Abstract
Lung cancer demands innovative approaches for early detection and targeted treatment. In addressing this urgent need, exosomes play a pivotal role in revolutionizing both the early detection and targeted treatment of lung cancer. Their remarkable capacity to encapsulate a diverse range of biomolecules, traverse biological barriers, and be engineered with specific targeting molecules makes them highly promising for both diagnostic markers and precise drug delivery to cancer cells. Furthermore, an in-depth analysis of exosomal content and biogenesis offers crucial insights into the molecular profile of lung tumors. This knowledge holds significant potential for the development of targeted therapies and innovative diagnostic strategies for cancer. Despite notable progress in this field, challenges in standardization and cargo loading persist. Collaborative research efforts are imperative to maximize the potential of exosomes and advance the field of precision medicine for the benefit of lung cancer patients.
1. Introduction
Cancer, as stated by the World Health Organization (WHO), stands as a leading cause of mortality worldwide [1]. Specifically, lung cancer is among the most reported cancer-related deaths globally [2,3], constituting 11.6% of new cancer diagnoses and accounting for 19.8% of cancer-related fatalities [4,5].
Regrettably, despite a decline in lung cancer mortality rates, a significant proportion of patients continue to receive diagnoses in advanced or metastatic stages, resulting in poor prognoses [6]. Advanced stages pose greater therapeutic challenges and exhibit a heightened propensity for developing resistance to treatment. Various treatment modalities have been employed in lung cancer care, each with its own advantages and drawbacks. Surgery remains a viable option for early-stage cases [7]. Chemotherapy, radiotherapy, and immunotherapy, either individually or in combination, are typical approaches for both early and advanced stages [8,9,10]. The differential factor lies in the responsiveness of lung cancer cells to these treatments, with a higher sensitivity observed during the early stages.
Hence, the study of tumorigenesis and therapeutic strategies against cancers, particularly lung cancer, is underscored. Recent research has yielded fresh insights into lung cancer biology, yielding notable progress in the realm of targeted therapies focused on novel biomarkers. These include molecular therapies targeting the epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), proto-oncogene B-Raf (BRAF), and immunotherapies employing checkpoint inhibitors against the programmed cell death 1 (PD-1) and programmed death-ligand 1 (PD-L1) pathways [8,11,12]. Nonetheless, lung cancer still lags behind with one of the lowest 5-year survival rates, standing at a mere 18%, among all cancer types [9].
Extracellular vesicles (EVs), which are a collective term covering exosomes, microvesicles (MVs), microparticles, ectosomes, oncosomes, and apoptotic bodies, were first discovered by Pan and Johnson during the maturation of sheep reticulocytes in 1983 [13]. EVs are defined as heterogeneous entities of phospholipid bilayer membrane-bound vesicles without any means of replication [14]. They were initially thought to be “cellular dust” or served as a mechanism by which cells actively dispose of their own waste [15]. In 1996, exosomes were found to play a role in intercellular communication [16]. Due to the heterogeneity between and within exosome types and the overlap in characteristics between exosomes and other EVs, it is difficult to define exosomes in a way that distinguishes them from other EVs. Over the past few years, it has been discovered that exosomes, which contain proteins and miRNAs with biological effects, have the ability to specifically transport and deliver these bioactive cargoes to tumor cells [15].
Exosomes, ranging from 30 to 150 nanometers in diameter, emerge as crucial biological nano-scale lipid bilayer vesicles [17,18]. These vesicles are secreted by various cell types, including dendritic cells, macrophages, B cells, T cells, mesenchymal stem cells, endothelial cells, epithelial cells, and several cancer cells. They float within a sucrose density gradient solution at a density of 1.13–1.19 g mL−1 and carry an array of biomolecules, such as proteins (either membrane-bound or encapsulated within the vesicle), RNA (comprising coding mRNA or diverse non-coding RNAs), DNA (both double-stranded and single-stranded), as well as glycans [16,19,20]. They have been reported in all biological fluids, and the composition of the complex cargo of exosomes is readily accessible via the sampling of biological fluids (liquid biopsies) [21,22]. Cumulative evidence has revealed that exosomes play an irreplaceable role in prognostic, diagnostic, and even therapeutic aspects [21,23]. Nanotechnology has been employed for drug delivery for increasing the bioavailability of therapeutic agents. Unfortunately, drug nanoformulations often lead to toxicity and are usually rapidly cleared through the mononuclear phagocytic system (MPS) [24,25].
EVs can be categorized, based on their size, into microvesicles (MVs; 100–1000 nm) and exosomes (EXOs; 30–150 nm). They play pivotal roles in immune responses, angiogenesis, neuronal regeneration, anti-inflammation, coagulation, and in the intracellular degradation pathways [14,26].
Exosomes’ unique properties make them promising tools for therapeutically targeting diseases, including neurodegenerative conditions and various cancers [27]. They have shown promise as non-invasive biomarkers for chronic pain mechanisms [28] and as potential carriers for biomarkers and drugs in conditions such as human immunodeficiency virus (HIV) [29]. They can cross biological barriers, including the blood–brain barrier, and have neuroprotective and tissue repair effects [30,31]. It has been demonstrated that exosomes have potential applications in a wide range of fields, from oncology (lung, liver, pancreatic, colorectal, gastric, kidney, bladder, prostate, breast, ovarian, cervical, head and neck, thyroid, glioma, melanoma, and hematological malignancies) to neurodegenerative disorders, including Alzheimer’s disease, Parkinson’s disease, Huntington’s diseases, and amyotrophic lateral sclerosis (ALS), mental health conditions, cardiovascular diseases, diabetes, and inflammatory/autoimmune disorders [27].
Numerous studies have investigated exosomes from diverse angles in the context of lung cancer. These encompass experimental techniques and isolation methods, detailed characterization, insights into biogenesis and secretion, understanding molecular composition and functions, communication within the tumor environment, impact on tumor progression, and the potential clinical applications in cancer treatment, among other varied perspectives.
2. The Biogenesis of Exosomes
Exosomes are small vesicles generated from late endosomes, which are formed via the inward budding of the multivesicular body (MVB) membrane. They are different from other vesicles called MVs, which are larger and generated via outward budding from the plasma membrane [32]. Exosomes have an approximate size range from 30 nm to 150 nm [17,18] and contain proteins, mRNAs, microRNAs, and lipids. They are derived from various cellular sources and have biomarkers such as TSG101, CHMP2A, RAB11B, CD63, and CD81 [33]. Exosomes play a crucial role in cell-to-cell communication and are involved in various biological processes and diseases [34]. They can escape clearance by the immune system and have a longer circulation time compared to other vesicles [32]. The formation of exosomes involves the invagination of late endosomal membranes, resulting in the formation of intraluminal vesicles (ILVs) within MVBs [35]. ILVs are eventually secreted as exosomes through fusion of MVBs with the plasma membrane [36]. The content of exosomes depends on the cell type they originate from and can include proteins and components from various cellular compartments [33].
Exosomes have a distinct shape, appearing biconcave or cup-like when dried and spheroid when observed under a microscope [32]. Exosomes can form early-sorting endosomes (ESEs) de novo or merge with existing ESEs. The trans-Golgi network and endoplasmic reticulum contribute to ESE formation. ESEs mature into late-sorting endosomes (LSEs) and eventually generate MVBs, also known as multivesicular endosomes [20]. During maturation, the content of endosomes becomes acidic, and markers such as Rab5 are replaced by Rab7, Rab9, and the mannose-6-phosphate receptor to indicate late endosomes [35]. The influence of specific markers or cargo on these pathways is still unclear [37].
Several mechanisms contribute to exosome generation. The endosomal sorting complexes required for transport (ESCRT-0, -I, -II, and -III) and the ATPase Vps4 complex are important in this process [38]. SNARE proteins and RAB GTPases are also involved in exosome secretion [39,40]. Even when key subunits of ESCRTs are depleted, ESCRT-independent ILV biogenesis can still occur [41]. Tetraspanins and lipids play significant roles in exosome generation and release [42]. Identifying molecules that regulate exosome biogenesis could lead to therapeutic targets for controlling exosome secretion and modulating related signaling pathways in conditions such as metastasis and inflammation [43,44]. Cholesterol treatment in hepatoma cells reduces MVBs and increases exosome secretion with the ability to induce specific immune responses. Statins, known for lowering cholesterol, have been shown to reduce exosome release, suggesting their potential as therapeutic agents to control exosome production in specific cells [42].
Exosome biogenesis and release are influenced by various molecules, including components of the ESCRT machinery, Rab GTPases, tetraspanins, and the adaptor protein syntenin. The ESCRT mechanism comprises four separate protein complexes (ESCRT-0 through III) that work together to facilitate the formation of MVBs, vesicle budding, and sorting of protein cargo. Ubiquitinated proteins are recognized and sequestered to specific domains of the endosomal membrane by the ubiquitin-binding subunits of ESCRT-0, initiating the process. ESCRT-I and ESCRT-II complexes interact with ESCRT-III to promote budding. The ESCRT-III complex then separates from the MVB membrane, aided by the Vps4 sorting protein, after cleaving the buds to form ILVs [32,45].
In a study using HeLa cells, an RNAi screen targeting twenty-three ESCRT and ESCRT-associated proteins identified seven proteins that impacted exosome secretion. The depletion of ESCRT-0 proteins (Hrs and TSG101) and the ESCRT-I protein STAM1 reduced exosome secretion, while the knockdown of ESCRT-III and its associated proteins (CHMP4C, VPS4B, VTA1, and ALIX) increased exosome secretion. Further analysis verified the role of these proteins, with Hrs, TSG101, and STAM1 depletion decreasing exosome secretion and VPS4B knockdown increasing it. ALIX depletion appeared to affect the protein composition of exosomes rather than secretion, suggesting it may impact cargo loading and the types of MVBs destined for secretion [46,47].
The interaction between syntenin and ALIX is important for sorting syndecans, membrane proteins with heparan sulfate chains, into exosomes. Syndecan sorting and ILV formation are facilitated by syntenin, which binds to both syndecans and ALIX [47,48]. Heparanases trim the heparan sulfate chains, promoting the formation of syndecan clusters that enhance binding to syntenin. Heparanase also stimulates the sorting of CD63, indicating a potential relationship between the sorting of these two molecules [47,49]. The syndecan–syntenin–ALIX mechanism is estimated to control around 50% of secreted vesicles in MCF-7 cells, suggesting the involvement of different sorting mechanisms in exosomal molecule sorting [47,50]. These findings suggest that ESCRT function plays a role in exosomal biogenesis.
Cellular metabolic status, including ceramide metabolism, ER stress, autophagy, and intracellular calcium levels, can impact exosome production. Adiponectin, a protein secreted by adipocytes, can stimulate exosome biogenesis in certain cell types [38,51]. Glucagon has been found to regulate exosome production in endothelial cells in adipose tissues [38,52]. The interaction between the exosome biogenesis pathway and the other molecular pathways involved in intracellular vesicle trafficking can complicate the interpretation of functional studies. Different cell types, culture conditions, and cell health can also affect the regulation of exosome biogenesis [20,53]. Inconsistencies in identifying regulatory elements may arise from variations in the methods used for exosome production, enrichment, and concentration [20,54].
