R434. [Google Scholar] [CrossRef]
- Blomberg, O.S.; Spagnuolo, L.; Garner, H.; Voorwerk, L.; Isaeva, O.I.; van Dyk, E.; Bakker, N.; Chalabi, M.; Klaver, C.; Duijst, M.; et al. IL-5-producing CD4+ T cells and eosinophils cooperate to enhance response to immune checkpoint blockade in breast cancer. Cancer Cell 2023, 41, 106–123. [Google Scholar] [CrossRef]
- Chavey, C.; Bibeau, F.; Gourgou-Bourgade, S.; Burlinchon, S.; Boissière, F.; Laune, D.; Roques, S.; Lazennec, G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007, 9, R15. [Google Scholar] [CrossRef]
- Bolognese, A.C.; Sharma, A.; Yang, W.-L.; Nicastro, J.; Coppa, G.F.; Wang, P. Cold-inducible RNA-binding protein activates splenic T cells during sepsis in a TLR4-dependent manner. Cell. Mol. Immunol. 2018, 15, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, L.; Silva, L.; Llopiz, D.; Ruiz, M.; Iglesias, T.; Lozano, T.; Casares, N.; Hervas-Stubbs, S.; Rodríguez, M.J.; Carrascosa, J.L.; et al. The Toll like receptor 4 ligand cold-inducible RNA-binding protein as vaccination platform against cancer. OncoImmunology 2017, 7, e1409321. [Google Scholar] [CrossRef] [PubMed]
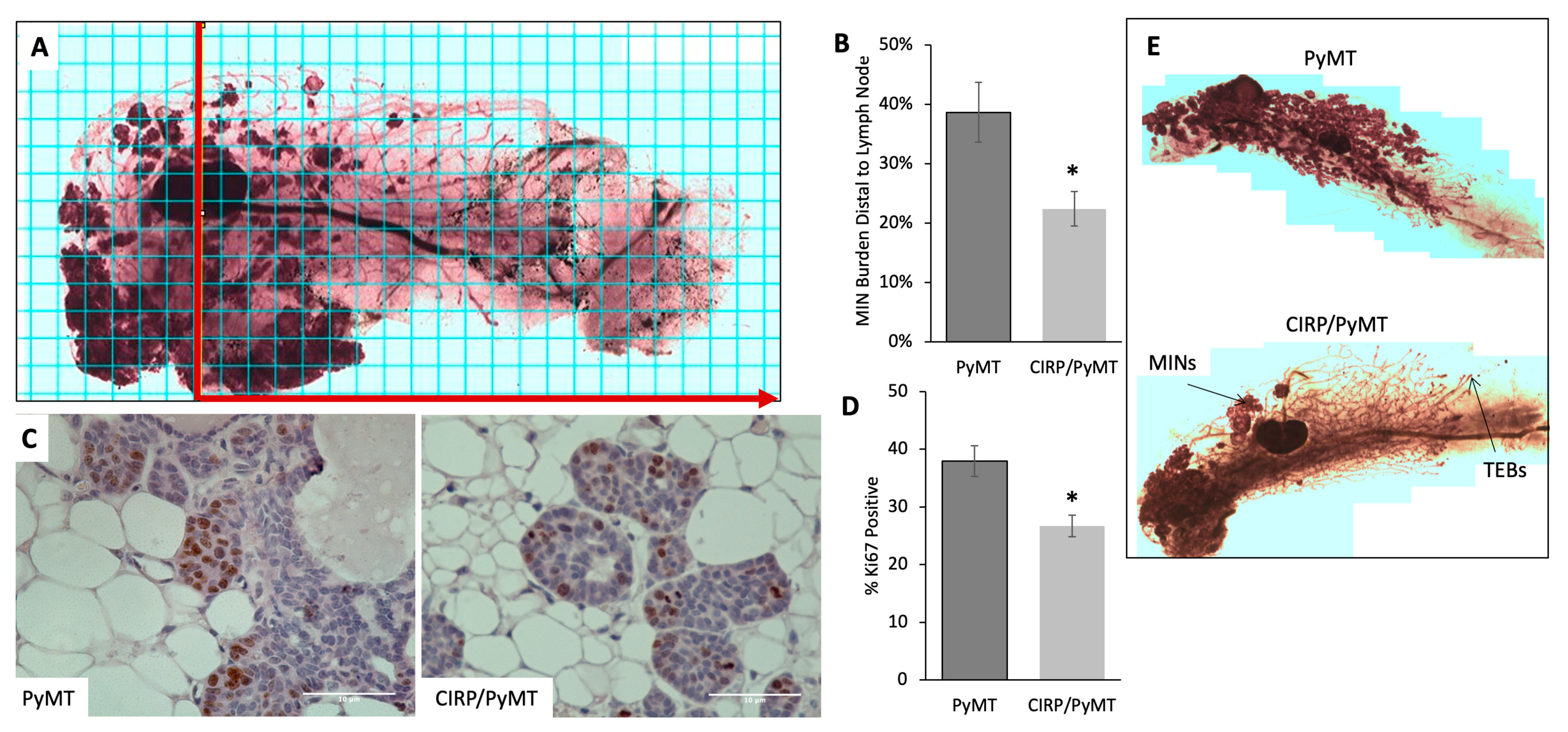
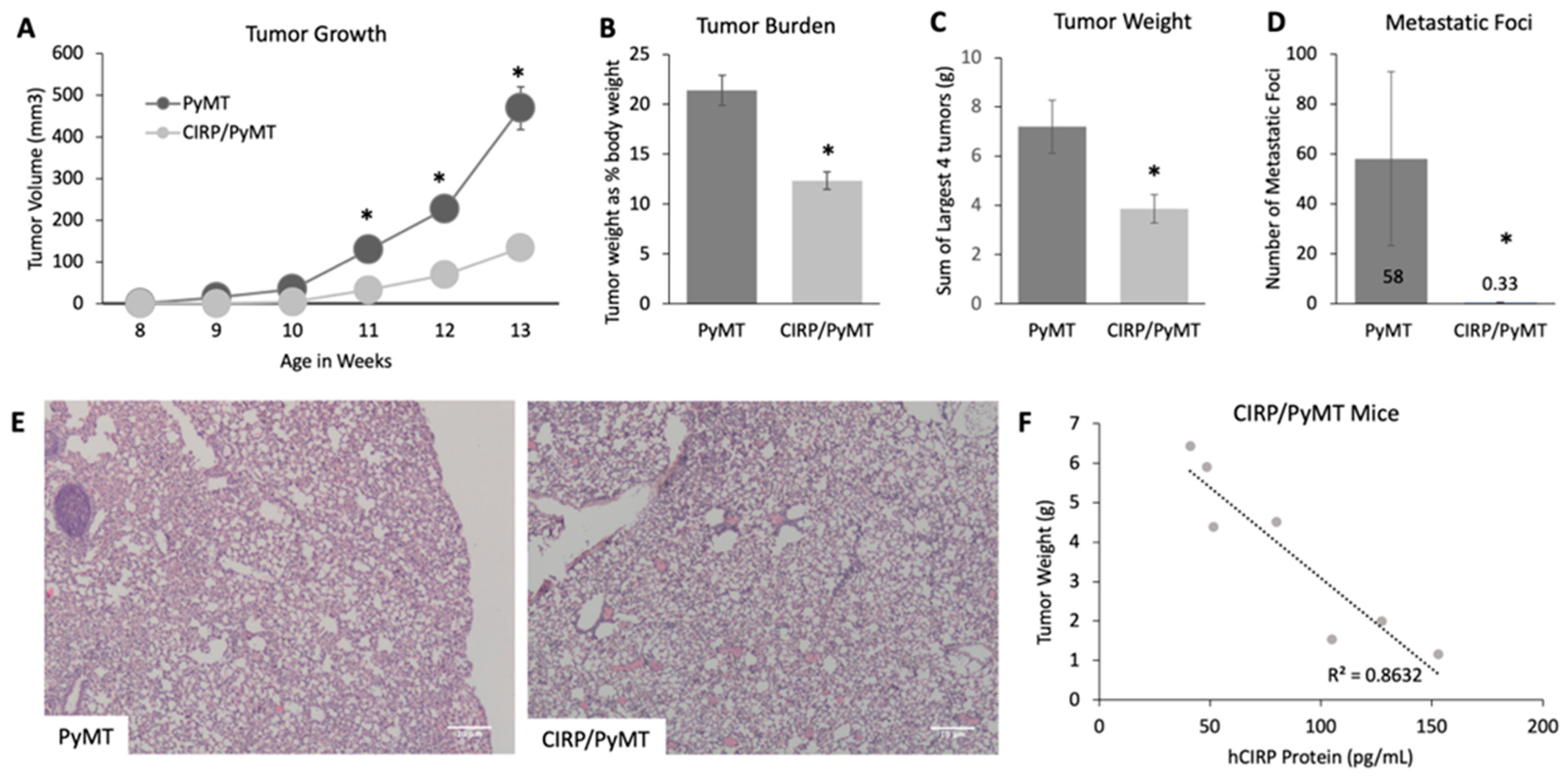
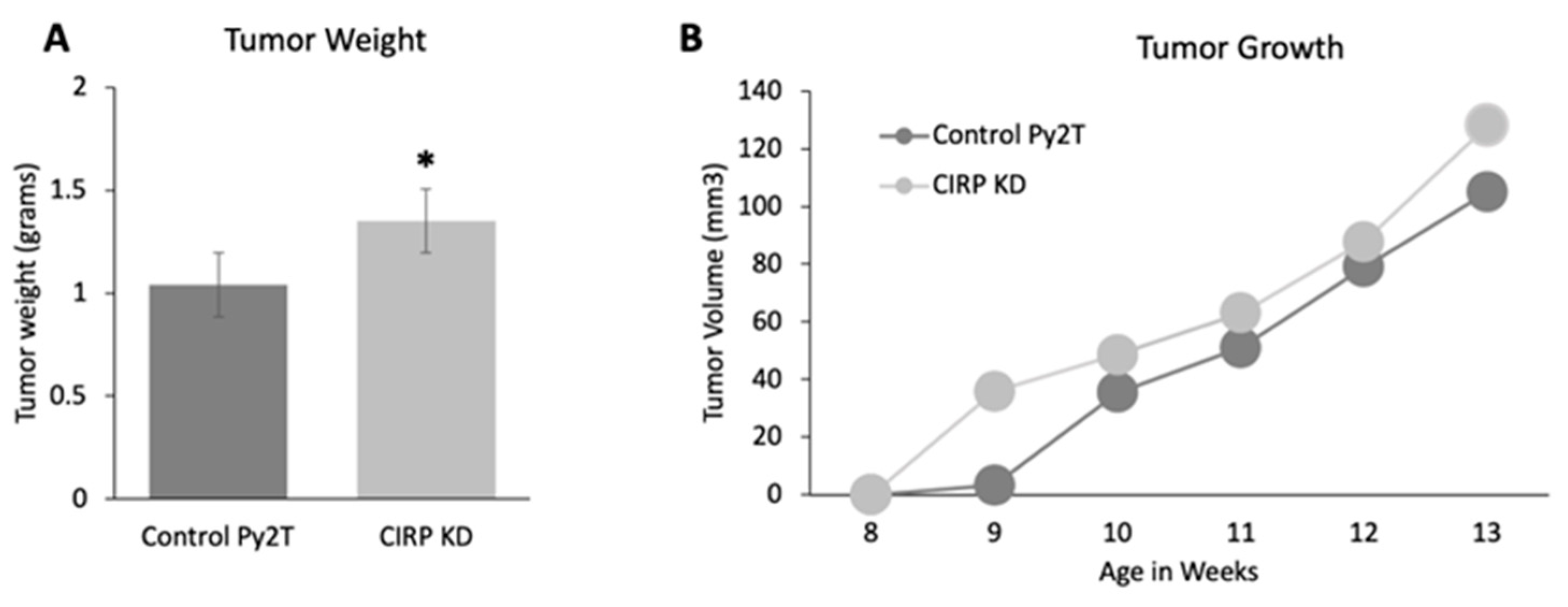
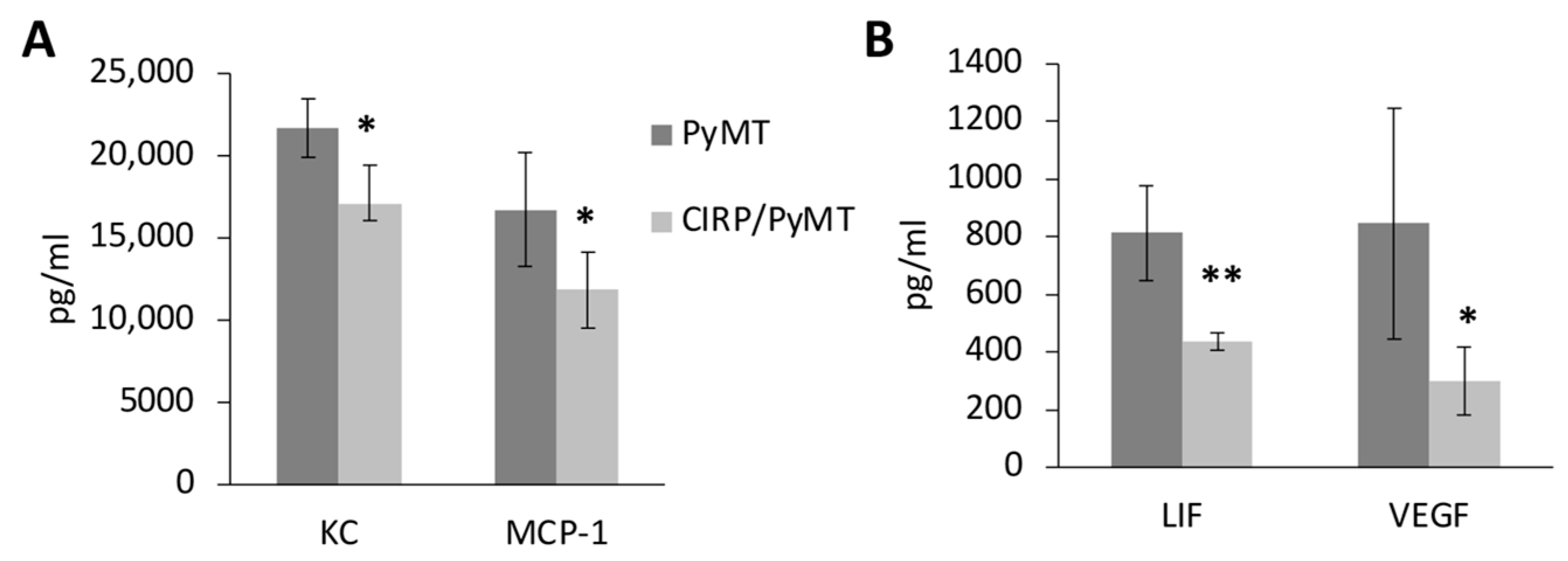
| PyMT | CIRP/PyMT | |
|---|---|---|
| Primary Lesions | Larger, less clearly demarcated | Smaller, more clearly demarcated |
| Secondary Duct Involvement | Considerably greater | Less secondary duct involvement |
