Supporting Machine Learning Model in the Treatment of Chronic Pain
Abstract
1. Introduction
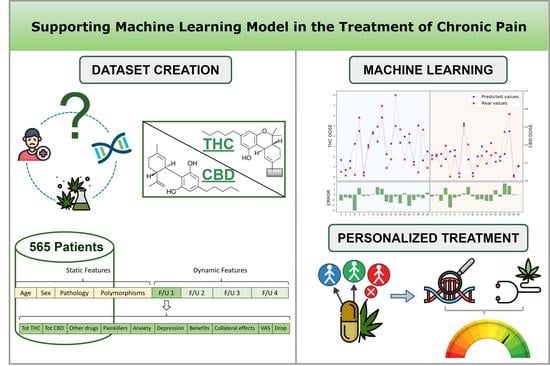
2. Materials and Methods
2.1. Data Collection
2.2. Gene Analysis
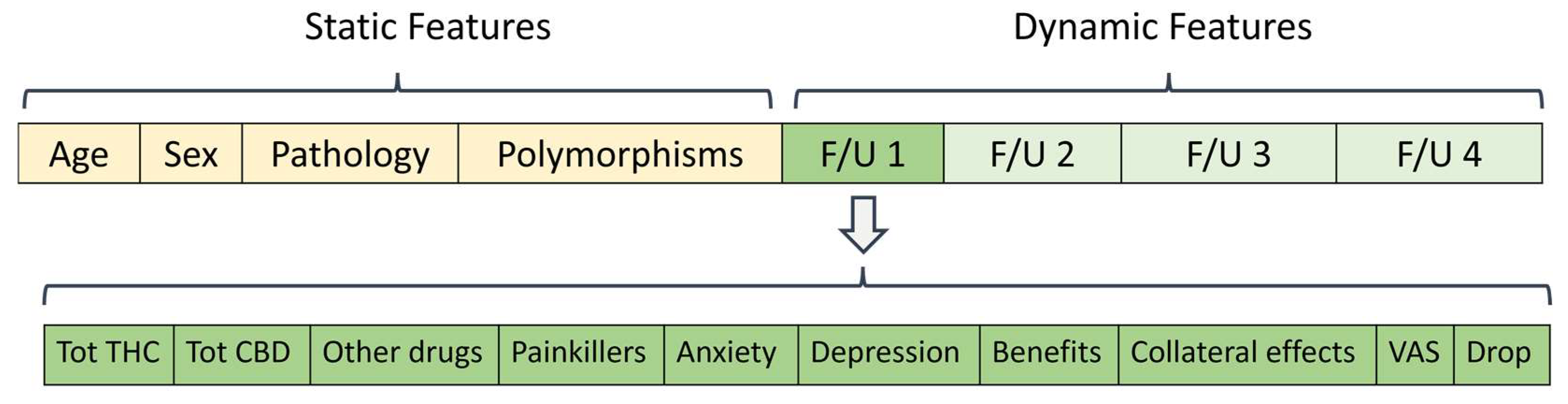
2.3. Clinical Dataset
2.4. Machine Learning Method
3. Results
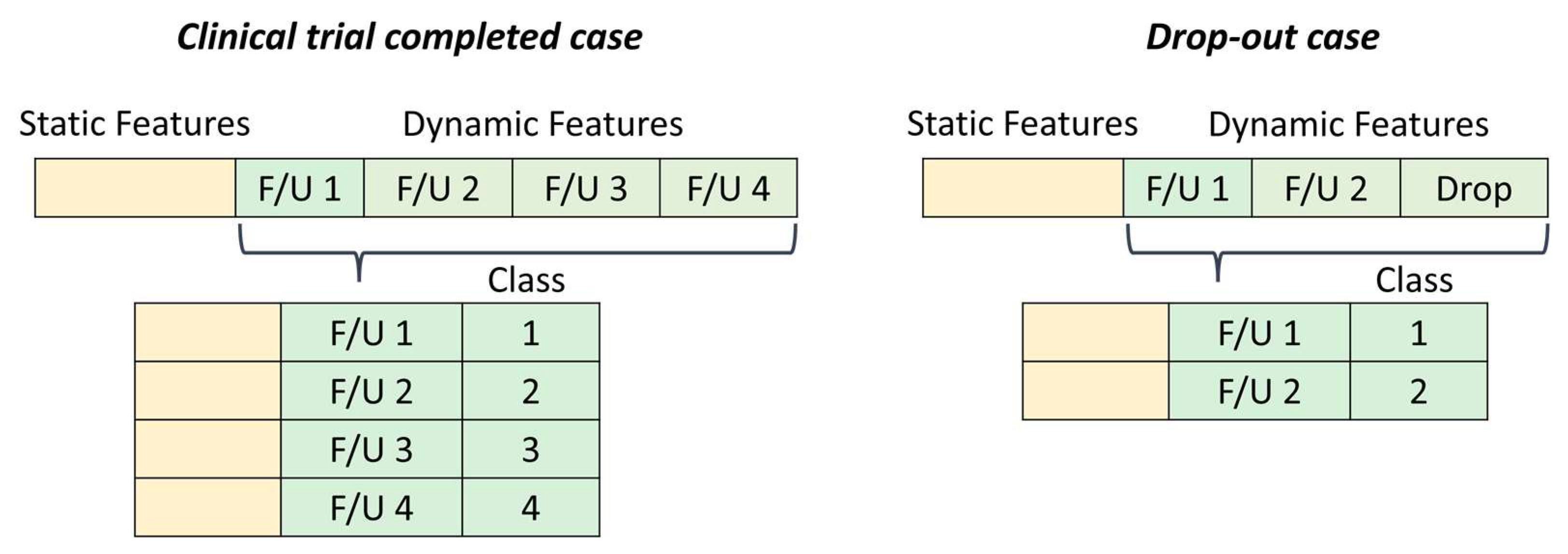
3.1. Data Pre-Processing
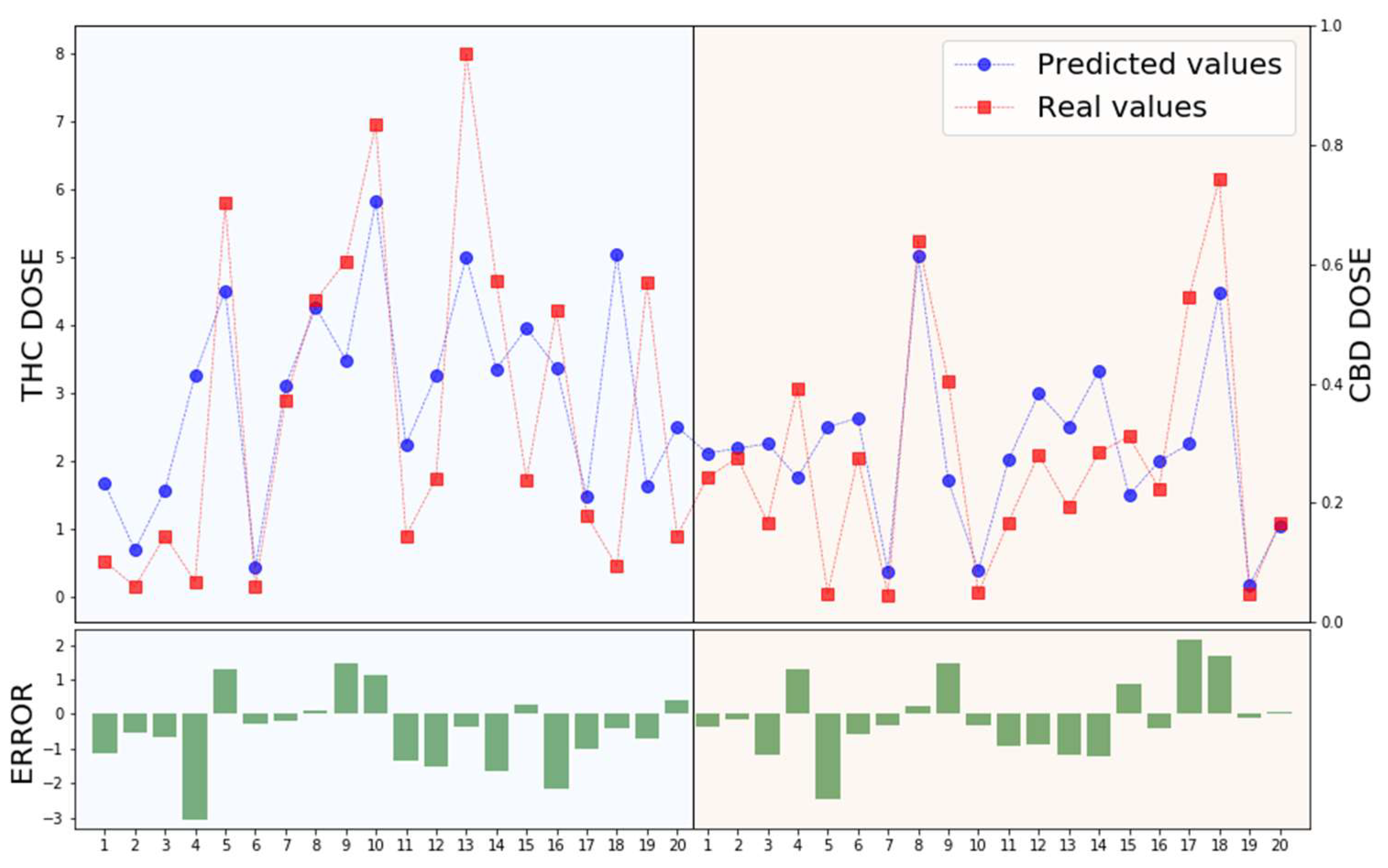
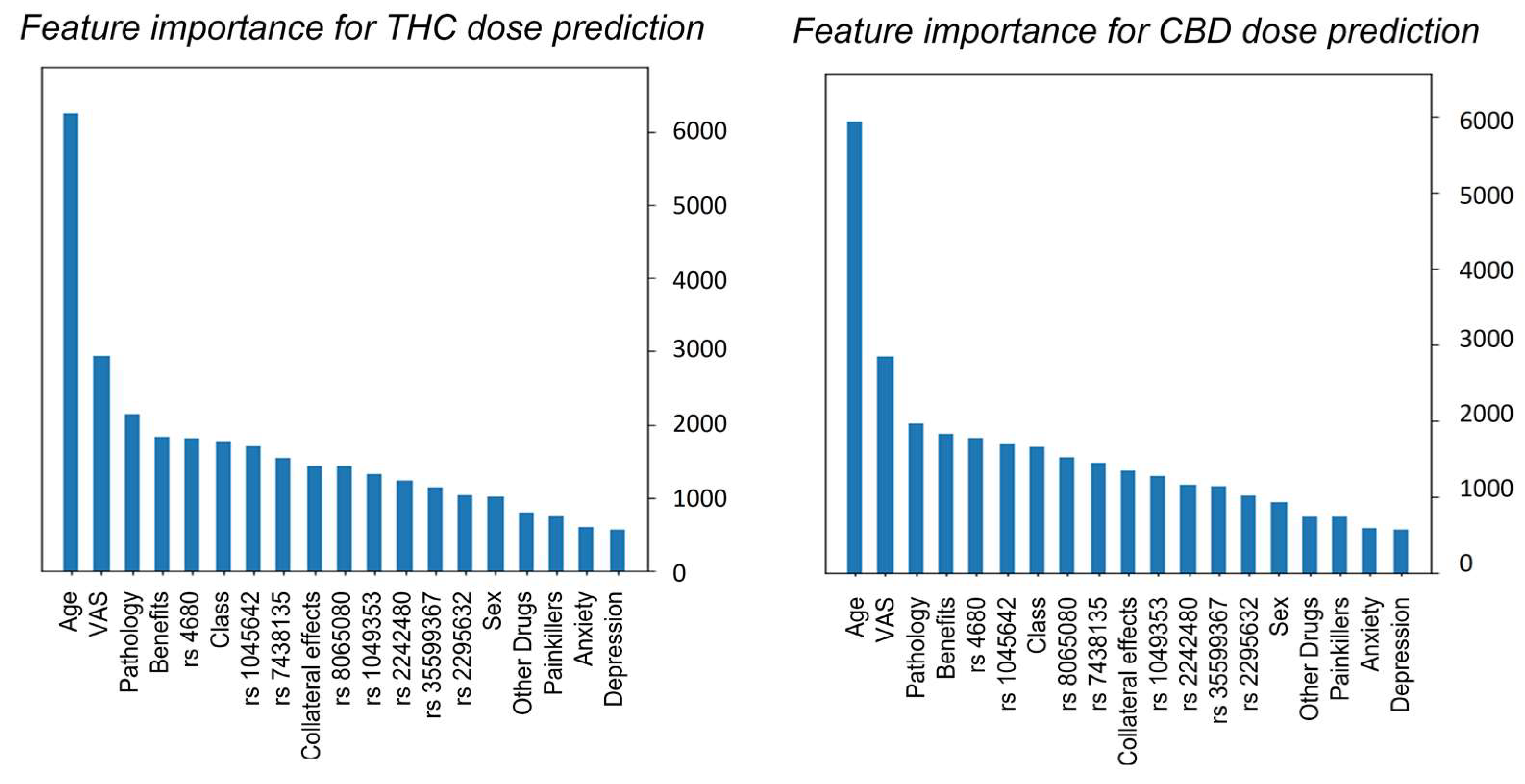
3.2. XGBoost Prediction
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dahlhamer, J.; Lucas, J.; Zelaya, C.; Nahin, R.; Mackey, S.; DeBar, L.; Kerns, R.; Korff, M.; Porter, L.; Helmick, C. Prevalence of Chronic Pain and High-Impact Chronic Pain Among Adults—United States, 2016. MMWR Morb. Mortal. Wkly. Rep. 2018, 67, 1001–1006. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare. Chronic Pain in Australia; Cat. no. PHE 267; Australian Institute of Health and Welfare: Canberra, Australia, 2020.
- Breivik, H.; Collett, B.; Ventafridda, V.; Cohen, R.; Gallacher, D. Survey of Chronic Pain in Europe: Prevalence, Impact on Daily Life, and Treatment. Eur. J. Pain 2006, 10, 287–333. [Google Scholar] [CrossRef]
- Fillingim, R.B. Chapter 49—Sex, Gender, and Pain. In Principles of Gender-Specific Medicine, 4th ed.; Legato, M.J., Ed.; Academic Press: Cambridge, MA, USA, 2023; pp. 769–792. ISBN 978-0-323-88534-8. [Google Scholar]
- Reid, M.C.; Eccleston, C.; Pillemer, K. Management of Chronic Pain in Older Adults. BMJ 2015, 350, h532. [Google Scholar] [CrossRef]
- Jackson, T.; Thomas, S.; Stabile, V.; Han, X.; Shotwell, M.; McQueen, K. Prevalence of Chronic Pain in Low-Income and Middle-Income Countries: A Systematic Review and Meta-Analysis. Lancet 2015, 385, S10. [Google Scholar] [CrossRef]
- Hadi, M.A.; McHugh, G.A.; Closs, S.J. Impact of Chronic Pain on Patients’ Quality of Life: A Comparative Mixed-Methods Study. J. Patient Exp. 2019, 6, 133–141. [Google Scholar] [CrossRef]