The specific case of exosome biogenesis in human syncytiotrophoblasts has been studied at the ultrastructural level. Certain molecules, such as MICA/B, ULBP 1–5, FasL, TRAIL, and PD-L1, are present on the membranes of MVBs and nanosized vesicles within MVBs, but not on the plasma membrane of syncytiotrophoblasts. These molecules may be sorted from the Golgi apparatus to the MVBs. MICA and MICB are expressed on the apical membrane of syncytiotrophoblasts and within exosomes contained within MVBs [35,55].
ESCRTs are not only involved in exosome release but also play a role in packaging biomolecules into exosomes [56]. Heparanase modulates the syndecan–syntenin–ALIX pathway, leading to endosomal membrane budding and subsequent exosome formation by trimming heparan sulfate chains on syndecans [49,56]. However, previous research indicated that cargo molecules within exosomes are segregated into distinct subdomains on the endosomal membrane [56,57]. Additionally, the transfer of exosome-associated domains into the endosomal lumen does not rely on ESCRT function but is induced by sphingolipid ceramide. Purified exosomes contain ceramides, and the inhibition of neutral sphingomyelinases reduces exosome release, suggesting the regulatory role of lipids in exosome secretion [56].
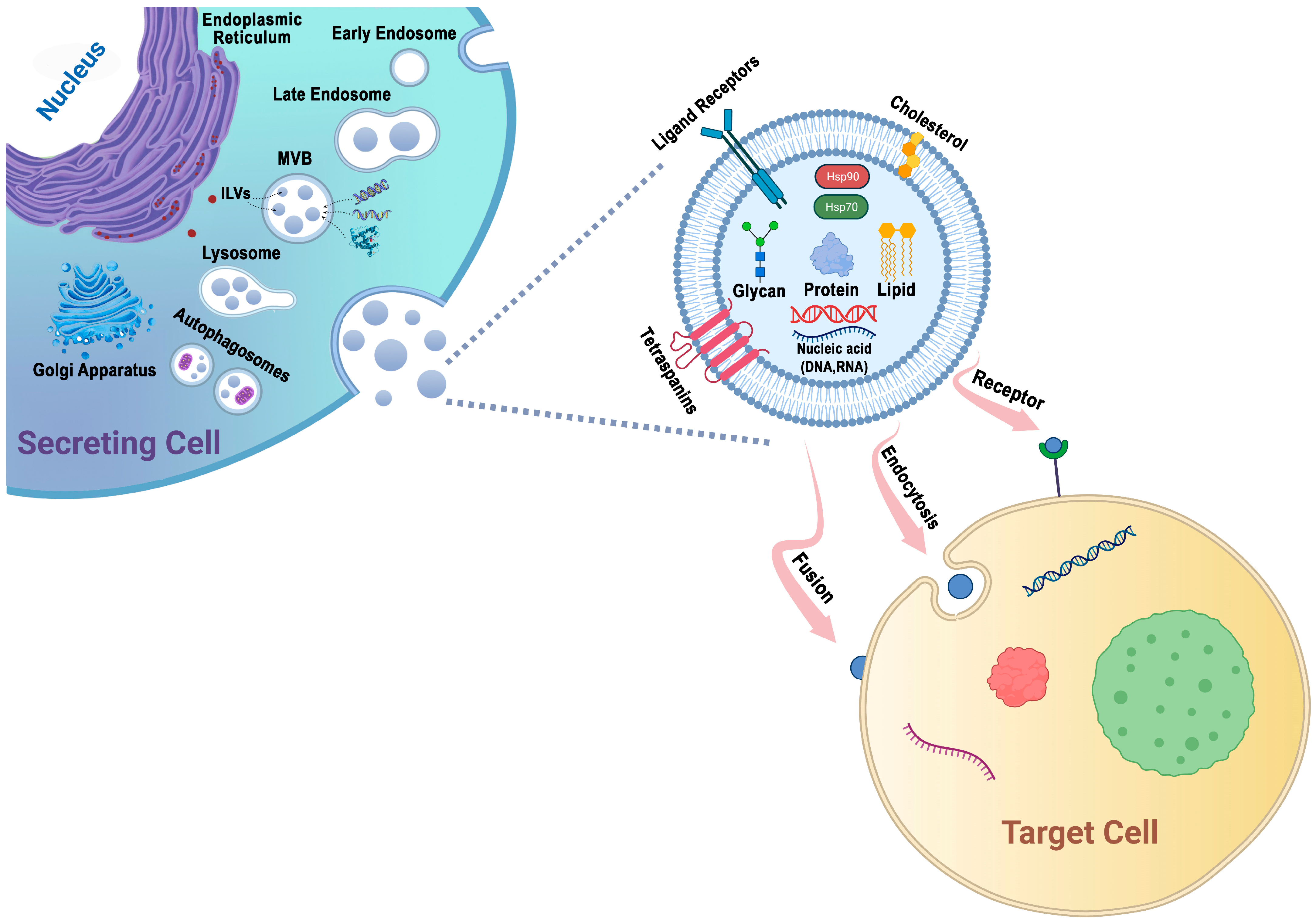
Protein sorting within MVBs, the precursors of exosomes, can occur through the ESCRT-dependent and ESCRT-independent pathways. These pathways may work in synergy, and different subpopulations of exosomes may depend on distinct machinery [47,58] (Figure 1).
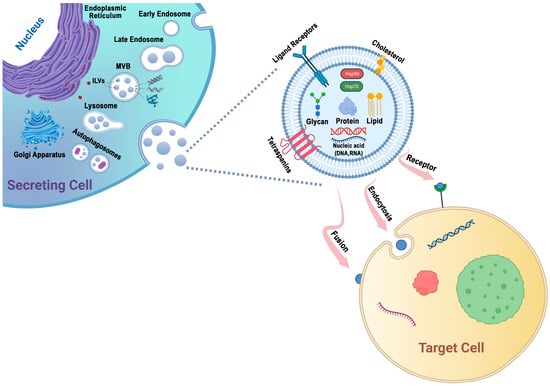
Figure 1.
Exosomes are generated through a series of biogenetic processes, starting with the maturation of early endosomes into late endosomes. These late endosomes then transform into MVBs. Within the MVBs, ILVs are formed via the inward budding of the endosomal membrane, which selectively captures proteins, nucleic acids, glycans, and lipids. The MVBs have two main fates: they can fuse with lysosomes for degradation or fuse with the plasma membrane, leading to the release of ILVs as exosomes into the extracellular space. The generation and release of exosomes are influenced by molecules such as tetraspanins and lipids, including cholesterol. These components play important roles in facilitating exosome formation and release. Exosomes possess the capability to adhere to surface receptors present on the target cell, promote endocytosis, or engage in direct fusion with the plasma membrane of the target cell.
3. The Isolation of Exosomes
One of the main challenges in exosome research is the lack of an efficient and standardized isolation strategy for specific exosome subpopulations, leading to heterogeneity in their size and molecular contents. However, advances in nanotechnologies and microfluidics have shown promise in incorporating microfluidics into exosome isolation, offering potential tools for future research [59].
Exosomes, although playing a crucial role in early detection and treatment, present challenges due to their small size, low density, and similarity to other EVs in body fluids [60]. Separating the exosomes from cell debris and other EVs is essential, and different isolation strategies based on their size and affinity have been developed [61]. These include ultracentrifugation, size-based techniques, immunoaffinity capture, EV precipitation, and microfluidics-based techniques. Each method has its advantages and disadvantages, and the appropriate choice depends on the research purpose [62,63]. Microfluidics, in particular, offers an integrated platform with properties such as high purity, high throughput, and low sample volumes [59].
While traditional methods, like ultracentrifugation, are widely used, they have drawbacks, such as large sample consumption, potential damage to exosomes, low purity, and lengthy procedures. Common exosome isolation technologies, such as ultrafiltration, immunoaffinity, and ultracentrifugation, can be expensive and result in low isolation efficiency, sample loss, and contamination [59,64].
In summary, the development of efficient and standardized isolation strategies for specific exosome subpopulations remains a challenge. However, advancements in microfluidics and other techniques offer promising avenues for future research, with microfluidics showing potential due to its integrated platform and desirable properties [59]. The choice of isolation method should consider the research purpose, and while traditional methods are widely used, they have limitations that hinder meeting the increasing demands of scientific research [63].
4. Exosomes: Effective Drug Delivery Vehicles
The development of drug delivery systems (DDSs) with enhanced therapeutic effectiveness has garnered significant attention [65,66]. Nanoscale drug carriers offer advantages over traditional chemotherapeutic agents, including targeted accumulation in tumors and reduced toxicity in normal cells [67]. Active targeting can be achieved by binding nanocarriers to molecules that bind to overexpressed antigens [68]. While most drug delivery vehicles are chemically synthesized using lipids or lipid-like molecules, natural nanoparticles derived from bacteria, viruses, red blood cells, and lymphocytes have emerged as potential alternatives, as they exhibit advantages such as evasion of the host immune system and efficient cellular entry [69].
Exosomes, a type of natural nanoparticle, have been investigated as direct therapeutic approaches for treating diseases or injuries [30]. They possess the ability to transfer proteins and nucleic acids between cells, making them suitable for drug delivery. Exosomes offer benefits, such as tissue specificity, safety, and stability, as they can deliver cargo across plasma membranes into the correct cellular compartments to exert a functional response [69,70]. They also protect their cargo from clearance or damage by complement fixation or macrophages due to their double-layered membrane and nanoscale size, thereby prolonging circulation and improving biological activity [71]. Additionally, exosomes display intrinsic homing capabilities, enabling them to specifically target tissues [72].
Although the field of exosome-based therapeutics is still in its early stages, progress has been made in engineering exosomes to display specific proteins, incorporate nucleic acid and protein cargo, and load therapeutic agents [73]. The size and stability of exosomes make them favorable for drug delivery systems, as they exhibit reduced aggregation and precipitation. For example, milk-derived exosomes can be stored for periods of time without causing a loss of activity. Overall, exosomal drug delivery holds the potential to enhance drug efficacy by prolonging circulation times and facilitating cellular uptake [74].
5. Approaches for Drug Loading on Exosomes
Methods for loading cargo into exosomes can be categorized into cell-based and non-cell-based approaches. In the cell-based approach, cargo is delivered to donor cells, which then package it into EVs for secretion and therapeutic use. The non-cell-based approach involves directly loading drugs into isolated EVs using techniques like electroporation, extrusion, sonication, and bio-conjugation. However, the loading efficiencies of these methods are not always desirable. The drug encapsulation process can occur through postloading, preloading, or fusion methods, and active loading and passive methods are employed to overcome the challenges of incorporating drugs into the lipid bilayer membrane of exosomes [75,76].
To enhance the therapeutic potential of exosomes, various targeting strategies are employed. These strategies can involve cellular modification through engineering techniques, altering the surface expression molecules and cargo of exosomes [73]. Modifying exosomal tetraspanin complexes can improve target cell selection and enhance tissue and cell type-specific targeting [77]. Lentiviral vectors can be utilized to create exosomes with modified membrane proteins for in vivo and cell uptake studies [78]. Alternatively, direct modifications can be achieved through passive loading techniques that leverage spontaneous membrane interactions or physical methods that temporarily disrupt the integrity of the membranes to facilitate cargo loading into exosomes [75].
6. Lung Cancer and the Role of Exosomes in Its Diagnosis and Treatment
Lung cancer is classified into two primary categories: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC) [79].
SCLC is an extremely aggressive neuroendocrine carcinoma. The prognosis for SCLC patients is extremely poor, with a 5-year survival rate of less than 5% and an average overall survival period of only 2–4 months for untreated patients. SCLC represents approximately 15% of all lung cancer cases [80,81].
