Lightweight Visual Transformers Outperform Convolutional Neural Networks for Gram-Stained Image Classification: An Empirical Study
Abstract
1. Introduction
2. Background
3. Materials and Methods
3.1. Data Set
3.2. Study Design
3.3. Metrics
3.4. Apparatus
4. Results
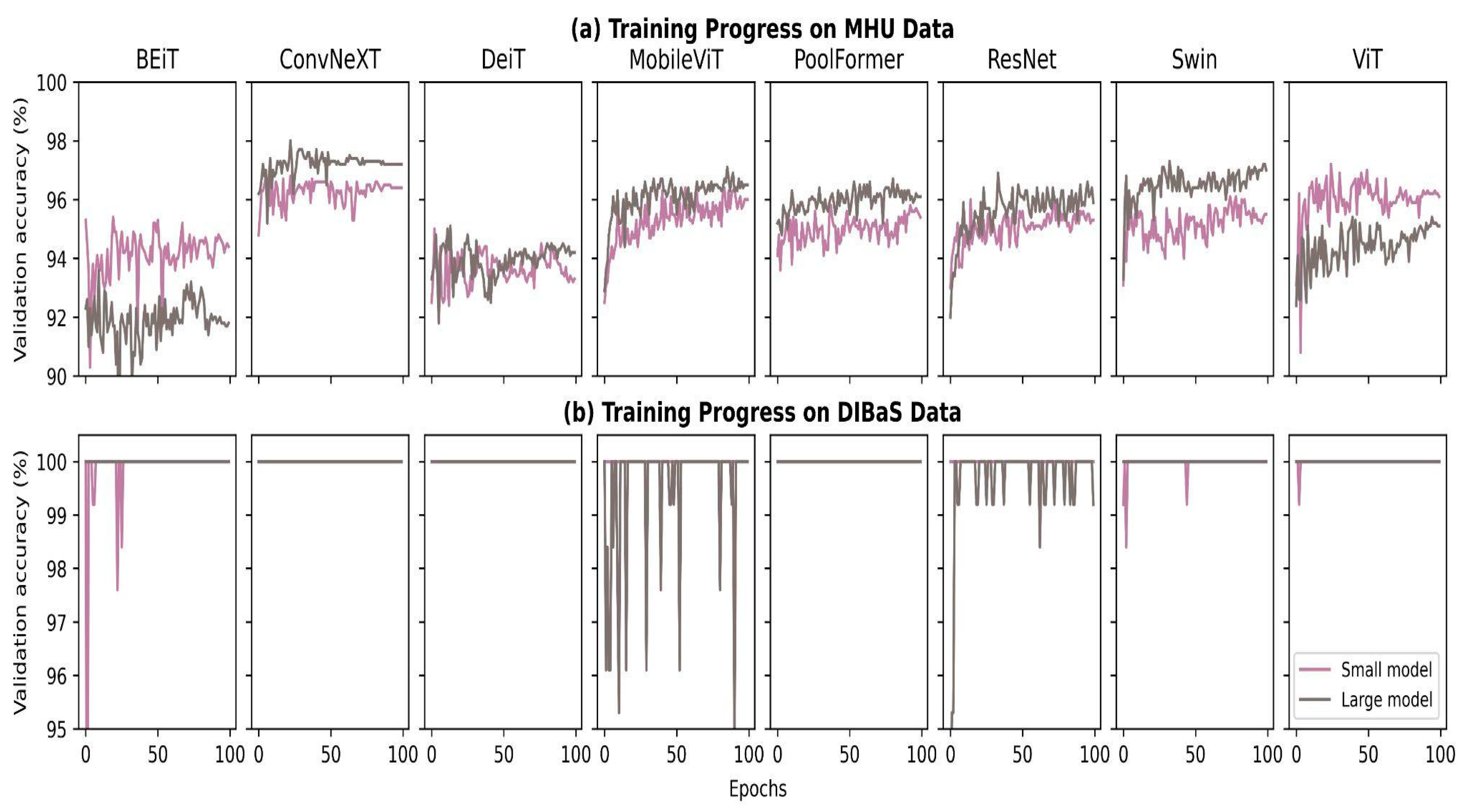
4.1. Fine-Tuning Progress
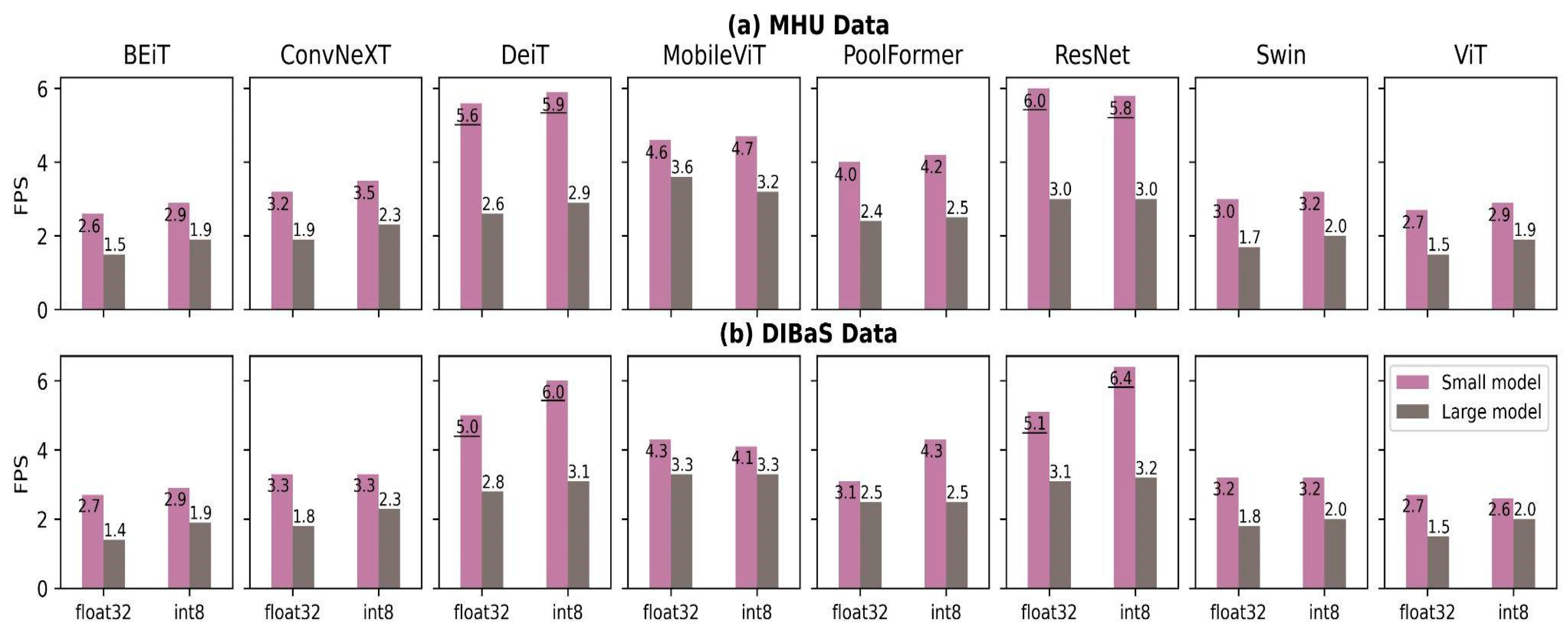
4.2. Accuracy and Quantization
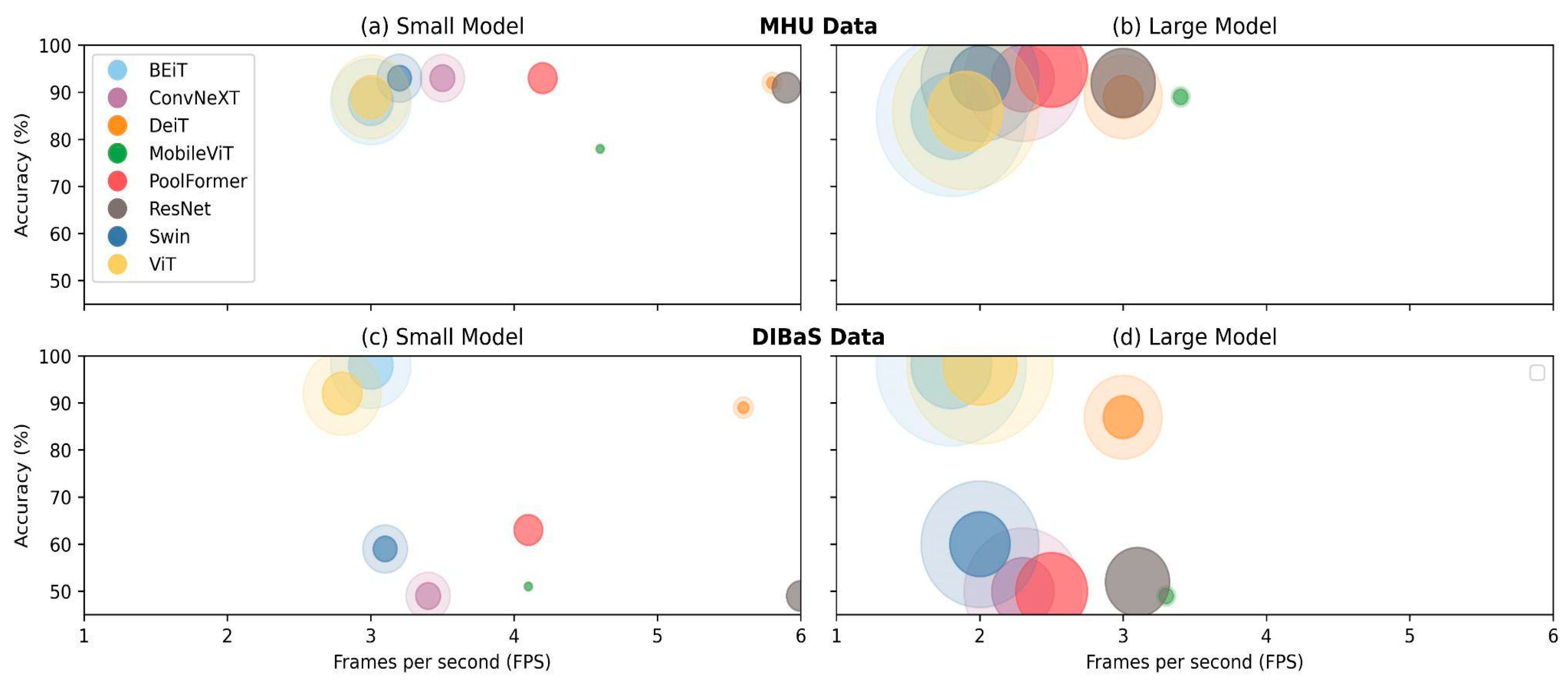
4.3. Time, Size and Trade-Offs
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. F1-score

References
- Kim, H.E.; Cosa-Linan, A.; Santhanam, N.; Jannesari, M.; Maros, M.E.; Ganslandt, T. Transfer Learning for Medical Image Classification: A Literature Review. BMC Med. Imaging 2022, 22, 69. [Google Scholar] [CrossRef] [PubMed]
- Pitkänen, H.; Raunio, L.; Santavaara, I.; Ståhlberg, T. European Medical Device Regulations MDR & IVDR; Business Finland: Helsinki, Finland, 2020; pp. 7–110. [Google Scholar]
- Ahmad, M.A.; Eckert, C.; Teredesai, A. Interpretable Machine Learning in Healthcare. In Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Washington, DC, USA, 29 August–1 September 2018; pp. 559–560. [Google Scholar]
- Ryoo, J.; Rizvi, S.; Aiken, W.; Kissell, J. Cloud Security Auditing: Challenges and Emerging Approaches. IEEE Secur. Priv. 2013, 12, 68–74. [Google Scholar] [CrossRef]
- Seymour, C.W.; Gesten, F.; Prescott, H.C.; Friedrich, M.E.; Iwashyna, T.J.; Phillips, G.S.; Lemeshow, S.; Osborn, T.; Terry, K.M.; Levy, M.M. Time to Treatment and Mortality during Mandated Emergency Care for Sepsis. N. Engl. J. Med. 2017, 376, 2235–2244. [Google Scholar] [CrossRef] [PubMed]
- Coico, R. Gram Staining. Curr. Protocol. Microbiol. 2006, A–3C. [Google Scholar] [CrossRef] [PubMed]
- Centner, F.-S.; Oster, M.E.; Dally, F.-J.; Sauter-Servaes, J.; Pelzer, T.; Schoettler, J.J.; Hahn, B.; Fairley, A.-M.; Abdulazim, A.; Hackenberg, K.A.M. Comparative Analyses of the Impact of Different Criteria for Sepsis Diagnosis on Outcome in Patients with Spontaneous Subarachnoid Hemorrhage. J. Clin. Med. 2022, 11, 3873. [Google Scholar] [CrossRef]
