Exome Profiling Suggests Combined Effect of Myeloperoxidase, Toll-Like Receptors, and Metallopeptidase in Hidradenitis Suppurativa
Abstract
1. Introduction
2. Materials and Methods
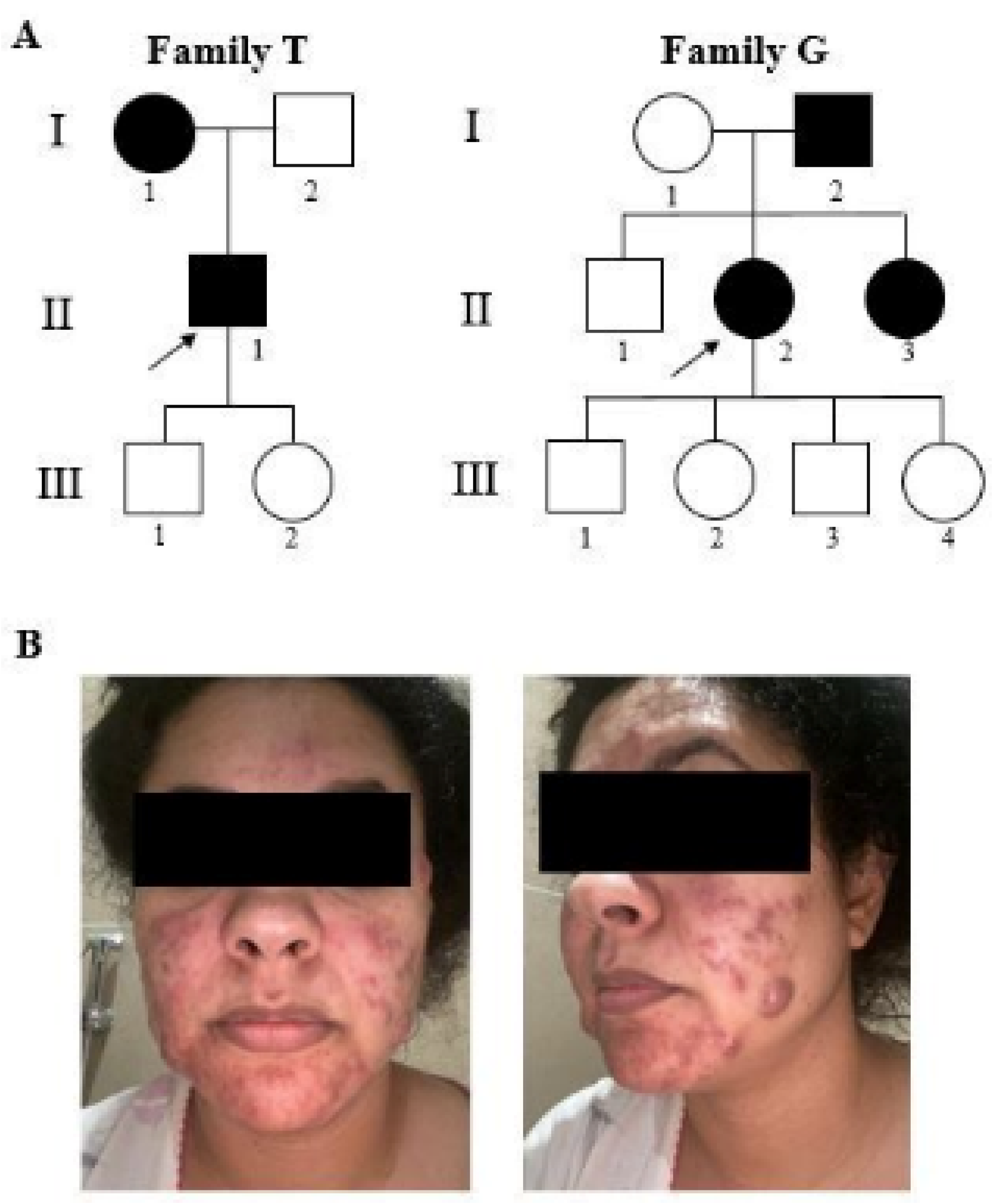
2.1. Clinical Features of Patients
2.2. Whole-Exome Sequencing
2.3. Direct Sanger Sequencing
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Revuz, J. Hidradenitis Suppurativa. J. Eur. Acad. Dermatol. Venereol. 2009, 23, 985–998. [Google Scholar] [CrossRef] [PubMed]
- Sabat, R.; Jemec, G.B.E.; Matusiak, Ł.; Kimball, A.B.; Prens, E.; Wolk, K. Hidradenitis suppurativa. Nat. Rev. Dis. Prim. 2020, 6, 1–20. [Google Scholar] [CrossRef]
- Boer, J.; Nazary, M.; Riis, P.T. The Role of Mechanical Stress in Hidradenitis Suppurativa. Dermatol. Clin. 2016, 34, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Nelson, A.M.; Cong, Z.; Gettle, S.L.; Longenecker, A.L.; Kidacki, M.; Kirby, J.S.; Adams, D.R.; Stairs, D.B.; Danby, F.W. E-cadherin and p120ctn protein expression are lost in hidradenitis suppurativa lesions. Exp. Dermatol. 2019, 28, 867–871. [Google Scholar] [CrossRef]
- Von Laffert, M.; Helmbold, P.; Wohlrab, J.; Fiedler, E.; Stadie, V.; Marsch, W.C. Hidradenitis suppurativa (acne inversa): Early inflammatory events at terminal follicles and at interfollicular epidermis. Exp. Dermatol. 2010, 19, 533–537. [Google Scholar] [CrossRef]
- Kokolakis, G.; Sabat, R. Distinguishing Mild, Moderate, and Severe Hidradenitis Suppurativa. JAMA Dermatol. 2018, 154, 971–972. [Google Scholar] [CrossRef]
- Gao, M.; Wang, P.-G.; Cui, Y.; Yang, S.; Zhang, Y.-H.; Lin, D.; Zhang, K.-Y.; Liang, Y.-H.; Sun, L.-D.; Yan, K.-L.; et al. Inversa Acne (Hidradenitis Suppurativa): A Case Report and Identification of the Locus at Chromosome 1p21.1–1q25.3. J. Investig. Dermatol. 2006, 126, 1302–1306. [Google Scholar] [CrossRef] [PubMed]
- Wolkenstein, P.; Loundou, A.; Barrau, K.; Auquier, P.; Revuz, J.; Quality of Life Group of the French Society of Dermatology. Quality of Life Impairment in Hidradenitis Suppurativa: A Study of 61 Cases. J. Am. Acad. Dermatol. 2007, 56, 621–623. [Google Scholar] [CrossRef]
- Jansen, T.; Altmeyer, P.; Plewig, G. Acne inversa (alias hidradenitis suppurativa). J. Eur. Acad. Dermatol. Venereol. 2001, 15, 532–540. [Google Scholar] [CrossRef]
- Sartorius, K.; Emtestam, L.; Jemec, G.; Lapins, J. Objective scoring of hidradenitis suppurativa reflecting the role of tobacco smoking and obesity. Br. J. Dermatol. 2009, 161, 831–839. [Google Scholar] [CrossRef]
- Moltrasio, C.; Tricarico, P.M.; Romagnuolo, M.; Marzano, A.V.; Crovella, S. Hidradenitis Suppurativa: A Perspective on Genetic Factors Involved in the Disease. Biomedicines 2022, 10, 2039. [Google Scholar] [CrossRef] [PubMed]