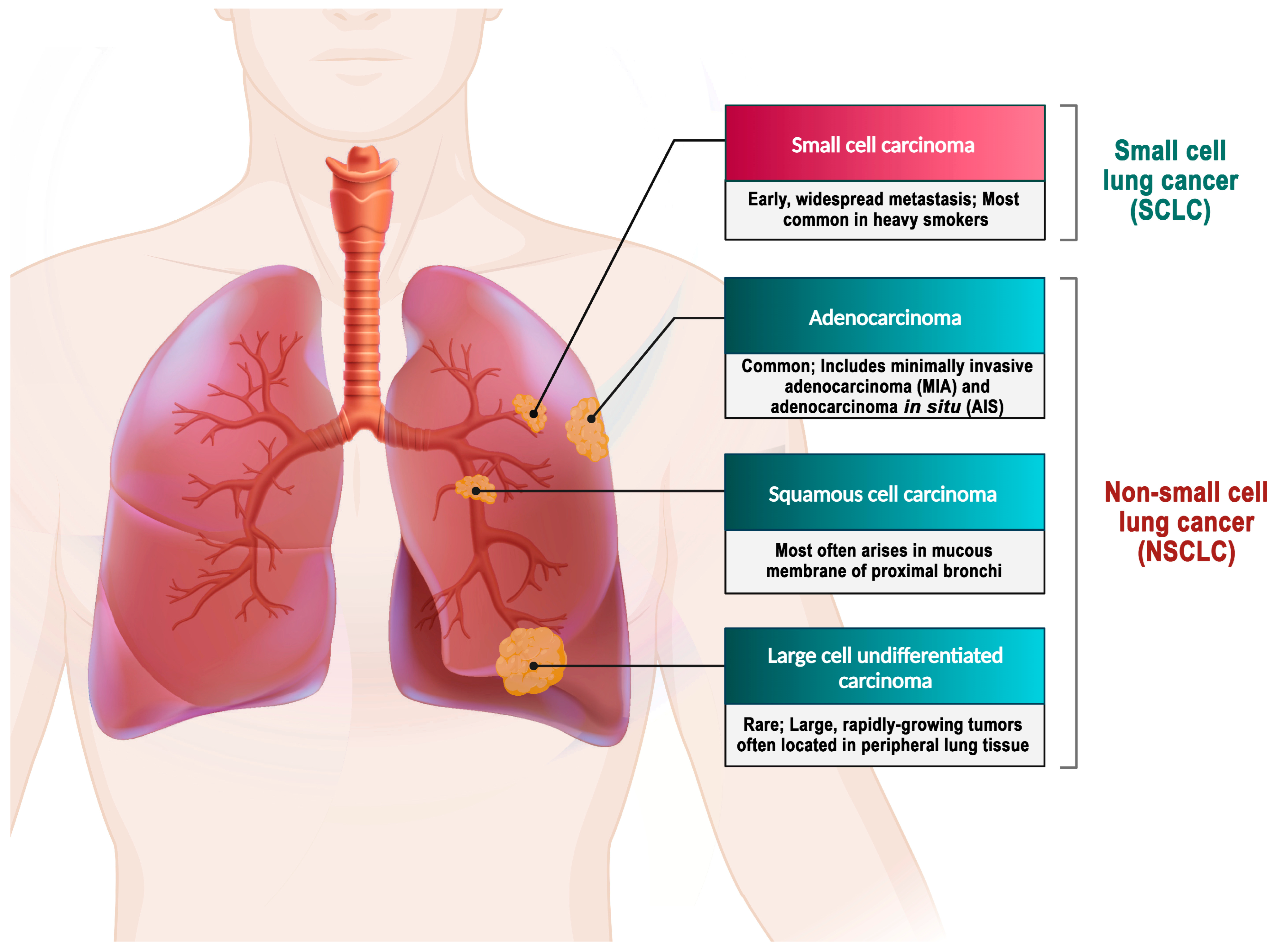
In contrast, NSCLC comprises approximately 85% of all lung cancer cases [82]. Histologically, NSCLC is classified into different types, namely adenocarcinoma, large-cell carcinoma, and squamous cell carcinoma. The stages of this disease are further categorized based on specific criteria [3,79]. Adenocarcinoma represents 38.5% of all lung cancer cases, while squamous-cell carcinoma accounts for 20%, and large-cell carcinoma accounts for 2.9% [4] (Figure 2).
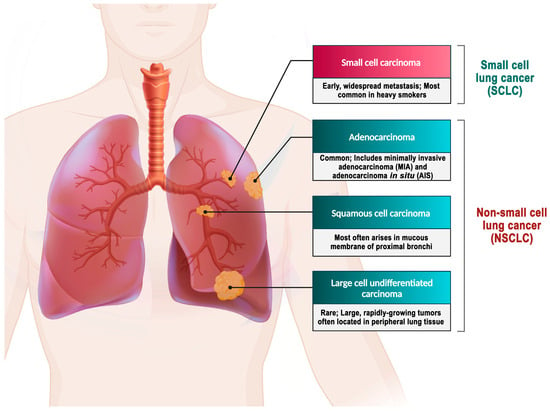
Figure 2.
Types of lung cancer. Adenocarcinoma is the most prevalent form of lung cancer, accounting for approximately 40% of all cases [83]. A mutation in the EGFR has been identified as one of the early detectable mutations in lung adenocarcinoma, suggesting its potential as a biomarker for the early detection of this disease [84]. About 25–30% of all lung cancer cases are squamous cell carcinoma, which develops from the premature forms of squamous cells in the airway epithelial cells located in the bronchial tubes at the center of the lungs. This type of NSCLC is highly associated with smoking cigarettes [85]. Large-cell carcinoma is an uncommon form of malignant lung tumor, accounting for 2.9% of lung cancer cases. Despite being categorized as NSCLC, large-cell carcinoma is more severe and malignant than other NSCLCs and shares similar biological characteristics with SCLC. The majority of large-cell carcinoma patients are elderly males who smoke, and their clinical symptoms are not specific [4,86].
6.1. Role of Exosomes in Lung Cancer Diagnosis
Exosomes are emerging as valuable candidates for both diagnosing and predicting lung cancer due to their detectability in various bodily fluids, such as plasma, serum, and alveolar lavage fluid. Traditional methods, like low-dose computed tomography (LDCT), for lung cancer screening can result in cumulative radiation exposure and side effects. Tissue sample analysis, the gold standard, is hindered by sampling bias and invasiveness. In contrast, exosomes provide a non-invasive liquid biopsy option with the potential for early cancer detection. They carry unique proteins, receptors, signaling molecules, and lipids that can be compared with healthy controls to identify cellular abnormalities. Exosomes hold promise as diagnostic tools for chronic diseases and are particularly valuable in biomarker research due to their stability, cargo of proteins and RNA, and ability to present antigens [87,88,89].
There are several clinical trials that have been conducted on lung cancer diagnosis via exosome detection in clinicaltrials.gov/, (accessed on 27 November 2023) (Table 1). Nano-sized exosomes in circulation offer components, like proteins, miRNAs, and lncRNAs, for cancer diagnosis and monitoring. They contain mRNA, miRNAs, dsDNA, and proteins, making them valuable predictive and prognostic biomarkers for different tumor types. Their role in intercellular communication and protection of membrane bilayers and contents from degradation adds to their potential [88,89,90].

Table 1.
Clinical trials on lung cancer diagnosis and treatment via exosome detection since 2015.
Researchers have identified exosomal miRNAs as promising biomarkers for early lung cancer diagnosis due to their stability, accessibility, and specificity [87]. One study highlighted miR-1268b and miR-6075 as promising serum markers for lung cancer diagnosis. Elevated levels of these biomarkers were detected even during early-stage lung cancer, indicating their potential as an early diagnostic indicator. When combined, miR-1268b and miR-6075 showed a high accuracy rate in distinguishing lung cancer patients from non-cancerous individuals, offering high sensitivity and specificity [91]. Moreover, research has indicated that exosomal miR-205 expression can differentiate between squamous and non-squamous NSCLCs, even in poorly differentiated tumors [92].
Another study discovered heightened miR-96 levels in lung cancer patients, notably in high-grade cancers, observed in tissues, serum, and isolated exosomes. This finding suggested its potential as a diagnostic biomarker for lung cancer. Elevated exosomal miR-96 correlated with increased lung cancer risk, higher stages, and metastatic tumors in the lymph nodes, indicating its involvement in cancer progression. It holds promise for diagnosing and predicting outcomes in NSCLC. The detection of miR-96 in serum could facilitate early identification, accurately assess cancer aggressiveness, and predict patient survival [93]. A study that focused on miR-451a in NSCLC patients post-surgery found it to be a promising biomarker for diagnosing cancer and predicting recurrence and prognosis across different disease stages (I–III). Higher levels of exosomal miR-451a correlated with lymph node metastasis, vascular invasion, and disease stage. This study hinted that these miRNAs in the plasma likely originate from the tumor tissue itself, supported by the correlation observed between the levels of exosomal miR-451a and its expression in the primary tumor tissues of the same patients [94].
Moreover, deregulation in the expression of miR-21, miR-143, and miR-181a has been correlated with clinical pathology and patient prognosis in NSCLC, indicating their usefulness as diagnostic or prognostic markers [95]. Additionally, studies have also linked miR-21 and miR-4257 expression with recurrence and poor survival in lung cancer cases [96]. Exosomal miRNAs, such as miR-181-5p, miR-30a-3p, miR-30e-3p, and miR-320b, have also been identified as biomarkers in NSCLC to differentiate between adenocarcinoma and squamous-cell carcinoma [97,98].
A pivotal breakthrough in the realm of lung cancer biomarkers took place in 2009 with the discovery of a group of 12 miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-199, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214). These miRNAs effectively distinguished NSCLC patients from their healthy counterparts. Remarkably, these miRNAs were not only detectable in lung cancer tissues but also in circulating exosomes, suggesting their potential as diagnostic indicators. Moreover, the levels of these miRNAs in the exosomes from NSCLC patients exhibited a substantial elevation when compared to those from healthy donors. This discovery held immense promise for more precise and timely diagnoses of lung cancer [99,100]. Further research has expanded these findings, leading to the development of miRNA profiles for lung cancer screening and diagnosis. A study introduced two miRNA profiles based on four miRNAs for screening and six miRNAs for diagnosis [101]. These profiles demonstrated high sensitivity and specificity, indicating their potential utility in identifying lung adenocarcinomas. Another study established a miRNA profile consisting of four miRNAs with high diagnostic accuracy for NSCLC, further highlighting the potential of exosomal miRNAs as diagnostic biomarkers [90,98].
Notably, the association of specific miRNAs with prognostic outcomes in NSCLC reveals their potential as valuable indicators for disease progression and treatment response. For instance, the downregulation of miR-503 in NSCLC tissues associates with advanced tumor stages and a poor prognosis, hinting at its potential as a prognostic indicator [102]. Additionally, exosomal miRNAs like miR-146-5p, miR-23b-3p, miR-10b-5p, and miR-21-5p have shown connections to survival rates and treatment responses in NSCLC. Combinations of these miRNAs and clinical variables enhance predictive models [98,103]. Moreover, miR-1246, miR-208a, and miR-100-5p have been linked to tumor proliferation, radiotherapy resistance, and mediating resistance to cisplatin, underscoring the potential of exosomal miRNAs in understanding and foreseeing treatment resistance in NSCLC [104,105,106].