- Komorowski, M.; Celi, L.A.; Badawi, O.; Gordon, A.C.; Faisal, A.A. The Artificial Intelligence Clinician Learns Optimal Treatment Strategies for Sepsis in Intensive Care. Nat. Med. 2018, 24, 1716–1720. [Google Scholar] [CrossRef]
- Liu, K.; Ke, Z.; Chen, P.; Zhu, S.; Yin, H.; Li, Z.; Chen, Z. Classification of Two Species of Gram-Positive Bacteria through Hyperspectral Microscopy Coupled with Machine Learning. Biomed. Opt. Express 2021, 12, 7906–7916. [Google Scholar] [CrossRef]
- Smith, K.P.; Kang, A.D.; Kirby, J.E. Automated Interpretation of Blood Culture Gram Stains by Use of a Deep Convolutional Neural Network. J. Clin. Microbiol. 2018, 56, e01521-17. [Google Scholar] [CrossRef]
- Kim, H.E.; Maros, M.E.; Siegel, F.; Ganslandt, T. Rapid Convolutional Neural Networks for Gram-Stained Image Classification at Inference Time on Mobile Devices: Empirical Study from Transfer Learning to Optimization. Biomedicines 2022, 10, 2808. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. In Advances in Neural Information Processing Systems 25; Pereira, F., Burges, C.J.C., Bottou, L., Weinberger, K.Q., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; pp. 1097–1105. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Proceedings of the 31st Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June2016; pp. 770–778. [Google Scholar]
- Raghu, M.; Unterthiner, T.; Kornblith, S.; Zhang, C.; Dosovitskiy, A. Do Vision Transformers See Like Convolutional Neural Networks? In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2021; Volume 34, pp. 12116–12128. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin Transformer: Hierarchical Vision Transformer Using Shifted Windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Wang, W.; Xie, E.; Li, X.; Fan, D.-P.; Song, K.; Liang, D.; Lu, T.; Luo, P.; Shao, L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 568–578. [Google Scholar]