| Secondary Lesions | More advanced | Less advanced |
| Atypia | No difference | |
| Vascularity | ||
| Inflammatory Cell Infiltrate | ||
| Mean Mitotic Figures | 2.2 figures per field | 0.7 figures per field |
| Mean Apoptotic Figures | 1.15 figures per field | 4.1 figures per field |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lujan, D.A.; Ochoa, J.L.; Beswick, E.J.; Howard, T.A.; Hathaway, H.J.; Perrone-Bizzozero, N.I.; Hartley, R.S. Cold-Inducible RNA Binding Protein Impedes Breast Tumor Growth in the PyMT Murine Model for Breast Cancer. Biomedicines 2024, 12, 340. https://doi.org/10.3390/biomedicines12020340
Lujan DA, Ochoa JL, Beswick EJ, Howard TA, Hathaway HJ, Perrone-Bizzozero NI, Hartley RS. Cold-Inducible RNA Binding Protein Impedes Breast Tumor Growth in the PyMT Murine Model for Breast Cancer. Biomedicines. 2024; 12(2):340. https://doi.org/10.3390/biomedicines12020340
Chicago/Turabian StyleLujan, Daniel A., Joey L. Ochoa, Ellen J. Beswick, Tamara A. Howard, Helen J. Hathaway, Nora I. Perrone-Bizzozero, and Rebecca S. Hartley. 2024. "Cold-Inducible RNA Binding Protein Impedes Breast Tumor Growth in the PyMT Murine Model for Breast Cancer" Biomedicines 12, no. 2: 340. https://doi.org/10.3390/biomedicines12020340
APA StyleLujan, D. A., Ochoa, J. L., Beswick, E. J., Howard, T. A., Hathaway, H. J., Perrone-Bizzozero, N. I., & Hartley, R. S. (2024). Cold-Inducible RNA Binding Protein Impedes Breast Tumor Growth in the PyMT Murine Model for Breast Cancer. Biomedicines, 12(2), 340. https://doi.org/10.3390/biomedicines12020340