Tumor-derived exosomal miRNAs and proteins play intricate roles in lung cancer metastasis, influencing various pathways and cellular processes. Exosomal miR-619-5p promotes metastasis by targeting the tumor suppressor RCAN1.4. miRNAs such as miR-433 and miR-1260b interact with the Wnt/β-catenin pathway, influencing NSCLC progression and metastasis [107]. Several trials using exosomes with miRNA cargo for lung cancer diagnosis are outlined in Table 2.

Table 2.
Clinical use of exosomes as biomarkers.
Table 2.
Clinical use of exosomes as biomarkers.
| Molecules Detected/Cargo Used | Sample | Function | Reference | |
|---|---|---|---|---|
| 1 | miR-375, miR-429, miR-203a-3p, miR-141-3p, miR-205-5p, miR-483-5p, miR-200c-3p, miR-200b-3p, and miR-200a-3p | Pleural effusion | Diagnostic biomarkers for distinguishing lung adenocarcinoma over tuberculosis and other benign lesions | [108] |
| 2 | miR-30a-3p, miR-100, miR-139-5p, miR-151a-5p, miR-379, miR-154-3p, miR-200b-5p, miR-378a, and miR-629 | Plasma | Diagnosis | [101] |
| 3 | miR-9-3p, miR-205-5p, miR-210-5p, and miR-1269a | Serum | Diagnosis | [109] |
| 4 | miR-34b, miR-125b, miR-200b, miR-203, miR-429, and miR-205 | Serum | Diagnosis (including early stage) | [110] |
| 5 | miR-19a, miR-19b, miR-20a, miR-30b, and miR-205 | Plasma | Diagnostic biomarkers of lung squamous-cell carcinoma (SCC) | [111] |
| 6 | miR-328-3p, miR-423-3p, and miR-574-5p | Plasma | Bone metastasis detection | [112] |
| 7 | miR-10b-5p, miR-21-5p, and miR-23b-3p | Plasma | Prognostic biomarkers for overall survival | [103] |
| 8 | miR-19-3p, miR-221-3p, and miR-21-5p | Plasma | Diagnostic biomarkers for lung adenocarcinoma | [113] |
| 9 | miR-21 and miR-4257 | Plasma | Prognostic biomarkers for disease-free survival (recurrence) | [114] |
| 10 | miR-20b-5p and miR-3187-5p | Serum | Diagnosis (including early stage) | [115] |
| 11 | miR-182 and miR-210 | Pleural effusion | Diagnostic biomarker for malignant pleural effusion (lung adenocarcinoma) over benign (non-neoplastic) pleural effusion | [116] |
| 12 | miR-320a, miR-622 | Plasma | Metastasis detection | [117] |
| 13 | miR-200b and miR-205-5p | Pleural effusion | Diagnostic biomarker for lung cancer | [118] |
| 14 | miR-29a-3p and miR-150-5p | Plasma | predictive biomarker for unexpected responses to radiation therapy | [119] |
| 15 | miR-200 | Pleural effusion | Diagnostic biomarker for malignant pleural effusion (lung adenocarcinoma) over benign (non-neoplastic) pleural effusion | [120] |
| 16 | miR-5684 | Serum | Diagnosis | [121] |
| 17 | miR-125b-5p | Serum | Diagnosis Prognosis: metastasis detection and survival Therapy monitoring | [121] |
| 18 | miR-23b-3p | Serum | Diagnosis Prognosis: tumor size, depth of invasion, liver metastasis, and TNM stage | [122] |
| 19 | miR-21 | Serum | Diagnosis (including versus benign and inflammatory lung diseases) | [123] |
| 20 | miRNA-205 | Urine and saliva | Diagnosis | [124] |
| 21 | miR-146a-5p | Serum | Prognostic biomarker for recurrence | [125] |
| 22 | miR-1268b and miR-6075 | Serum | Early detection of resectable lung cancer | [91] |
| 23 | miR-96 | Serum | Early detection of lung cancer | [93] |
| 24 | miR-451a | Plasma | Diagnostic and prognostic: early detection, lung cancer aggressiveness, and patient survival | [94] |
| 25 | miR-181 | Tissue | Prognostic NSCLC patients with stage I, II, or III cancer | [95] |
| 26 | miR-503 | Tissue | Prognostic biomarker in NSCLC. | [102] |
| 27 | miR-181-5p, miR-30a-3p, miR-30e-3p, and miR-320b | Plasma | Differentiating adenocarcinoma and squamous-cell carcinoma in NSCLC | [98] |
| 28 | miR-17-3p, miR-21, miR-106a, miR-146, miR-155, miR-199, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214 | Plasma | Diagnostic and prognostic biomarkers of adenocarcinoma | [99,100] |
| 29 | miR-146-5p, miR-23b-3p, miR-10b-5p, and miR-21-5p | Predicting survival rates and treatment responses in NSCLC | [124,126] | |
| 30 | miR-619-5p, miR-433, and miR-1260b | Progression and metastasis detection | [127] |
Exosomal proteins are being explored as potential biomarkers for NSCLC, similar to miRNAs. Exosomal proteins present a promising avenue in the quest for effective biomarkers in NSCLC diagnosis and prognosis, akin to the role played by miRNAs. Among these proteins, EGFR, KRAS, EMMPRIN, claudins, and RAB family proteins have emerged as key players associated with lung cancer [90,128]. Notably, exosomal EGFR, a membrane protein, has been notably prevalent in NSCLC exosomes. Proteomic analyses have unearthed a repertoire of differentially expressed proteins in NSCLC exosomes in contrast to their normal counterparts, highlighting their potential diagnostic value [129]. Additionally, specific markers, such as CD91 and CD317, have been proposed for NSCLC detection. Beyond diagnosis, the prognostic landscape has been explored, with membrane-bound proteins like NYESO-1 and PLAP linked to poorer outcomes. Excitingly, multi-marker models leveraging exosomal proteins have shown substantial promise in effectively discerning lung cancer patients from their controls. Notably, CD151, CD171, and tetraspanin 8 were identified as strong markers for differentiating lung cancer histological subtypes [130].
6.2. Role of Exosomes in Lung Cancer Treatment
Exosomes have shown great potential for revolutionizing lung cancer treatment. They serve as nanocarriers, offering advantages over traditional drugs by providing high payloads, precisely targeting tumors, and enhancing treatment effectiveness. Studies have indicated that exosomes induce G2 phase cell cycle arrest, apoptosis, and DNA damage, suggesting preferential toxicity to tumor cells. Moreover, exosomes derived from antigen-stimulated cells show their potential for vaccination [126,131,132,133].
Exosomes have gained prominence in immune regulation and hold promise as tools for cancer immunotherapy. Their capacity to present tumor antigens to immune cells positions them as potential solutions for addressing the immunogenicity challenge in cancer treatment. Tumor-derived exosomes loaded with antigens have been found to stimulate immune responses against tumors. Many investigations have been conducted on exosomes released by activated antigen-presenting cells (APCs), like dendritic cells (DCs), macrophages, T cells, and B cells [3,127,134,135,136]. Immune cells primed with antigens can package cancer cell components into exosomes, further promoting immune responses. In mouse models, exosomes secreted by DCs that are exposed to tumor antigens have shown promise in eradicating tumors. They induced tumor-sensitized T cells to secrete high levels of interferon-γ (IFN-γ), a cytokine that is important for immune responses [137].
Lung cancer cells use exosomes to communicate with immune cells, creating an environment that suppresses the immune response and supports tumor growth. Understanding this form of communication paves the way for developing targeted lung cancer immunotherapies by influencing the interactions mediated by exosomes. Lung cancer-derived exosomes carry specific surface markers that immune cells, particularly macrophages, take in. Consequently, these exosomes induce alterations in the macrophages’ characteristics, fostering an environment that promotes tumor growth and suppresses the immune response within the tumor microenvironment [138].
In a study, exosomes derived from lung tumor cells engineered with the CD40L gene (CD40L-EXO) were proven to be more effective in activating DCs and stimulating anti-tumor T cell immunity compared to standard exosomes. In lung cancer, DCs often transition from activating the immune system to fostering tolerance. CD40L-EXO demonstrated the ability to activate DCs, potentially reversing their tolerance status. When combined with the ability of heat-stressed tumor cell-derived exosomes to attract DCs to the tumor site, the use of CD40L-EXO appears to be promising for lung cancer treatment [139].
Exosomes released by NSCLC cells carry program death-ligand 1 (PD-L1) on their surface, playing a vital role in supporting tumor growth by hampering the function of CD8+ T cells, crucial for anti-tumor immunity, and triggering cell death in these T cells. These exosomes essentially aid the evasion of the immune system in NSCLC. Understanding the significance of PD-L1-positive exosomes is crucial in maximizing the effectiveness of PD-1/PD-L1 immunotherapy. Targeting these exosomes could offer a new approach to bolster the immune response against NSCLC, potentially improving the outcomes of immunotherapy treatments [140].
Tumor-derived exosomes possess immunosuppressive effects by hindering the maturation and migration of DCs, as well as the secretion of pro-inflammatory factors. The interaction between tumor-derived exosomes and DCs results in the upregulation of PD-L1, an immune checkpoint protein, which ultimately leads to the inhibition of the immune response. Blocking PD-L1 has shown potential in partially restoring the suppressed immune functions caused by exosome-treated DCs. This highlights the importance of considering both the cell surface and exosome presentation of PD-L1 in therapeutic strategies in combination with current immune checkpoint therapies [10,141,142].
In recent studies, the intricate connection between exosomes and immune responses in the context of cancer has been a subject of intense research [140,142,143]. Exosomes have demonstrated a significant influence on immune modulation and potential applications in cancer immunotherapy [144,145]. However, further research is needed to fully understand their mechanisms and optimize their use in clinical settings.
Building on their potential in cancer treatment, exosomes derived from tumor cells and innate immune cells have been explored for their use in vaccine development. These exosomes carry molecules such as MHC and costimulatory molecules that enhance immune cell responses against cancer. Exosome-based vaccines have exhibited positive outcomes against infectious diseases and are being considered for cancer immunotherapies [146].
In advanced NSCLC, a study revealed a significant decline in natural killer cell (NK cell) function, particularly in NKp30 expression, suggesting impairment. Soluble BAG6 levels were associated with weakened NK cell function in this context. Higher MHC-II and IFN-γ correlated with increased NK cell activity, particularly related to NKp30. IFN-γ use during production impacted BAG6 expression, restoring NKp30 function. Activating NK cells through NKp30 signaling using IFN-γ-Dex may be a promising immunotherapy, especially for NSCLC patients with NKp30-specific defects. These Dex, exosomes derived from dendritic cells, are engineered to elicit immune responses against cancer cells in NSCLC patients [147]. Dex demonstrate significant potential as vaccines for cancer treatment. Yet, their production requires specialized facilities, stringent manufacturing practices, and expensive growth factors and maturation agents [148].
7. Application of Exosomes in Drug Delivery
Nanomedicine, an innovative drug delivery technology, has brought about a paradigm shift in the conventional approach to treating severe illnesses, especially cancers [149,150,151,152]. Exosomes, being nano-sized and equipped with cell surface molecules, possess strong interstitial penetration and targeting capabilities. This makes them efficient for targeted delivery of chemotherapeutics. Novel exosome applications involve delivering drugs directly to specific tissues, particularly tumor tissues, showing promise as natural drug carriers [3,131,133,153].