- Yang, J.; Li, C.; Zhang, P.; Dai, X.; Xiao, B.; Yuan, L.; Gao, J. Focal Self-Attention for Local-Global Interactions in Vision Transformers. arXiv 2021, arXiv:2107.00641. [Google Scholar]
- Lin, T.-Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 936–944. [Google Scholar]
- Lin, T.-Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. arXiv 2018, arXiv:1708.02002. [Google Scholar]
- Hassani, A.; Shi, H. Dilated Neighborhood Attention Transformer. arXiv 2022, arXiv:2209.15001. [Google Scholar]
- Ren, S.; Gao, Z.; Hua, T.; Xue, Z.; Tian, Y.; He, S.; Zhao, H. Co-Advise: Cross Inductive Bias Distillation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16773–16782. [Google Scholar]
- Lin, S.; Xie, H.; Wang, B.; Yu, K.; Chang, X.; Liang, X.; Wang, G. Knowledge Distillation via the Target-Aware Transformer. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 10915–10924. [Google Scholar]
- Devlin, J.; Chang, M.-W.; Lee, K.; Toutanova, K. Bert: Pre-Training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Chen, M.; Radford, A.; Child, R.; Wu, J.; Jun, H.; Luan, D.; Sutskever, I. Generative Pretraining from Pixels. In Proceedings of the International Conference on Machine Learning, Online, 13–18 July 2020; pp. 1691–1703. [Google Scholar]
- Radford, A.; Wu, J.; Child, R.; Luan, D.; Amodei, D.; Sutskever, I. Language Models Are Unsupervised Multitask Learners. OpenAI Blog 2019, 1, 9. [Google Scholar]
- Bao, H.; Dong, L.; Piao, S.; Wei, F. Beit: Bert Pre-Training of Image Transformers. arXiv 2021, arXiv:2106.08254. [Google Scholar]
- Ramachandran, P.; Parmar, N.; Vaswani, A.; Bello, I.; Levskaya, A.; Shlens, J. Stand-Alone Self-Attention in Vision Models. In Proceedings of the 33rd Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 8–9 December 2019. [Google Scholar]
- Srinivas, A.; Lin, T.-Y.; Parmar, N.; Shlens, J.; Abbeel, P.; Vaswani, A. Bottleneck Transformers for Visual Recognition. arXiv 2021, arXiv:2101.11605. [Google Scholar]