- Mathias, J.L.; Cant, M.L.; Burke, A.L.J. Sleep Disturbances and Sleep Disorders in Adults Living with Chronic Pain: A Meta-Analysis. Sleep Med. 2018, 52, 198–210. [Google Scholar] [CrossRef]
- Institute of Medicine (US) Committee on Advancing Pain Research, Care, and Education. Relieving Pain in America: A Blueprint for Transforming Prevention, Care, Education, and Research; The National Academies Press: Washington, DC, USA, 2011; ISBN 978-0-309-25627-8. [Google Scholar]
- Tang, N.; Crane, C. Suicidality in Chronic Pain: A Review of the Prevalence, Risk Factors and Psychological Links. Psychol. Med. 2006, 36, 575–586. [Google Scholar] [CrossRef]
- Hoehe, M.R.; Caenazzo, L.; Martinez, M.M.; Hsieh, W.T.; Modi, W.S.; Gershon, E.S.; Bonner, T.I. Genetic and Physical Mapping of the Human Cannabinoid Receptor Gene to Chromosome 6q14-Q15. New Biol. 1991, 3, 880–885. [Google Scholar]
- Gérard, C.; Mollereau, C.; Vassart, G.; Parmentier, M. Nucleotide Sequence of a Human Cannabinoid Receptor CDNA. Nucleic Acids Res. 1990, 18, 7142. [Google Scholar] [CrossRef]
- Galiègue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carrière, D.; Carayon, P.; Bouaboula, M.; Shire, D.; LE Fur, G.; Casellas, P. Expression of Central and Peripheral Cannabinoid Receptors in Human Immune Tissues and Leukocyte Subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef]
- Pattnaik, F.; Nanda, S.; Mohanty, S.; Dalai, A.K.; Kumar, V.; Ponnusamy, S.K.; Naik, S. Cannabis: Chemistry, Extraction and Therapeutic Applications. Chemosphere 2022, 289, 133012. [Google Scholar] [CrossRef]
- Russo, E. Cannabis Therapeutics and the Future of Neurology. Front. Integr. Neurosci. 2018, 12, 51. [Google Scholar] [CrossRef]
- Billnitzer, A.; Jankovic, J. Current Management of Tics and Tourette Syndrome: Behavioral, Pharmacologic, and Surgical Treatments. Neurotherapeutics 2020, 17, 1681–1693. [Google Scholar] [CrossRef]