- Fitzsimmons, J.S.; Guilbert, P.R.; Fitzsimmons, E.M. Evidence of genetic factors in hidradenitis suppurativa. Br. J. Dermatol. 1985, 113, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Pink, A.E.; Simpson, M.A.; Desai, N.; Dafou, D.; Hills, A.; Mortimer, P.; Smith, C.H.; Trembath, R.C.; Barker, J.N.W. Mutations in the γ-Secretase Genes NCSTN, PSENEN, and PSEN1 Underlie Rare Forms of Hidradenitis Suppurativa (Acne In-versa). J. Investig. Dermatol. 2012, 132, 2459–2461. [Google Scholar] [CrossRef]
- Nomura, T. Hidradenitis Suppurativa as a Potential Subtype of Autoinflammatory Keratinization Disease. Front. Immunol. 2020, 11, 847. [Google Scholar] [CrossRef]
- Shi, T.; Bai, N.; Zhang, J.; Lu, F.; Chen, X.; Kong, X.; Yu, J. Mutations in the γ-Secretase Genes PSEN1, PSENEN, and NCSTN in a Family with Acne Inversa. Eur. J. Dermatol. 2018, 28, 374–376. [Google Scholar] [CrossRef]
- Riis, P.T.; Loft, I.; Yazdanyar, S.; Andersen, R.K.; Pedersen, O.; Ring, H.; Huber, R.; Sultan, M.; Loesche, C.; Saunte, D.; et al. Full exome sequencing of 11 families with Hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2020, 35, 1203–1211. [Google Scholar] [CrossRef]
- Jfri, A.H.; O’brien, E.A.; Litvinov, I.V.; Alavi, A.; Netchiporouk, E. Hidradenitis Suppurativa: Comprehensive Review of Predisposing Genetic Mutations and Changes. J. Cutan. Med. Surg. 2019, 23, 519–527. [Google Scholar] [CrossRef]
- Mintoff, D.; Borg, I.; Pace, N.P. The Clinical Relevance of the Microbiome in Hidradenitis Suppurativa: A Systematic Review. Vaccines 2021, 9, 1076. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Nogueira da Costa, A.; Makrantonaki, E.; Hou, X.X.; Almansouri, D.; Dudley, J.T.; Edwards, H.; Readhead, B.; Balthasar, O.; Jemec, G.B.; et al. Alterations in innate immunity and epithelial cell differentiation are the molecular pillars of hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 846–861. [Google Scholar] [CrossRef]
- Nikolakis, G.; Kokolakis, G.; Kaleta, K.; Wolk, K.; Hunger, R.; Sabat, R.; Zouboulis, C.C. Pathogenesis of Hidradenitis Suppurativa/Acne Inversa. Der Hautarzt 2021, 72, 658–665. [Google Scholar] [CrossRef]
- Goecks, J.; Nekrutenko, A.; Taylor, J.; The Galaxy Team. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015, 10, 1556–1566. [Google Scholar] [CrossRef] [PubMed]
- Stelzer, G.; Plaschkes, I.; Oz-Levi, D.; Alkelai, A.; Olender, T.; Zimmerman, S.; Twik, M.; Belinky, F.; Fishilevich, S.; Nudel, R.; et al. VarElect: The phenotype-based variation prioritizer of the GeneCards Suite. BMC Genom. 2016, 17, 195–206. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef]
- Rentzsch, P.; Witten, D.; Cooper, G.M.; Shendure, J.; Kircher, M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019, 47, D886–D894. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef] [PubMed]