Additionally, exosomes can be engineered to carry ligands that bind to cancer cell receptors, enhancing their specificity and efficacy. Their small size and lipid bilayer enable fusion with target cell membranes, bypassing endosomal–lysosomal pathways and improving drug delivery efficiency. Moreover, modified exosomes can feature specific targeting ligands, boosting their affinity for particular cell types. In cancer treatment, exosomes hold their potential for delivering chemotherapeutic agents into cancer cells [132,146,154,155,156,157,158].
Exosomes have been explored in various studies, showcasing their potential in transporting chemotherapeutics into tumor tissues, thereby reducing tumor growth without observing any adverse effects [126,131,159,160]. In vivo and in vitro analyses have shown drugs such as paclitaxel and doxorubicin encapsulated in exosomes that are able of stimulating therapeutic responses in lung cancer [161]. One study investigated using exosomes to deliver the drug doxorubicin into lung cancer cells. These researchers created nanosomes, which are exosomes loaded with stimuli-responsive nanoparticles, by loading doxorubicin attached to gold nanoparticles onto exosomes, forming NanoDox. They exhibited a slower and more consistent release of Dox, leading to prolonged effectiveness in killing tumor cells. These exosomes exhibited delayed and sustained drug release, inducing cell cycle arrest, apoptosis, and DNA damage [131].
Additionally, exosomes offer a promising avenue for targeted therapies in lung cancer through the delivery of specific miRNAs and siRNAs. Certain miRNAs carried by exosomes, such as miR-200c, miR-193a-3p, and miR-193a-5p, demonstrate the potential to inhibit NSCLC cell behavior, including proliferation, migration, and invasion [87,162]. Additionally, re-expressing miR-451 in docetaxel-resistant lung adenocarcinoma cells shows promise in reversing radioresistance through apoptosis promotion and DNA damage induction [163]. These findings underline miRNAs’ crucial roles in cancer and therapeutic potential.
Moreover, exosomes are identified as valuable therapeutic agents capable of targeting multiple pathways involved in lung cancer progression. For instance, exosomal miRNAs like miR-200a show promise in inhibiting tumor growth and metastasis [164,165]. They combat chemotherapeutic resistance through pathways, such as miR-1 targeting ATG3 [166], and miR-181b targeting Notch2 [167]. These observations position exosomes as potential carriers for enhancing lung cancer treatment. Notably, miR-148b stands out as a tumor suppressor in NSCLC, acting on the MAPK/JNK pathway [168]. As studies continue to unveil the diverse roles of exosomes, further investigation and clinical validation become crucial to fully harness their potential in both lung cancer therapy and monitoring. While caution is advised regarding the use of tumor-derived exosomes for drug delivery due to the potential enhancement of proliferation, additional research is necessary to validate their efficacy and safety.
8. Conclusions
In conclusion, exosomes, small extracellular vesicles derived from diverse cell types, represent a burgeoning frontier in the realm of lung cancer diagnosis and therapy. These vesicles boast exceptional attributes that make them highly promising tools in the fight against lung cancer. Serving as potential drug carriers, exosomes exhibit the capacity to transport therapeutic cargo, such as chemotherapeutic drugs, siRNAs, and immune modulators, directly to cancer cells, facilitated by their ability to traverse biological barriers, including the blood–brain barrier. Moreover, engineered exosomes with specific targeting molecules demonstrate a precise delivery mechanism to cancer cells, minimizing off-target effects.
Notably, exosomes also offer invaluable insights for lung cancer diagnosis. Laden with a plethora of molecules, like proteins, nucleic acids, and metabolites, these vesicles serve as a rich source of biomarkers. Analyzing the contents of exosomes, including genetic mutations, microRNAs, and proteins, aids in unraveling the molecular profiles of lung tumors. This analysis supports the early detection, prognostic assessment, and monitoring of treatment responses.
In the realm of targeted drug delivery, exosomes stand as promising carriers, delivering therapeutics precisely to augment treatment efficacy while minimizing adverse effects on healthy tissues. This evolving landscape in exosome research holds the potential to usher in a new era of tailored, efficient, and minimally invasive interventions for managing lung cancer.
Nonetheless, challenges persist in the realm of exosome research. Establishing standardized isolation and purification techniques, along with comprehensive characterization of exosomal cargo, remains pivotal for precision and consistency. Delving deeper into the mechanisms of exosome-mediated drug delivery and refining targeting strategies can amplify the effectiveness and safety of therapies based on exosomes. Additionally, the potential development of advanced technologies capable of detecting and analyzing single exosomes stand poised to revolutionize diagnostic accuracy and offer profound insights into tumor biology, paving the way for personalized treatment strategies.
Author Contributions
S.R. visualization and writing—original draft. H.N. conceptualization and writing—original draft. B.A. writing—original draft. M.D. conceptualization, reviewing and editing the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The figures were created with the BioRender.com, (accessed on 17 September 2023–1 October 2023) free trial plan.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ajam-Hosseini, M.; Akhoondi, F.; Doroudian, M. Nano based-oncolytic viruses for cancer therapy. Crit. Rev. Oncol./Hematol. 2023, 185, 103980. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Doroudian, M.; Azhdari, M.H.; Goodarzi, N.; O’Sullivan, D.; Donnelly, S.C. Smart Nanotherapeutics and Lung Cancer. Pharmaceutics 2021, 13, 1972. [Google Scholar] [CrossRef]
- Dela Cruz, C.S.; Tanoue, L.T.; Matthay, R.A. Lung cancer: Epidemiology, etiology, and prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef]
- Shahrivar, R.Y.; Fakhr, Z.A.; Abbasgholinejad, E.; Doroudian, M. Smart Lipid-Based Nanoparticles in Lung Cancer Treatment: Current Status and Future Directions. Adv. Ther. 2023, 6, 2300275. [Google Scholar] [CrossRef]
- Ferro, A.; Sepulcri, M.; Schiavon, M.; Scagliori, E.; Mancin, E.; Lunardi, F.; Gennaro, G.; Frega, S.; Dal Maso, A.; Bonanno, L.; et al. The Multidisciplinary Approach in Stage III Non-Small Cell Lung Cancer over Ten Years: From Radiation Therapy Optimisation to Innovative Systemic Treatments. Cancers 2022, 14, 5700. [Google Scholar] [CrossRef]
- Uramoto, H.; Tanaka, F. Recurrence after surgery in patients with NSCLC. Transl. Lung Cancer Res. 2014, 3, 242–249. [Google Scholar] [CrossRef]
- Laine, A.M.; Westover, K.D.; Choy, H. Radiation Therapy as a Backbone of Treatment of Locally Advanced Non-Small Cell Lung Cancer. Semin. Oncol. 2014, 41, 57–68. [Google Scholar] [CrossRef]
- Petrella, F.; Rizzo, S.; Attili, I.; Passaro, A.; Zilli, T.; Martucci, F.; Bonomo, L.; Del Grande, F.; Casiraghi, M.; De Marinis, F.; et al. Stage III Non-Small-Cell Lung Cancer: An Overview of Treatment Options. Curr. Oncol. 2023, 30, 3160–3175. [Google Scholar] [CrossRef]
- Doroudian, M.; Zanganeh, S.; Abbasgholinejad, E.; Donnelly, S. Nanomedicine in Lung Cancer Immunotherapy. Front. Bioeng. Biotechnol. 2023, 11, 1144653. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.; Gao, Y.; Sugimura, H.; Minervini, F.; Uchino, J.; Halmos, B.; Yendamuri, S.; Velotta, J.B.; Li, M. Neoadjuvant immunotherapy or chemoimmunotherapy in non-small cell lung cancer: A systematic review and meta-analysis. Transl. Lung Cancer Res. 2022, 11, 277–294. [Google Scholar] [CrossRef]
- Tsuboi, M.; Weder, W.; Escriu, C.; Blakely, C.; He, J.; Dacic, S.; Yatabe, Y.; Zeng, L.; Walding, A.; Chaft, J.E. Neoadjuvant osimertinib with/without chemotherapy versus chemotherapy alone for EGFR-mutated resectable non-small-cell lung cancer: NeoADAURA. Future Oncol. 2021, 17, 4045–4055. [Google Scholar] [CrossRef]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, D.; Ma, X.; Wang, J.; Hou, W.; Zhang, W. Exosomes as drug carriers for cancer therapy and challenges regarding exosome uptake. Biomed. Pharmacother. 2020, 128, 110237. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Nahand, J.S.; Vandchali, N.R.; Darabi, H.; Doroudian, M.; Banafshe, H.R.; Moghoofei, M.; Babaei, F.; Salmaninejad, A.; Mirzaei, H. Exosomal microRNAs: Novel players in cervical cancer. Epigenomics 2020, 12, 1651–1660. [Google Scholar] [CrossRef]
- Hong, C.; Yang, S.; Ndukaife, J.C. Exosomes trapping, manipulation and size-based separation using opto-thermo-electrohydrodynamic tweezers. Nanoscale Adv. 2023, 5, 2973–2978. [Google Scholar] [CrossRef]
- Rajput, A.; Varshney, A.; Bajaj, R.; Pokharkar, V. Exosomes as New Generation Vehicles for Drug Delivery: Biomedical Applications and Future Perspectives. Molecules 2022, 27, 7289. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef]
- Hanjani, N.A.; Esmaelizad, N.; Zanganeh, S.; Gharavi, A.T.; Heidarizadeh, P.; Radfar, M.; Omidi, F.; MacLoughlin, R.; Doroudian, M. Emerging role of exosomes as biomarkers in cancer treatment and diagnosis. Crit. Rev. Oncol./Hematol. 2022, 169, 103565. [Google Scholar] [CrossRef]
- Farjadian, F.; Ghasemi, S.; Akbarian, M.; Hoseini-Ghahfarokhi, M.; Moghoofei, M.; Doroudian, M. Physically stimulus-responsive nanoparticles for therapy and diagnosis. Front. Chem. 2022, 10, 952675. [Google Scholar] [CrossRef]