- Fitzcharles, M.-A.; Baerwald, C.; Ablin, J.; Häuser, W. Efficacy, Tolerability and Safety of Cannabinoids in Chronic Pain Associated with Rheumatic Diseases (Fibromyalgia Syndrome, Back Pain, Osteoarthritis, Rheumatoid Arthritis). Der. Schmerz. 2016, 30, 47–61. [Google Scholar] [CrossRef]
- Scicluna, J.C.; Giovanni, G. Di Cannabinoids for Fibromyalgia: An Updated Systematic Review. medRxiv 2022. [Google Scholar] [CrossRef]
- Passani, A.; Posarelli, C.; Sframeli, A.T.; Perciballi, L.; Pellegrini, M.; Guidi, G.; Figus, M. Cannabinoids in Glaucoma Patients: The Never-Ending Story. J. Clin. Med. 2020, 9, 3978. [Google Scholar] [CrossRef]
- Dalavaye, N.; Erridge, S.; Nicholas, M.; Pillai, M.; Bapir, L.; Holvey, C.; Coomber, R.; Rucker, J.J.; Hoare, J.; Sodergren, M.H. The Effect of Medical Cannabis in Inflammatory Bowel Disease: Analysis from the UK Medical Cannabis Registry. Expert Rev. Gastroenterol. Hepatol. 2023, 17, 85–98. [Google Scholar] [CrossRef]
- Mucke, M.; Phillips, T.; Radbruch, L.; Petzke, F.; Hauser, W. Cannabis Based Medicines for Chronic Neuropathic Pain in Adults. Cochrane Database Syst. Rev. 2018, 3, CD012182. [Google Scholar] [CrossRef]
- Chung, M.; Kim, H.K.; Abdi, S. Update on Cannabis and Cannabinoids for Cancer Pain. Curr. Opin. Anaesthesiol. 2020, 33, 825–831. [Google Scholar] [CrossRef]
- ElSohly, M.A. Marijuana and the Cannabinoids; Humana: Totowa, NJ, USA, 2006. [Google Scholar]
- Demuth, D.G.; Molleman, A. Cannabinoid Signalling. Life Sci. 2006, 78, 549–563. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. The Health Effects of Cannabis and Cannabinoids: The Current State of Evidence and Recommendations for Research; National Academies Press (US): Washington, DC, USA, 2017. [Google Scholar]
- Babayeva, M.; Loewy, Z.G. Cannabis Pharmacogenomics: A Path to Personalized Medicine. Curr. Issues Mol. Biol. 2023, 45, 3479–3514. [Google Scholar] [CrossRef] [PubMed]