- Stappers, M.H.T.; Thys, Y.; Oosting, M.; Plantinga, T.S.; Ioana, M.; Reimnitz, P.; Mouton, J.W.; Netea, M.G.; Joosten, L.A.B.; Gyssens, I.C. TLR1, TLR2, and TLR6 Gene Polymorphisms Are Associated with Increased Susceptibility to Complicated Skin and Skin Structure Infections. J. Infect. Dis. 2014, 210, 311–318. [Google Scholar] [CrossRef][Green Version]
- Hunger, R.E.; Surovy, A.M.; Hassan, A.S.; Braathen, L.R.; Yawalkar, N. Toll-like receptor 2 is highly expressed in lesions of acne inversa and colocalizes with C-type lectin receptor. Br. J. Dermatol. 2008, 158, 691–697. [Google Scholar] [CrossRef]
- Etokebe, G.E.; Skjeldal, F.; Nilsen, N.; Rodionov, D.; Knezevic, J.; Bulat-Kardum, L.; Espevik, T.; Bakke, O.; Dembic, Z. Toll-Like Receptor 2 (P631H) Mutant Impairs Membrane Internalization and is a Dominant Negative Allele. Scand. J. Immunol. 2010, 71, 369–381. [Google Scholar] [CrossRef] [PubMed]
- Vergnano, M.; Mockenhaupt, M.; Benzian-Olsson, N.; Paulmann, M.; Grys, K.; Mahil, S.K.; Chaloner, C.; Barbosa, I.A.; August, S.; Burden, A.D.; et al. Loss-of-Function Myeloperoxidase Mutations Are Associated with Increased Neutrophil Counts and Pustular Skin Disease. Am. J. Hum. Genet. 2020, 107, 539–543. [Google Scholar] [CrossRef] [PubMed]
- Haskamp, S.; Bruns, H.; Hahn, M.; Hoffmann, M.; Gregor, A.; Löhr, S.; Hahn, J.; Schauer, C.; Ringer, M.; Flamann, C.; et al. Myeloperoxidase Modulates Inflammation in Generalized Pustular Psoriasis and Additional Rare Pustular Skin Diseases. Am. J. Hum. Genet. 2020, 107, 527–538. [Google Scholar] [CrossRef]
- Marchetti, C.; Patriarca, P.; Solero, G.P.; Baralle, F.E.; Romano, M. Genetic Characterization of Myeloperoxidase Deficiency in Italy. Hum. Mutat. 2004, 23, 496–505. [Google Scholar] [CrossRef]
- Marzano, A.V.; Genovese, G.; Moltrasio, C.; Tricarico, P.M.; Gratton, R.; Piaserico, S.; Garcovich, S.; Boniotto, M.; Brandão, L.; Moura, R.; et al. Whole-Exome Sequencing in 10 Unrelated Patients with Syndromic Hidradenitis Suppurativa: A Preliminary Step for a Genotype-Phenotype Correlation. Dermatology 2022, 238, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Allam, J.-P.; Wenzel, J.; Jaksche, A.; Friedrichs, N.; Bieber, T.; Novak, N. Keratitis-Ichthyosis-Deafness Syndrome in Association with Follicular Occlusion Triad. Eur. J. Dermatol. 2005, 15, 347–352. [Google Scholar]
- Montgomery, J.R.; White, T.W.; Martin, B.L.; Turner, M.L.; Holland, S.M. A Novel Connexin 26 Gene Mutation Associated with Features of the Keratitis-Ichthyosis-Deafness Syndrome and the Follicular Occlusion Triad. J. Am. Acad. Dermatol. 2004, 51, 377–382. [Google Scholar] [CrossRef]
- Lazic, T.; Li, Q.; Frank, M.; Uitto, J.; Zhou, L.H. Extending the Phenotypic Spectrum of Keratitis-ichthyosis-deafness Syndrome: Report of a Patient with GJB2 (G12R) Connexin 26 Mutation and Unusual Clinical Findings. Pediatr. Dermatol. 2012, 29, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.; Le Jan, S.; Muller, C.; François, C.; Renard, Y.; Durlach, A.; Bernard, P.; Reguiai, Z.; Antonicelli, F. Matrix Remodelling and MMP Expression/Activation Are Associated with Hidradenitis Suppurativa Skin Inflammation. Exp. Dermatol. 2019, 28, 593–600. [Google Scholar] [CrossRef]