- Guo, J.; Han, K.; Wu, H.; Tang, Y.; Chen, X.; Wang, Y.; Xu, C. CMT: Convolutional Neural Networks Meet Vision Transformers. arXiv 2021, arXiv:2107.06263. [Google Scholar]
- Wu, H.; Xiao, B.; Codella, N.; Liu, M.; Dai, X.; Yuan, L.; Zhang, L. CvT: Introducing Convolutions to Vision Transformers. arXiv 2021, arXiv:2103.15808. [Google Scholar]
- Graham, B.; El-Nouby, A.; Touvron, H.; Stock, P.; Joulin, A.; Jégou, H.; Douze, M. LeViT: A Vision Transformer in ConvNet’s Clothing for Faster Inference. arXiv 2021, arXiv:2104.01136. [Google Scholar]
- Xiao, T.; Singh, M.; Mintun, E.; Darrell, T.; Dollár, P.; Girshick, R. Early Convolutions Help Transformers See Better. arXiv 2021, arXiv:2106.14881. [Google Scholar]
- Mehta, S.; Rastegari, M. MobileViT: Light-Weight, General-Purpose, and Mobile-Friendly Vision Transformer. arXiv 2022, arXiv:2110.02178. [Google Scholar]
- Heo, B.; Yun, S.; Han, D.; Chun, S.; Choe, J.; Oh, S.J. Rethinking Spatial Dimensions of Vision Transformers. arXiv 2021, arXiv:2103.16302. [Google Scholar]
- Yu, W.; Luo, M.; Zhou, P.; Si, C.; Zhou, Y.; Wang, X.; Feng, J.; Yan, S. MetaFormer Is Actually What You Need for Vision. arXiv 2022, arXiv:2111.11418. [Google Scholar]
- Zieliński, B.; Plichta, A.; Misztal, K.; Spurek, P.; Brzychczy-Wloch, M.; Ochońska, D. Deep Learning Approach to Bacterial Colony Classification. PLoS ONE 2017, 12, e0184554. [Google Scholar] [CrossRef]
- Zheng, Z.; Cai, Y.; Li, Y. Oversampling Method for Imbalanced Classification. Comput. Inform. 2015, 34, 1017–1037. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A Convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Elharrouss, O.; Akbari, Y.; Almaadeed, N.; Al-Maadeed, S. Backbones-Review: Feature Extraction Networks for Deep Learning and Deep Reinforcement Learning Approaches. arXiv 2022, arXiv:2206.08016. [Google Scholar]