- Hryhorowicz, S.; Walczak, M.; Zakerska-Banaszak, O.; Słomski, R.; Skrzypczak-Zielińska, M. Pharmacogenetics of Cannabinoids. Eur. J. Drug Metab. Pharmacokinet. 2018, 43, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Delgado, D.A.; Lambert, B.S.; Boutris Nickolas and McCulloch, P.C.; Robbins, A.B.; Moreno Michael, R.; Harris, J.D. Validation of Digital Visual Analog Scale Pain Scoring With a Traditional Paper-Based Visual Analog Scale in Adults. JAAOS Glob. Res. Rev. 2018, 2, e088. [Google Scholar] [CrossRef] [PubMed]
- Poli, P.; Peruzzi, L.; Maurizi, P.; Mencucci, A.; Scocca, A.; Carnevale, S.; Spiga, O.; Santucci, A. The Pharmacogenetics of Cannabis in the Treatment of Chronic Pain. Genes 2022, 13, 1832. [Google Scholar] [CrossRef]
- Cilluffo, G.; Fasola, S.; Ferrante, G.; Malizia, V.; Montalbano, L. Machine Learning: An Overview and Applications in Pharmacogenetics. Genes 2021, 12, 1511. [Google Scholar] [CrossRef]
- Zigmond, A.S.; Snaith, R.P. The Hospital Anxiety and Depression Scale. Acta Psychiatr. Scand. 1983, 67, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; p. 10. [Google Scholar]
- Romero-Sandoval, E.A.; Fincham, J.E.; Kolano, A.L.; Sharpe, B.N.; Alvarado-Vázquez, P.A. Cannabis for Chronic Pain: Challenges and Considerations. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2018, 38, 651–662. [Google Scholar] [CrossRef]
- Harris, M.; Erridge, S.; Ergisi, M.; Nimalan, D.; Kawka, M.; Salazar, O.; Ali, R.; Loupasaki, K.; Holvey, C.; Coomber, R.; et al. UK Medical Cannabis Registry: An Analysis of Clinical Outcomes of Medicinal Cannabis Therapy for Chronic Pain Conditions. Expert Rev. Clin. Pharmacol. 2022, 15, 473–485. [Google Scholar] [CrossRef]
- Blanton, H.L.; Barnes, R.C.; McHann, M.C.; Bilbrey, J.A.; Wilkerson, J.L.; Guindon, J. Sex Differences and the Endocannabinoid System in Pain. Pharmacol. Biochem. Behav. 2021, 202, 173107. [Google Scholar] [CrossRef]
- Fattore, L.; Fratta, W. How Important Are Sex Differences in Cannabinoid Action? Br. J. Pharmacol. 2010, 160, 544–548. [Google Scholar] [CrossRef]