- Mozeika, E.; Pilmane, M.; Nürnberg, B.M.; Jemec, G.B. Tumour Necrosis Factor-alpha and Matrix Metalloproteinase-2 are Expressed Strongly in Hidradenitis Suppurativa. Acta Derm. Venereol. 2013, 93, 301–304. [Google Scholar] [CrossRef]
- Savva, A.; Kanni, T.; Damoraki, G.; Kotsaki, A.; Giatrakou, S.; Grech, I.; Katoulis, A.; Papadavid, E.; Giamarellos-Bourboulis, E. Impact of Toll-like receptor-4 and tumour necrosis factor gene polymorphisms in patients with hidradenitis suppurativa. Br. J. Dermatol. 2013, 168, 311–317. [Google Scholar] [CrossRef] [PubMed]
| Gene | Position † | RefSeq Gene and HGVS Nomenclature †† | MAF | CADD | Mutation Taster | SIFT | Polyphen | GERP | |
|---|---|---|---|---|---|---|---|---|---|
| II-1T | TLR2 | chr4:154625951 | NM_001318789: c.1892C>A:p.(Pro631His) | 7944/282,314 | 25.2 | Uncertain | deleterious | Probably damaging | 5.65 |
| MPO | chr17:56355397 | NM_000250: c.995G>A:p.(Ala332Val) | 3566/282,336 | 23.9 | Benign moderate | deleterious | Benign | 5.32 | |
| II-2G | MMP2 | chr16:55532218 | NM_004530: c.1627T>C p.(Tyr543His) | 31/282,540 | 30 | Uncertain | deleterious | Probably damaging | 6.07 |
| GJB2 | chr13:20763243 | NM_004004: c.478C>T p.(Gly160Ser) | 170/282,496 | 25.2 | Uncertain | tolerated | Probably damaging | 5.47 | |
| TLR4 | chr9:120476597 | NM_138554: c.2191C>T p.(Arg731Ter) | 13/277,454 | 40 | Disease causing | na | na | 6.03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azzarà, A.; Cassano, I.; Lintas, C.; Gurrieri, F. Exome Profiling Suggests Combined Effect of Myeloperoxidase, Toll-Like Receptors, and Metallopeptidase in Hidradenitis Suppurativa. Biomedicines 2024, 12, 2498. https://doi.org/10.3390/biomedicines12112498
Azzarà A, Cassano I, Lintas C, Gurrieri F. Exome Profiling Suggests Combined Effect of Myeloperoxidase, Toll-Like Receptors, and Metallopeptidase in Hidradenitis Suppurativa. Biomedicines. 2024; 12(11):2498. https://doi.org/10.3390/biomedicines12112498
Chicago/Turabian StyleAzzarà, Alessia, Ilaria Cassano, Carla Lintas, and Fiorella Gurrieri. 2024. "Exome Profiling Suggests Combined Effect of Myeloperoxidase, Toll-Like Receptors, and Metallopeptidase in Hidradenitis Suppurativa" Biomedicines 12, no. 11: 2498. https://doi.org/10.3390/biomedicines12112498
APA StyleAzzarà, A., Cassano, I., Lintas, C., & Gurrieri, F. (2024). Exome Profiling Suggests Combined Effect of Myeloperoxidase, Toll-Like Receptors, and Metallopeptidase in Hidradenitis Suppurativa. Biomedicines, 12(11), 2498. https://doi.org/10.3390/biomedicines12112498