- Sasaki, Y.; Fellow, R. The Truth of the F-Measure; Manchester: MIB-School of Computer Science, University of Manchester: Manchester, UK, 2007. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Huggingface’s transformers: State-of-the-art natural language processing. arXiv 2019, arXiv:1910.03771. [Google Scholar]
- Krogh, A.; Hertz, J. A Simple Weight Decay Can Improve Generalization. In Proceedings of the 4th International Conference on Neural Information Processing Systems, San Francisco, CA, USA, 2–5 December 1991. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv 2015, arXiv:1502.03167. [Google Scholar]
- Salimans, T.; Kingma, D.P. Weight Normalization: A Simple Reparameterization to Accelerate Training of Deep Neural Networks. Available online: https://arxiv.org/abs/1602.07868v3 (accessed on 2 April 2023).
- Prechelt, L. Early Stopping—But When. In Neural Networks: Tricks of the Trade, 2nd Edition; Springer: Berlin/Heidelberg, Germany, 2012; pp. 53–67. [Google Scholar]
- Wu, H.; Judd, P.; Zhang, X.; Isaev, M.; Micikevicius, P. Integer Quantization for Deep Learning Inference: Principles and Empirical Evaluation. arXiv 2020, arXiv:2004.09602. [Google Scholar]
- Chicco, D.; Jurman, G. The Advantages of the Matthews Correlation Coefficient (MCC) over F1 Score and Accuracy in Binary Classification Evaluation. BMC Genom. 2020, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Demšar, J. Statistical Comparisons of Classifiers over Multiple Data Sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- Herbold, S. Autorank: A Python Package for Automated Ranking of Classifiers. J. Open Source Softw. 2020, 5, 2173. [Google Scholar] [CrossRef]
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Comput. Surv. (CSUR) 2022, 54, 200. [Google Scholar] [CrossRef]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. arXiv 2020, arXiv:2005.12872. [Google Scholar]
- Fang, Y.; Liao, B.; Wang, X.; Fang, J.; Qi, J.; Wu, R.; Niu, J.; Liu, W. You Only Look at One Sequence: Rethinking Transformer in Vision through Object Detection. arXiv 2021, arXiv:2106.00666. [Google Scholar]