- Nowell, W.B.; Gavigan, K.; Silverman, S.L. Cannabis for Rheumatic Disease Pain: A Review of Current Literature. Curr. Rheumatol. Rep. 2022, 24, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, Y.T.; McMahon, L.R.; Wilkerson, J.L. Medicinal Cannabis and Central Nervous System Disorders. Front. Pharmacol. 2022, 13, 881810. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.M.; Saklecha, A.; Patel, A.A.; Divi, S.N. Analyzing the Impact of Cannabinoids on the Treatment of Spinal Disorders. Curr. Rev. Musculoskelet. Med. 2022, 15, 133–142. [Google Scholar] [CrossRef]
- Chayasirisobhon, S. Cannabis and Neuropsychiatric Disorders: An Updated Review. Acta Neurol Taiwan 2019, 28, 27–39. [Google Scholar]
- EI-Mallakh, R.S. Marijuana and Migraine. Headache J. Head Face Pain 1987, 27, 442–443. [Google Scholar] [CrossRef]
- Chen, Q.; Lin, W.; Yang, J.; Lin, M.; Lin, X.; Weng, Y.; Chen, Y. Prognostic Value of Two Polymorphisms, Rs1045642 and Rs1128503, in ABCB1 Following Taxane-Based Chemotherapy: A Meta-Analysis. Asian Pac. J. Cancer Prev. 2021, 22, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Rao, Q.; Lin, K.; He, Y.; Cai, J.; Yang, M.; Xu, Y.; Hou, L.; Lin, Y.; Liu, H. CYP2C19-Rs4986893 Confers Risk to Major Depressive Disorder and Bipolar Disorder in the Han Chinese Population Whereas ABCB1-Rs1045642 Acts as a Protective Factor. BMC Psychiatry 2023, 23, 69. [Google Scholar] [CrossRef]
- Benyamina, A.; Bonhomme-Faivre, L.; Picard, V.; Sabbagh, A.; Richard, D.; Blecha, L.; Rahioui, H.; Karila, L.; Lukasiewicz, M.; Farinotti, R.; et al. Association between ABCB1 C3435T Polymorphism and Increased Risk of Cannabis Dependence. Prog. Neuropsychopharmacol. Biol. Psychiatry 2009, 33, 1270–1274. [Google Scholar] [CrossRef]
- Aziz, M.A.; Islam, M.S. The Role of ABCB1 Gene Polymorphisms in Steroid-Resistant Nephrotic Syndrome: Evidence from a Meta-Analysis of Steroid-Receiving Patients. J. Gene Med. 2022, 24, e3436. [Google Scholar] [CrossRef]