- Manzoor, T.; Saleem, A.; Farooq, N.; Dar, L.A.; Nazir, J.; Saleem, S.; Ismail, S.; Gugjoo, M.B.; Shiekh, P.A.; Ahmad, S.M. Extracellular vesicles derived from mesenchymal stem cells—A novel therapeutic tool in infectious diseases. Inflamm. Regen. 2023, 43, 17. [Google Scholar] [CrossRef]
- Doroudian, M.; Armstrong, M.E.; Donnelly, S.C. Nano-Based Therapies for Acute and Chronic Lung Diseases. In Biotechnology Applied to Inflammatory Diseases: Cellular Mechanisms and Nanomedicine; Ribeiro de Araujo, D., Carneiro-Ramos, M., Eds.; Springer Nature: Singapore, 2023; pp. 271–286. [Google Scholar] [CrossRef]
- Aheget, H.; Tristán-Manzano, M.; Mazini, L.; Cortijo-Gutierrez, M.; Galindo-Moreno, P.; Herrera, C.; Martin, F.; Marchal, J.A.; Benabdellah, K. Exosome: A New Player in Translational Nanomedicine. J. Clin. Med. 2020, 9, 2380. [Google Scholar] [CrossRef]
- Fan, Y.; Chen, Z.; Zhang, M. Role of exosomes in the pathogenesis, diagnosis, and treatment of central nervous system diseases. J. Transl. Med. 2022, 20, 291. [Google Scholar] [CrossRef]
- D’Agnelli, S.; Gerra, M.C.; Bignami, E.; Arendt-Nielsen, L. Exosomes as a new pain biomarker opportunity. Mol. Pain. 2020, 16, 1744806920957800. [Google Scholar] [CrossRef]
- Chen, J.; Li, C.; Li, R.; Chen, H.; Chen, D.; Li, W. Exosomes in HIV infection. Curr. Opin. HIV AIDS 2021, 16, 262–270. [Google Scholar] [CrossRef]
- Yu, T.; Xu, Y.; Ahmad, M.A.; Javed, R.; Hagiwara, H.; Tian, X. Exosomes as a Promising Therapeutic Strategy for Peripheral Nerve Injury. Curr. Neuropharmacol. 2021, 19, 2141–2151. [Google Scholar] [CrossRef]
- Yousefi Dehbidi, M.; Goodarzi, N.; Azhdari, M.H.; Doroudian, M. Mesenchymal stem cells and their derived exosomes to combat COVID-19. Rev. Med. Virol. 2022, 32, e2281. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kang, M.H.; Jeyaraj, M.; Qasim, M.; Kim, J.H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, F.; Zhong, J.Y.; Lin, X.; Shan, S.K.; Guo, B.; Zheng, M.H.; Yuan, L.Q. Exosomes as Mediators of Cell-to-Cell Communication in Thyroid Disease. Int. J. Endocrinol. 2020, 2020, 4378345. [Google Scholar] [CrossRef]
- Burkova, E.E.; Sedykh, S.E.; Nevinsky, G.A. Human Placenta Exosomes: Biogenesis, Isolation, Composition, and Prospects for Use in Diagnostics. Int. J. Mol. Sci. 2021, 22, 2158. [Google Scholar] [CrossRef]
- Wei, H.; Chen, Q.; Lin, L.; Sha, C.; Li, T.; Liu, Y.; Yin, X.; Xu, Y.; Chen, L.; Gao, W.; et al. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021, 17, 163–177. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal 2021, 19, 47. [Google Scholar] [CrossRef]
- Kita, S.; Maeda, N.; Shimomura, I. Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Investig. 2019, 129, 4041–4049. [Google Scholar] [CrossRef]
- Wang, T.; Li, L.; Hong, W. SNARE proteins in membrane trafficking. Traffic 2017, 18, 767–775. [Google Scholar] [CrossRef]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Wei, D.; Zhan, W.; Gao, Y.; Huang, L.; Gong, R.; Wang, W.; Zhang, R.; Wu, Y.; Gao, S.; Kang, T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021, 31, 157–177. [Google Scholar] [CrossRef]
- Donoso-Quezada, J.; Ayala-Mar, S.; González-Valdez, J. The role of lipids in exosome biology and intercellular communication: Function, analytics and applications. Traffic 2021, 22, 204–220. [Google Scholar] [CrossRef]
- Osaki, M.; Okada, F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef]
- Li, K.L.; Huang, H.Y.; Ren, H.; Yang, X.L. Role of exosomes in the pathogenesis of inflammation in Parkinson’s disease. Neural Regen. Res. 2022, 17, 1898–1906. [Google Scholar] [CrossRef]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef]
- Colombo, M.; Moita, C.; van Niel, G.; Kowal, J.; Vigneron, J.; Benaroch, P.; Manel, N.; Moita, L.F.; Théry, C.; Raposo, G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci. 2013, 126, 5553–5565. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef]
- Friand, V.; David, G.; Zimmermann, P. Syntenin and syndecan in the biogenesis of exosomes. Biol. Cell 2015, 107, 331–341. [Google Scholar] [CrossRef]
- Obata, Y.; Kita, S.; Koyama, Y.; Fukuda, S.; Takeda, H.; Takahashi, M.; Fujishima, Y.; Nagao, H.; Masuda, S.; Tanaka, Y.; et al. Adiponectin/T-cadherin system enhances exosome biogenesis and decreases cellular ceramides by exosomal release. JCI Insight 2018, 3, e99680. [Google Scholar] [CrossRef]
- Crewe, C.; Joffin, N.; Rutkowski, J.M.; Kim, M.; Zhang, F.; Towler, D.A.; Gordillo, R.; Scherer, P.E. An Endothelial-to-Adipocyte Extracellular Vesicle Axis Governed by Metabolic State. Cell 2018, 175, 695–708.e13. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Willms, E.; Cabañas, C.; Mäger, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef]
- Mincheva-Nilsson, L.; Baranov, V. The role of placental exosomes in reproduction. Am. J. Reprod. Immunol. 2010, 63, 520–533. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, C.; Mai, C.; Hu, X.; Cheng, N.; Chen, W.; Peng, D.; Wang, L.; Ji, Z.; Xie, Y. The Biogenesis, Biological Functions, and Applications of Macrophage-Derived Exosomes. Front. Mol. Biosci. 2021, 8, 715461. [Google Scholar] [CrossRef]
- Hunt, S.D.; Townley, A.K.; Danson, C.M.; Cullen, P.J.; Stephens, D.J. Microtubule motors mediate endosomal sorting by maintaining functional domain organization. J. Cell Sci. 2013, 126, 2493–2501. [Google Scholar] [CrossRef]
- Azhdari, M.H.; Goodarzi, N.; Doroudian, M.; MacLoughlin, R. Molecular Insight into the Therapeutic Effects of Stem Cell-Derived Exosomes in Respiratory Diseases and the Potential for Pulmonary Delivery. Int. J. Mol. Sci. 2022, 23, 6273. [Google Scholar] [CrossRef]
- Chen, J.; Li, P.; Zhang, T.; Xu, Z.; Huang, X.; Wang, R.; Du, L. Review on Strategies and Technologies for Exosome Isolation and Purification. Front. Bioeng. Biotechnol. 2021, 9, 811971. [Google Scholar] [CrossRef]
- Cui, S.; Cheng, Z.; Qin, W.; Jiang, L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer 2018, 116, 46–54. [Google Scholar] [CrossRef]
- Zhu, L.; Sun, H.T.; Wang, S.; Huang, S.L.; Zheng, Y.; Wang, C.Q.; Hu, B.Y.; Qin, W.; Zou, T.T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Meng, W.; He, C.; Hao, Y.; Wang, L.; Li, L.; Zhu, G. Prospects and challenges of extracellular vesicle-based drug delivery system: Considering cell source. Drug Deliv. 2020, 27, 585–598. [Google Scholar] [CrossRef]
- LeBleu, V.S.; Kalluri, R. Exosomes as a Multicomponent Biomarker Platform in Cancer. Trends Cancer 2020, 6, 767–774. [Google Scholar] [CrossRef]
- Lee, J.; Lee, J.H.; Chakraborty, K.; Hwang, J.; Lee, Y.K. Exosome-based drug delivery systems and their therapeutic applications. RSC Adv. 2022, 12, 18475–18492. [Google Scholar] [CrossRef]
- Mohammad, D.; Ronan, M.; Fergus, P.; Adriele, P.-M.; Seamas, C.D. Nanotechnology based therapeutics for lung disease. Thorax 2019, 74, 965. [Google Scholar] [CrossRef]
- Xin, Y.; Huang, Q.; Tang, J.Q.; Hou, X.Y.; Zhang, P.; Zhang, L.Z.; Jiang, G. Nanoscale drug delivery for targeted chemotherapy. Cancer Lett. 2016, 379, 24–31. [Google Scholar] [CrossRef]
- Peer, D.; Karp, J.M.; Hong, S.; Farokhzad, O.C.; Margalit, R.; Langer, R. Nanocarriers as an emerging platform for cancer therapy. Nat. Nanotechnol. 2007, 2, 751–760. [Google Scholar] [CrossRef]
- Moon, B.; Chang, S. Exosome as a Delivery Vehicle for Cancer Therapy. Cells 2022, 11, 316. [Google Scholar] [CrossRef]
- Heydari, R.; Koohi, F.; Rasouli, M.; Rezaei, K.; Abbasgholinejad, E.; Bekeschus, S.; Doroudian, M. Exosomes as Rheumatoid Arthritis Diagnostic Biomarkers and Therapeutic Agents. Vaccines 2023, 11, 687. [Google Scholar] [CrossRef]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef]
- Herrmann, I.K.; Wood, M.J.A.; Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 2021, 16, 748–759. [Google Scholar] [CrossRef]
- Liu, C.; Su, C. Design strategies and application progress of therapeutic exosomes. Theranostics 2019, 9, 1015–1028. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Han, Y.; Jones, T.W.; Dutta, S.; Zhu, Y.; Wang, X.; Narayanan, S.P.; Fagan, S.C.; Zhang, D. Overview and Update on Methods for Cargo Loading into Extracellular Vesicles. Processes 2021, 9, 356. [Google Scholar] [CrossRef]
- Zeng, H.; Guo, S.; Ren, X.; Wu, Z.; Liu, S.; Yao, X. Current Strategies for Exosome Cargo Loading and Targeting Delivery. Cells 2023, 12, 1416. [Google Scholar] [CrossRef]
- Jankovičová, J.; Sečová, P.; Michalková, K.; Antalíková, J. Tetraspanins, More than Markers of Extracellular Vesicles in Reproduction. Int. J. Mol. Sci. 2020, 21, 7568. [Google Scholar] [CrossRef]
- Campanella, C.; Caruso Bavisotto, C.; Logozzi, M.; Marino Gammazza, A.; Mizzoni, D.; Cappello, F.; Fais, S. On the Choice of the Extracellular Vesicles for Therapeutic Purposes. Int. J. Mol. Sci. 2019, 20, 236. [Google Scholar] [CrossRef]
- Nooreldeen, R.; Bach, H. Current and Future Development in Lung Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 8661. [Google Scholar] [CrossRef]
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-cell lung cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, Z.; Wang, Q. Emerging therapies for small cell lung cancer. J. Hematol. Oncol. 2019, 12, 47. [Google Scholar] [CrossRef]
- Neumann, J.M.; Freitag, H.; Hartmann, J.S.; Niehaus, K.; Galanis, M.; Griesshammer, M.; Kellner, U.; Bednarz, H. Subtyping non-small cell lung cancer by histology-guided spatial metabolomics. J. Cancer Res. Clin. Oncol. 2022, 148, 351–360. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef]
- Scheff, R.J.; Schneider, B.J. Non-small-cell lung cancer: Treatment of late stage disease: Chemotherapeutics and new frontiers. Semin. Interv. Radiol. 2013, 30, 191–198. [Google Scholar] [CrossRef]