- Xie, E.; Wang, W.; Yu, Z.; Anandkumar, A.; Alvarez, J.M.; Luo, P. SegFormer: Simple and Efficient Design for Semantic Segmentation with Transformers. arXiv 2021, arXiv:2105.15203. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked Autoencoders Are Scalable Vision Learners. arXiv 2021, arXiv:2111.06377. [Google Scholar]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in Medical Imaging: A Survey. arXiv 2022, arXiv:2201.09873. [Google Scholar] [CrossRef]
- Tanwani, A.K.; Barral, J.; Freedman, D. RepsNet: Combining Vision with Language for Automated Medical Reports. In Medical Image Computing and Computer Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2022; Volume 13435, pp. 714–724. [Google Scholar]
- Saase, V.; Wenz, H.; Ganslandt, T.; Groden, C.; Maros, M.E. Simple Statistical Methods for Unsupervised Brain Anomaly Detection on MRI Are Competitive to Deep Learning Methods. arXiv 2020, arXiv:2011.12735. [Google Scholar]
- Maros, M.E.; Cho, C.G.; Junge, A.G.; Kämpgen, B.; Saase, V.; Siegel, F.; Trinkmann, F.; Ganslandt, T.; Groden, C.; Wenz, H. Comparative Analysis of Machine Learning Algorithms for Computer-Assisted Reporting Based on Fully Automated Cross-Lingual RadLex Mappings. Sci. Rep. 2021, 11, 5529. [Google Scholar] [CrossRef] [PubMed]
- Montemurro, N.; Condino, S.; Carbone, M.; Cattari, N.; D’Amato, R.; Cutolo, F.; Ferrari, V. Brain Tumor and Augmented Reality: New Technologies for the Future. Int. J. Environ. Res. Public Health 2022, 19, 6347. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.-L.; Chen, S.-H.; Chang, T.-C.; Lee, W.-K.; Hsu, H.-T.; Chang, H.-H. Continual Learning of a Transformer-Based Deep Learning Classifier Using an Initial Model from Action Observation EEG Data to Online Motor Imagery Classification. Bioengineering 2023, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Ganslandt, T.; Miethke, T.; Neumaier, M.; Kittel, M. Deep Learning Frameworks for Rapid Gram Stain Image Data Interpretation: Protocol for a Retrospective Data Analysis. JMIR Res. Protoc. 2020, 9, e16843. [Google Scholar] [CrossRef] [PubMed]