- Tapanee, P.; Tidwell, D.K.; Schilling, M.W.; Peterson, D.G.; Tolar-Peterson, T. Genetic Variation in Taste Receptor Genes (SCNN1B, TRPV1) and Its Correlation with the Perception of Saltiness in Normotensive and Hypertensive Adults. Int. J. Hypertens 2021, 2021, 5559831. [Google Scholar] [CrossRef]
- Dias, A.G.; Rousseau, D.; Duizer, L.; Cockburn, M.; Chiu, W.; Nielsen, D.; El-Sohemy, A. Genetic Variation in Putative Salt Taste Receptors and Salt Taste Perception in Humans. Chem. Senses 2013, 38, 137–145. [Google Scholar] [CrossRef]
- Binder, A.; May, D.; Baron, R.; Maier, C.; Tölle, T.R.; Treede, R.-D.; Berthele, A.; Faltraco, F.; Flor, H.; Gierthmühlen, J.; et al. Transient Receptor Potential Channel Polymorphisms Are Associated with the Somatosensory Function in Neuropathic Pain Patients. PLoS ONE 2011, 6, e17387. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.-N.; Ho, I.-K.; Tsou, H.-H.; Fang, C.-P.; Hsiao, C.-F.; Chen, C.-H.; Tan, H.K.-L.; Lin, L.; Wu, C.-S.; Su, L.-W.; et al. UGT2B7 Genetic Polymorphisms Are Associated with the Withdrawal Symptoms in Methadone Maintenance Patients. Pharmacogenomics 2012, 13, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Verdejo-García, A.; Fagundo, A.B.; Cuenca, A.; Rodriguez, J.; Cuyás, E.; Langohr, K.; de Sola Llopis, S.; Civit, E.; Farré, M.; Peña-Casanova, J.; et al. COMT Val158met and 5-HTTLPR Genetic Polymorphisms Moderate Executive Control in Cannabis Users. Neuropsychopharmacology 2013, 38, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.; Vieira-Coelho, M.A. Cannabis Induced Psychosis: A Systematic Review on the Role of Genetic Polymorphisms. Pharmacol. Res. 2022, 181, 106258. [Google Scholar] [CrossRef] [PubMed]
- Gerra, M.C.; Manfredini, M.; Cortese, E.; Antonioni, M.C.; Leonardi, C.; Magnelli, F.; Somaini, L.; Jayanthi, S.; Cadet, J.L.; Donnini, C. Genetic and Environmental Risk Factors for Cannabis Use: Preliminary Results for the Role of Parental Care Perception. Subst. Use Misuse 2019, 54, 670–680. [Google Scholar] [CrossRef]