- Zappa, C.; Mousa, S.A. Non-small cell lung cancer: Current treatment and future advances. Transl. Lung Cancer Res. 2016, 5, 288–300. [Google Scholar] [CrossRef]
- Yang, L.; Fan, Y.; Lu, H. Pulmonary Large Cell Neuroendocrine Carcinoma. Pathol. Oncol. Res. 2022, 28, 1610730. [Google Scholar] [CrossRef]
- Xu, K.; Zhang, C.; Du, T.; Gabriel, A.N.A.; Wang, X.; Li, X.; Sun, L.; Wang, N.; Jiang, X.; Zhang, Y. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed. Pharmacother. 2021, 134, 111111. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Lu, J.; Ng, I.O.-L. Exosomes and cancer—Diagnostic and prognostic biomarkers and therapeutic vehicle. Oncogenesis 2022, 11, 54. [Google Scholar] [CrossRef]
- Huda, M.N.; Nafiujjaman, M.; Deaguero, I.G.; Okonkwo, J.; Hill, M.L.; Kim, T.; Nurunnabi, M. Potential Use of Exosomes as Diagnostic Biomarkers and in Targeted Drug Delivery: Progress in Clinical and Preclinical Applications. ACS Biomater. Sci. Eng. 2021, 7, 2106–2149. [Google Scholar] [CrossRef]
- Reclusa, P.; Taverna, S.; Pucci, M.; Durendez, E.; Calabuig, S.; Manca, P.; Serrano, M.J.; Sober, L.; Pauwels, P.; Russo, A.; et al. Exosomes as diagnostic and predictive biomarkers in lung cancer. J. Thorac. Dis. 2017, 9, S1373–S1382. [Google Scholar] [CrossRef]
- Asakura, K.; Kadota, T.; Matsuzaki, J.; Yoshida, Y.; Yamamoto, Y.; Nakagawa, K.; Takizawa, S.; Aoki, Y.; Nakamura, E.; Miura, J.; et al. A miRNA-based diagnostic model predicts resectable lung cancer in humans with high accuracy. Commun. Biol. 2020, 3, 134. [Google Scholar] [CrossRef]
- Lebanony, D.; Benjamin, H.; Gilad, S.; Ezagouri, M.; Dov, A.; Ashkenazi, K.; Gefen, N.; Izraeli, S.; Rechavi, G.; Pass, H.; et al. Diagnostic Assay Based on hsa-miR-205 Expression Distinguishes Squamous from Nonsquamous Non–Small-Cell Lung Carcinoma. J. Clin. Oncol. 2009, 27, 2030–2037. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhou, J.; Mei, S.; Wu, D.; Mu, Z.; Chen, B.; Xie, Y.; Ye, Y.; Liu, J. Circulating exosomal microRNA-96 promotes cell proliferation, migration and drug resistance by targeting LMO7. J. Cell. Mol. Med. 2017, 21, 1228–1236. [Google Scholar] [CrossRef] [PubMed]
- Kanaoka, R.; Iinuma, H.; Dejima, H.; Sakai, T.; Uehara, H.; Matsutani, N.; Kawamura, M. Usefulness of Plasma Exosomal MicroRNA-451a as a Noninvasive Biomarker for Early Prediction of Recurrence and Prognosis of Non-Small Cell Lung Cancer. Oncology 2018, 94, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Yu, Y.; Cao, H.; Shen, H.; Li, X.; Pan, S.; Shu, Y. Deregulated expression of miR-21, miR-143 and miR-181a in non small cell lung cancer is related to clinicopathologic characteristics or patient prognosis. Biomed. Pharmacother. 2010, 64, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Xu, X.; Qian, Z.; Zhang, C.; Niu, Y.; Wang, Z.; Sun, J.; Zhang, X.; Yu, Y. The biological functions and clinical applications of exosomes in lung cancer. Cell. Mol. Life Sci. 2019, 76, 4613–4633. [Google Scholar] [CrossRef]
- Xu, Z.-H.; Miao, Z.-W.; Jiang, Q.-Z.; Gan, D.-X.; Wei, X.-G.; Xue, X.-Z.; Li, J.-Q.; Zheng, F.; Qin, X.-X.; Fang, W.-G.; et al. Brain microvascular endothelial cell exosome–mediated S100A16 up-regulation confers small-cell lung cancer cell survival in brain. FASEB J. 2019, 33, 1742–1757. [Google Scholar] [CrossRef]
- Jin, X.; Chen, Y.; Chen, H.; Fei, S.; Chen, D.; Cai, X.; Liu, L.; Lin, B.; Su, H.; Zhao, L.; et al. Evaluation of Tumor-Derived Exosomal miRNA as Potential Diagnostic Biomarkers for Early-Stage Non-Small Cell Lung Cancer Using Next-Generation Sequencing. Clin. Cancer Res. 2017, 23, 5311–5319. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gerçel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Ghasempour, E.; Hesami, S.; Movahed, E.; Keshel, S.H.; Doroudian, M. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy in the brain tumors. Stem Cell Res. Ther. 2022, 13, 527. [Google Scholar] [CrossRef]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef]
- Liu, L.; Qu, W.; Zhong, Z. Down-regulation of miR-503 expression predicate advanced mythological features and poor prognosis in patients with NSCLC. Int. J. Clin. Exp. Pathol. 2015, 8, 5609–5613. [Google Scholar] [PubMed]
- Liu, Q.; Yu, Z.; Yuan, S.; Xie, W.; Li, C.; Hu, Z.; Xiang, Y.; Wu, N.; Wu, L.; Bai, L.; et al. Circulating exosomal microRNAs as prognostic biomarkers for non-small-cell lung cancer. Oncotarget 2017, 8, 13048–13058. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Xu, J.; Wang, J.; Pan, Y.; Fu, J.; Bai, Y.; Zhang, J.; Shao, C. Extracellular miR-1246 promotes lung cancer cell proliferation and enhances radioresistance by directly targeting DR5. Oncotarget 2016, 7, 32707–32722. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Cui, Y.; Li, Z.; Jiao, Z.; Zhang, Y.; He, Y.; Chen, G.; Zhou, Q.; Wang, W.; Zhou, X.; et al. Erratum to: Radiation-induced miR-208a increases the proliferation and radioresistance by targeting p21 in human lung cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 20. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int. J. Nanomed. 2017, 12, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.N.; Ma, Y.; Nenkov, M.; Jin, L.; Schröder, D.C.; Westermann, M.; Gaßler, N.; Chen, Y. Tumor-derived exosomes: Key players in non-small cell lung cancer metastasis and their implication for targeted therapy. Mol. Carcinog. 2022, 61, 269–280. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.M.; Zou, Y.Q.; Lin, J.; Huang, B.; Liu, J.; Li, J.; Zhang, J.; Yang, W.M.; Min, Q.H.; et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Medicine 2017, 96, e8361. [Google Scholar] [CrossRef]
- Wang, X.; Jiang, X.; Li, J.; Wang, J.; Binang, H.; Shi, S.; Duan, W.; Zhao, Y.; Zhang, Y. Serum exosomal miR-1269a serves as a diagnostic marker and plays an oncogenic role in non-small cell lung cancer. Thorac. Cancer 2020, 11, 3436–3447. [Google Scholar] [CrossRef]
- Halvorsen, A.R.; Bjaanæs, M.; LeBlanc, M.; Holm, A.M.; Bolstad, N.; Rubio, L.; Peñalver, J.C.; Cervera, J.; Mojarrieta, J.C.; López-Guerrero, J.A.; et al. A unique set of 6 circulating microRNAs for early detection of non-small cell lung cancer. Oncotarget 2016, 7, 37250–37259. [Google Scholar] [CrossRef]
- Aushev, V.N.; Zborovskaya, I.B.; Laktionov, K.K.; Girard, N.; Cros, M.P.; Herceg, Z.; Krutovskikh, V. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS ONE 2013, 8, e78649. [Google Scholar] [CrossRef]
- Yang, X.R.; Pi, C.; Yu, R.; Fan, X.J.; Peng, X.X.; Zhang, X.C.; Chen, Z.H.; Wu, X.; Shao, Y.; Wu, Y.L.; et al. Correlation of exosomal microRNA clusters with bone metastasis in non-small cell lung cancer. Clin. Exp. Metastasis 2021, 38, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wen, W.; Shan, X.; Zhu, W.; Xu, J.; Guo, R.; Cheng, W.; Wang, F.; Qi, L.W.; Chen, Y.; et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget 2017, 8, 6513–6525. [Google Scholar] [CrossRef] [PubMed]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef]
- Zhang, Z.J.; Song, X.G.; Xie, L.; Wang, K.Y.; Tang, Y.Y.; Yu, M.; Feng, X.D.; Song, X.R. Circulating serum exosomal miR-20b-5p and miR-3187-5p as efficient diagnostic biomarkers for early-stage non-small cell lung cancer. Exp. Biol. Med. 2020, 245, 1428–1436. [Google Scholar] [CrossRef] [PubMed]
- Tamiya, H.; Mitani, A.; Saito, A.; Ishimori, T.; Saito, M.; Isago, H.; Jo, T.; Yamauchi, Y.; Tanaka, G.; Nagase, T. Exosomal MicroRNA Expression Profiling in Patients with Lung Adenocarcinoma-associated Malignant Pleural Effusion. Anticancer Res. 2018, 38, 6707–6714. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Guo, W.; Song, X.; Liu, L.; Niu, L.; Song, X.; Xie, L. Tumor-associated exosomal miRNA biomarkers to differentiate metastatic vs. nonmetastatic non-small cell lung cancer. Clin. Chem. Lab. Med. 2020, 58, 1535–1545. [Google Scholar] [CrossRef]
- Lin, J.; Wang, Y.; Zou, Y.Q.; Chen, X.; Huang, B.; Liu, J.; Xu, Y.M.; Li, J.; Zhang, J.; Yang, W.M.; et al. Differential miRNA expression in pleural effusions derived from extracellular vesicles of patients with lung cancer, pulmonary tuberculosis, or pneumonia. Tumour Biol. 2016, 37, 15835–15845. [Google Scholar] [CrossRef]
- Dinh, T.K.; Fendler, W.; Chałubińska-Fendler, J.; Acharya, S.S.; O’Leary, C.; Deraska, P.V.; D’Andrea, A.D.; Chowdhury, D.; Kozono, D. Circulating miR-29a and miR-150 correlate with delivered dose during thoracic radiation therapy for non-small cell lung cancer. Radiat. Oncol. 2016, 11, 61. [Google Scholar] [CrossRef]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Brandén, E.; Koyi, H.; Novak, M.; Kanter, L.; Hååg, P.; Hurley, J.; Tadigotla, V.; et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer 2018, 124, 45–52. [Google Scholar] [CrossRef]
- Zhang, Z.; Tang, Y.; Song, X.; Xie, L.; Zhao, S.; Song, X. Tumor-Derived Exosomal miRNAs as Diagnostic Biomarkers in Non-Small Cell Lung Cancer. Front. Oncol. 2020, 10, 560025. [Google Scholar] [CrossRef]
- Wang, J.; Xue, H.; Zhu, Z.; Gao, J.; Zhao, M.; Ma, Z. Expression of serum exosomal miR-23b-3p in non-small cell lung cancer and its diagnostic efficacy. Oncol. Lett. 2020, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wang, T.; Qu, X.; Chen, S.; Han, Z.; Chen, S.; Chen, M.; Lin, J.; Yu, S.; Gao, L.; et al. Exosomal miR-21/Let-7a ratio distinguishes non-small cell lung cancer from benign pulmonary diseases. Asia Pac. J. Clin. Oncol. 2020, 16, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Lu, F.; Wang, J.; Wang, K.; Liu, B.; Li, N.; Tang, B. A portable point-of-care testing system to diagnose lung cancer through the detection of exosomal miRNA in urine and saliva. Chem. Commun. 2020, 56, 8968–8971. [Google Scholar] [CrossRef] [PubMed]