| Model | Architecture Traits | Image Size | Patch Size | # Attention Heads | # Parameters (min) | # Parameters (max) |
|---|---|---|---|---|---|---|
| BEiT | Self-supervised VT | 224 | 16 | 12; 16 | 86 M | 307 M |
| ConvNeXT | CNN | 224 | N/A | N/A | 29 M | 198 M |
| DeiT | Knowledge distillation VT | 224 | 16 | 3; 12 | 5 M | 86 M |
| MobileViT | Hybrid | 256 | 2 | 4 | 1.3 M | 5.6 M |
| PoolFormer | Hybrid | 224 | 7, 3, 3, 3 | N/A | 11.9 M | 73.4 M |
| ResNet | CNN | 224 | N/A | N/A | 11 M | 60 M |
| Swin | Multi-stage VT | 224 | 4 | [3,6,12,24]; [4,8,16,32] | 29 M | 197 M |
| ViT | Original VT | 224 | 16 | 12; 16 | 86 M | 307 M |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, H.E.; Maros, M.E.; Miethke, T.; Kittel, M.; Siegel, F.; Ganslandt, T. Lightweight Visual Transformers Outperform Convolutional Neural Networks for Gram-Stained Image Classification: An Empirical Study. Biomedicines 2023, 11, 1333. https://doi.org/10.3390/biomedicines11051333
Kim HE, Maros ME, Miethke T, Kittel M, Siegel F, Ganslandt T. Lightweight Visual Transformers Outperform Convolutional Neural Networks for Gram-Stained Image Classification: An Empirical Study. Biomedicines. 2023; 11(5):1333. https://doi.org/10.3390/biomedicines11051333
Chicago/Turabian StyleKim, Hee E., Mate E. Maros, Thomas Miethke, Maximilian Kittel, Fabian Siegel, and Thomas Ganslandt. 2023. "Lightweight Visual Transformers Outperform Convolutional Neural Networks for Gram-Stained Image Classification: An Empirical Study" Biomedicines 11, no. 5: 1333. https://doi.org/10.3390/biomedicines11051333
APA StyleKim, H. E., Maros, M. E., Miethke, T., Kittel, M., Siegel, F., & Ganslandt, T. (2023). Lightweight Visual Transformers Outperform Convolutional Neural Networks for Gram-Stained Image Classification: An Empirical Study. Biomedicines, 11(5), 1333. https://doi.org/10.3390/biomedicines11051333