- Wang, L.; Bai, M.; Jin, T.; Zheng, J.; Wang, Y.; He, Y.; Yuan, D.; He, X. Effects of CYP3A4 Polymorphisms on Drug Addiction Risk Among the Chinese Han Population. Front. Public Health 2019, 7, 315. [Google Scholar] [CrossRef]
- Chen, C.-H.; Wang, S.-C.; Tsou, H.-H.; Ho, I.-K.; Tian, J.-N.; Yu, C.-J.; Hsiao, C.-F.; Chou, S.-Y.; Lin, Y.-F.; Fang, K.-C.; et al. Genetic Polymorphisms in CYP3A4 Are Associated with Withdrawal Symptoms and Adverse Reactions in Methadone Maintenance Patients. Pharmacogenomics 2011, 12, 1397–1406. [Google Scholar] [CrossRef]
- Wang, D.; Sadee, W. CYP3A4 Intronic SNP Rs35599367 (CYP3A4*22) Alters RNA Splicing. Pharmacogenet. Genom. 2016, 26, 40–43. [Google Scholar] [CrossRef]
- Kitzmiller, J.; (Talameh) Luzum, J.; Baldassarre, D.; Krauss, R.; Medina, M. CYP3A4z.Ast22 and CYP3A5z.Ast3 Are Associated with Increased Levels of Plasma Simvastatin Concentrations in the Cholesterol and Pharmacogenetics Study Cohort. Pharmacogenet. Genom. 2014, 24, 486–491. [Google Scholar] [CrossRef]
- Hopfer, C.; Young, S.; Purcell, S.; Crowley, T.; Stallings, M.; Corley, R.; Rhee, S.; Smolen, A.; Krauter, K.; Hewitt, J.; et al. Cannabis Receptor Haplotype Associated With Fewer Cannabis Dependence Symptoms In Adolescents. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006, 141B, 895–901. [Google Scholar] [CrossRef]
- Zhang, P.-W.; Ishiguro, H.; Ohtsuki, T.; Hess, J.; Carillo, F.; Walther, D.; Onaivi, E.S.; Arinami, T.; Uhl, G.R. Human Cannabinoid Receptor 1: 5′ Exons, Candidate Regulatory Regions, Polymorphisms, Haplotypes and Association with Polysubstance Abuse. Mol. Psychiatry 2004, 9, 916–931. [Google Scholar] [CrossRef]
- Pabalan, N.; Chaweeborisuit, P.; Tharabenjasin, P.; Tasanarong, A.; Jarjanazi, H.; Eiamsitrakoon, T.; Tapanadechopone, P. Associations of CB1 Cannabinoid Receptor (CNR1) Gene Polymorphisms with Risk for Alcohol Dependence. Medicine 2021, 100, e27343. [Google Scholar] [CrossRef]