- Yuwen, D.L.; Sheng, B.B.; Liu, J.; Wenyu, W.; Shu, Y.Q. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 2650–2658. [Google Scholar] [PubMed]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Yuan, D.; Deygen, I.; Klyachko, N.L.; Kabanov, A.V.; Batrakova, E.V. Engineering macrophage-derived exosomes for targeted paclitaxel delivery to pulmonary metastases: In vitro and in vivo evaluations. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zeng, S.; Gong, Z.; Yan, Y. Exosome-based immunotherapy: A promising approach for cancer treatment. Mol. Cancer 2020, 19, 160. [Google Scholar] [CrossRef] [PubMed]
- Hsu, M.-T.; Wang, Y.-K.; Tseng, Y.J. Exosomal Proteins and Lipids as Potential Biomarkers for Lung Cancer Diagnosis, Prognosis, and Treatment. Cancers 2022, 14, 732. [Google Scholar] [CrossRef]
- Yamashita, T.; Kamada, H.; Kanasaki, S.; Maeda, Y.; Nagano, K.; Abe, Y.; Inoue, M.; Yoshioka, Y.; Tsutsumi, Y.; Katayama, S.; et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013, 68, 969–973. [Google Scholar]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Bæk, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.H.; Rasmussen, T.R.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef]
- Srivastava, A.; Amreddy, N.; Babu, A.; Panneerselvam, J.; Mehta, M.; Muralidharan, R.; Chen, A.; Zhao, Y.D.; Razaq, M.; Riedinger, N.; et al. Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells. Sci. Rep. 2016, 6, 38541. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zuo, B.; Jing, R.; Gao, X.; Rao, Q.; Liu, Z.; Qi, H.; Guo, H.; Yin, H. Dendritic cell-derived exosomes elicit tumor regression in autochthonous hepatocellular carcinoma mouse models. J. Hepatol. 2017, 67, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Kalimuthu, S.; Gangadaran, P.; Oh, J.M.; Lee, H.W.; Baek, S.H.; Jeong, S.Y.; Lee, S.W.; Lee, J.; Ahn, B.C. Exosomes Derived From Natural Killer Cells Exert Therapeutic Effect in Melanoma. Theranostics 2017, 7, 2732–2745. [Google Scholar] [CrossRef] [PubMed]
- Neviani, P.; Wise, P.M.; Murtadha, M.; Liu, C.W.; Wu, C.H.; Jong, A.Y.; Seeger, R.C.; Fabbri, M. Natural Killer-Derived Exosomal miR-186 Inhibits Neuroblastoma Growth and Immune Escape Mechanisms. Cancer Res. 2019, 79, 1151–1164. [Google Scholar] [CrossRef] [PubMed]
- Romagnoli, G.G.; Zelante, B.B.; Toniolo, P.A.; Migliori, I.K.; Barbuto, J.A. Dendritic Cell-Derived Exosomes may be a Tool for Cancer Immunotherapy by Converting Tumor Cells into Immunogenic Targets. Front. Immunol. 2014, 5, 692. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, A.; Tousif, S.; Wang, Y.; Hough, K.; Khan, S.; Strenkowski, J.; Chacko, B.K.; Darley-Usmar, V.M.; Deshane, J.S. Lung Tumor Cell-Derived Exosomes Promote M2 Macrophage Polarization. Cells 2020, 9, 1303. [Google Scholar] [CrossRef]
- Wang, J.; Wang, L.; Lin, Z.; Tao, L.; Chen, M. More efficient induction of antitumor T cell immunity by exosomes from CD40L gene-modified lung tumor cells. Mol. Med. Rep. 2014, 9, 125–131. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, H.; Choi, Y.J.; Kim, S.Y.; Lee, J.E.; Sung, K.J.; Sung, Y.H.; Pack, C.G.; Jung, M.K.; Han, B.; et al. Exosomal PD-L1 promotes tumor growth through immune escape in non-small cell lung cancer. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Poggio, M.; Hu, T.; Pai, C.C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef]
- Ning, Y.; Shen, K.; Wu, Q.; Sun, X.; Bai, Y.; Xie, Y.; Pan, J.; Qi, C. Tumor exosomes block dendritic cells maturation to decrease the T cell immune response. Immunol. Lett. 2018, 199, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Barros, F.M.; Carneiro, F.; Machado, J.C.; Melo, S.A. Exosomes and Immune Response in Cancer: Friends or Foes? Front. Immunol. 2018, 9, 730. [Google Scholar] [CrossRef] [PubMed]
- Chalmin, F.; Ladoire, S.; Mignot, G.; Vincent, J.; Bruchard, M.; Remy-Martin, J.P.; Boireau, W.; Rouleau, A.; Simon, B.; Lanneau, D.; et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Investig. 2010, 120, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Gharavi, A.T.; Hanjani, N.A.; Movahed, E.; Doroudian, M. The role of macrophage subtypes and exosomes in immunomodulation. Cell. Mol. Biol. Lett. 2022, 27, 83. [Google Scholar] [CrossRef]
- Rezaie, J.; Feghhi, M.; Etemadi, T. A review on exosomes application in clinical trials: Perspective, questions, and challenges. Cell Commun. Signal 2022, 20, 145. [Google Scholar] [CrossRef]
- Besse, B.; Charrier, M.; Lapierre, V.; Dansin, E.; Lantz, O.; Planchard, D.; Le Chevalier, T.; Livartoski, A.; Barlesi, F.; Laplanche, A.; et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology 2016, 5, e1071008. [Google Scholar] [CrossRef]
- Viaud, S.; Théry, C.; Ploix, S.; Tursz, T.; Lapierre, V.; Lantz, O.; Zitvogel, L.; Chaput, N. Dendritic Cell-Derived Exosomes for Cancer Immunotherapy: What’s Next? Cancer Res. 2010, 70, 1281–1285. [Google Scholar] [CrossRef]
- Farjadian, F.; Faghih, Z.; Fakhimi, M.; Iranpour, P.; Mohammadi-Samani, S.; Doroudian, M. Glucosamine-Modified Mesoporous Silica-Coated Magnetic Nanoparticles: A “Raisin-Cake”-like Structure as an Efficient Theranostic Platform for Targeted Methotrexate Delivery. Pharmaceutics 2023, 15, 2491. [Google Scholar]
- Zanganeh, S.; Doroudian, M.; Nowzari, Z.R.; Nasirmoghadas, P.; Nahand, J.S.; Sepehrpour, S.; Moghoofei, M. Viral Nanoparticles-Mediated Delivery of Therapeutic Cargo. Adv. Ther. 2023, 6, 2300082. [Google Scholar] [CrossRef]
- Hajiaghapour Asr, M.; Dayani, F.; Saedi Segherloo, F.; Kamedi, A.; Neill, A.O.; MacLoughlin, R.; Doroudian, M. Lipid Nanoparticles as Promising Carriers for mRNA Vaccines for Viral Lung Infections. Pharmaceutics 2023, 15, 1127. [Google Scholar]
- Doroudian, M.; O’Neill, A.; O’Reilly, C.; Tynan, A.; Mawhinney, L.; McElroy, A.; Webster, S.S.; MacLoughlin, R.; Volkov, Y.; Armstrong, M.E.; et al. Aerosolized drug-loaded nanoparticles targeting migration inhibitory factors inhibit Pseudomonas aeruginosa-induced inflammation and biofilm formation. Nanomedicine 2020, 15, 2933–2953. [Google Scholar] [CrossRef] [PubMed]
- Hejabi, F.; Abbaszadeh, M.S.; Taji, S.; O’Neill, A.; Farjadian, F.; Doroudian, M. Nanocarriers: A novel strategy for the delivery of CRISPR/Cas systems. Front. Chem. 2022, 10, 957572. [Google Scholar] [PubMed]
- Jang, S.C.; Kim, O.Y.; Yoon, C.M.; Choi, D.-S.; Roh, T.-Y.; Park, J.; Nilsson, J.; Lötvall, J.; Kim, Y.-K.; Gho, Y.S. Bioinspired Exosome-Mimetic Nanovesicles for Targeted Delivery of Chemotherapeutics to Malignant Tumors. ACS Nano 2013, 7, 7698–7710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xu, Y.; Ma, L.; Yu, K.; Niu, Y.; Xu, X.; Shi, Y.; Guo, S.; Xue, X.; Wang, Y.; et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun. 2022, 42, 287–313. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Duan, L.; Lu, J.; Xia, J. Engineering exosomes for targeted drug delivery. Theranostics 2021, 11, 3183–3195. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.; Zaro, J.; Shen, Y. Advances in Exosome-Based Drug Delivery and Tumor Targeting: From Tissue Distribution to Intracellular Fate. Int. J. Nanomed. 2020, 15, 9355–9371. [Google Scholar] [CrossRef]
- Sadeghi, S.; Tehrani, F.R.; Tahmasebi, S.; Shafiee, A.; Hashemi, S.M. Exosome engineering in cell therapy and drug delivery. Inflammopharmacology 2023, 31, 145–169. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Kim, M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release 2015, 219, 396–405. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Li, M.-Y.; Liu, L.-Z.; Dong, M. Progress on pivotal role and application of exosome in lung cancer carcinogenesis, diagnosis, therapy and prognosis. Mol. Cancer 2021, 20, 22. [Google Scholar] [CrossRef]
- Qi, R.; Zhao, Y.; Guo, Q.; Mi, X.; Cheng, M.; Hou, W.; Zheng, H.; Hua, B. Exosomes in the lung cancer microenvironment: Biological functions and potential use as clinical biomarkers. Cancer Cell Int. 2021, 21, 333. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, D.Q.; Huang, J.Y.; Zhang, K.; Feng, B.; Pan, B.Z.; Chen, J.; De, W.; Chen, L.B. Acquisition of radioresistance in docetaxel-resistant human lung adenocarcinoma cells is linked with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling. Oncotarget 2014, 5, 6113–6129. [Google Scholar] [CrossRef] [PubMed]
- Wani, J.A.; Majid, S.; Imtiyaz, Z.; Rehman, M.U.; Alsaffar, R.M.; Shah, N.N.; Alshehri, S.; Ghoneim, M.M.; Imam, S.S. MiRNAs in Lung Cancer: Diagnostic, Prognostic, and Therapeutic Potential. Diagnostics 2022, 12, 1610. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, J.; Chen, H.; Wang, S.; Chen, L.; Xu, Y.; Su, F.; Lu, X. MicroRNA-200a suppresses migration and invasion and enhances the radiosensitivity of NSCLC cells by inhibiting the HGF/c-Met signaling pathway. Oncol. Rep. 2019, 41, 1497–1508. [Google Scholar] [CrossRef]
- Hua, L.; Zhu, G.; Wei, J. MicroRNA-1 overexpression increases chemosensitivity of non-small cell lung cancer cells by inhibiting autophagy related 3-mediated autophagy. Cell Biol. Int. 2018, 42, 1240–1249. [Google Scholar] [CrossRef]
- Wang, X.; Meng, Q.; Qiao, W.; Ma, R.; Ju, W.; Hu, J.; Lu, H.; Cui, J.; Jin, Z.; Zhao, Y.; et al. miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res. Ther. 2018, 9, 327. [Google Scholar] [CrossRef]
- Lu, L.; Liu, Q.; Wang, P.; Wu, Y.; Liu, X.; Weng, C.; Fang, X.; Li, B.; Cao, X.; Mao, H.; et al. MicroRNA-148b regulates tumor growth of non-small cell lung cancer through targeting MAPK/JNK pathway. BMC Cancer 2019, 19, 209. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).