- Tyndale, R.; Payne, J.; Gerber, A.; Sipe, J. The Fatty Acid Amide Hydrolase C385A (P129T) Missense Variant in Cannabis Users: Studies of Drug Use and Dependence in Caucasians. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2007, 144B, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Hosseinzadeh Anvar, L.; Ahmadalipour, A. Fatty Acid Amide Hydrolase C385A Polymorphism Affects Susceptibility to Various Diseases. BioFactors 2023, 49, 62–78. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.L.; Xia, H.; Caballero, M.Y.; Valtier, S.; Chaudry, G.J. A Military Community Cohort Study Reveals Single Nucleotide Polymorphisms in Inflammation Mediator Genes That Associate With Type 2 Diabetes. Mil. Med. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Lucas, C.J.; Galettis, P.; Schneider, J. The Pharmacokinetics and the Pharmacodynamics of Cannabinoids. Br. J. Clin. Pharmacol. 2018, 84, 2477–2482. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Visibelli, A.; Peruzzi, L.; Poli, P.; Scocca, A.; Carnevale, S.; Spiga, O.; Santucci, A. Supporting Machine Learning Model in the Treatment of Chronic Pain. Biomedicines 2023, 11, 1776. https://doi.org/10.3390/biomedicines11071776
Visibelli A, Peruzzi L, Poli P, Scocca A, Carnevale S, Spiga O, Santucci A. Supporting Machine Learning Model in the Treatment of Chronic Pain. Biomedicines. 2023; 11(7):1776. https://doi.org/10.3390/biomedicines11071776
Chicago/Turabian StyleVisibelli, Anna, Luana Peruzzi, Paolo Poli, Antonella Scocca, Simona Carnevale, Ottavia Spiga, and Annalisa Santucci. 2023. "Supporting Machine Learning Model in the Treatment of Chronic Pain" Biomedicines 11, no. 7: 1776. https://doi.org/10.3390/biomedicines11071776
APA StyleVisibelli, A., Peruzzi, L., Poli, P., Scocca, A., Carnevale, S., Spiga, O., & Santucci, A. (2023). Supporting Machine Learning Model in the Treatment of Chronic Pain. Biomedicines, 11(7), 1776. https://doi.org/10.3390/biomedicines11071776