Targeted DNA Demethylation: Vectors, Effectors and Perspectives
Abstract
1. Introduction
2. DNA Methylation as a Key Epigenetic Mechanism of Gene Function Control
3. Etiological Significance of DNA Methylation
4. Biochemical Mechanisms of Active DNA Demethylation
4.1. Enzymatic Removal of the Methyl Group of 5mC
4.2. BER Through Direct Excision of 5mC Converts
4.3. Potential for Enzymatic Demethylation without Excision and BER
5. Biological Effects of De-Repression of Epigenetically Silenced Genes
5.1. For Instructing Cells to Produce a Protective/Therapeutic Protein
5.2. In Targeting Specific Monogenic Epigenetic Aberrations That Single-Handedly Cause Disease
5.3. In Experimental Strategies Such as Transdifferentiation and Regeneration
5.3.1. Transdifferentiation
5.3.2. Proliferation
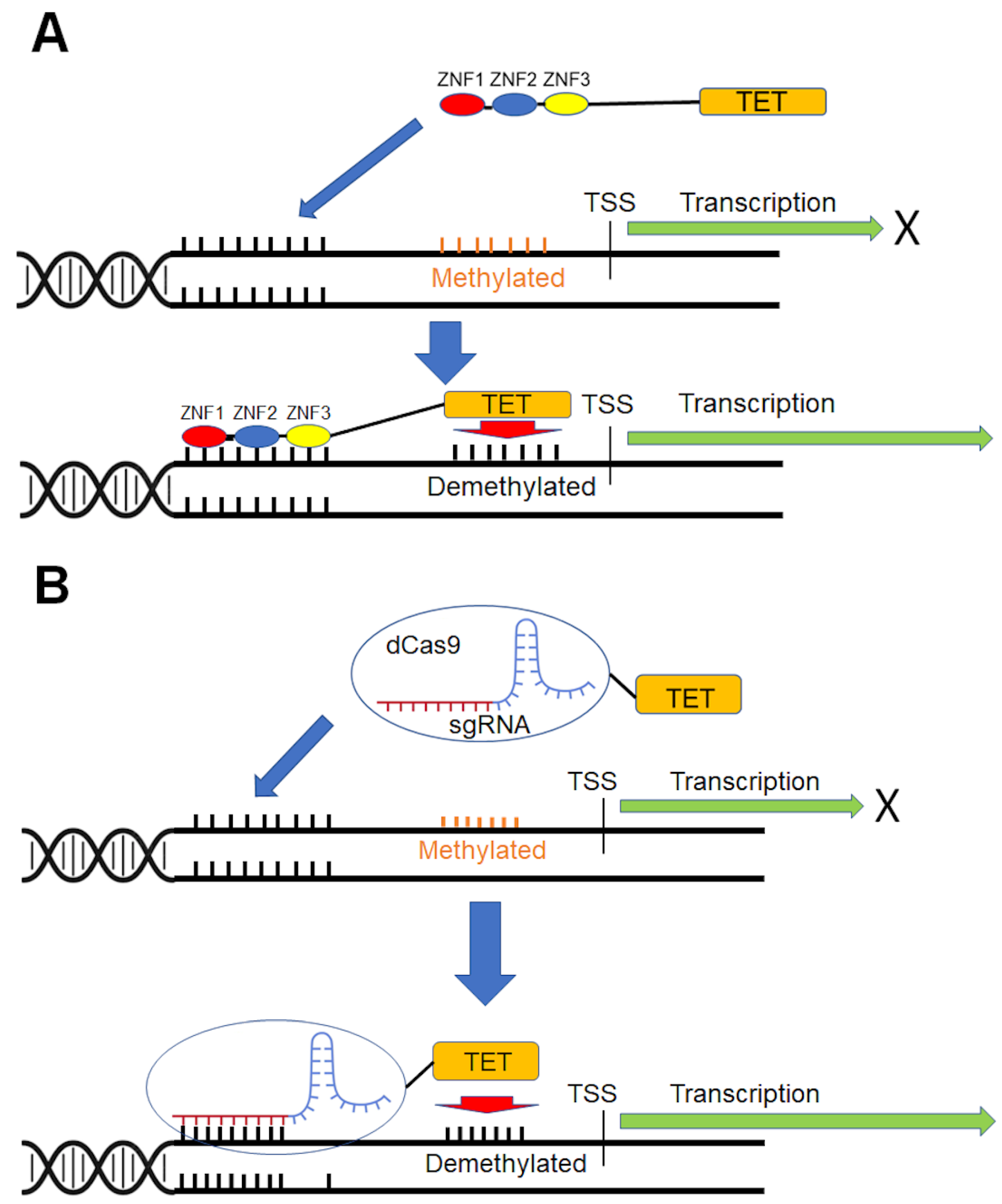
6. Gene-Specific DNA Demethylation
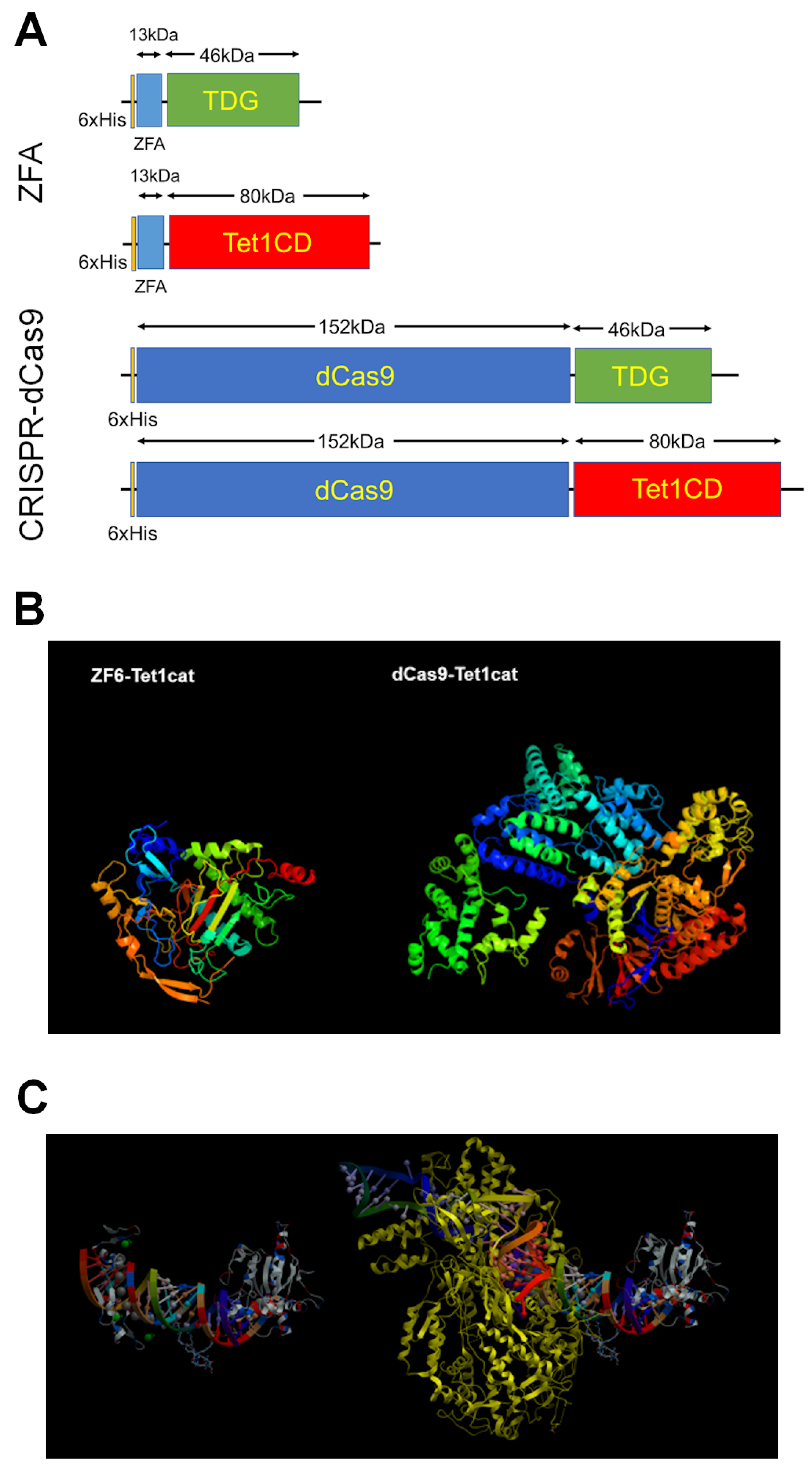
6.1. Selection of Demethylase for Targeted DNA Demethylation
6.2. Site-Specific DNA-Binding Domains
7. In Vivo Delivery
7.1. Viral Delivery
7.1.1. Adenoviral Vectors (AdVs)
7.1.2. Adeno-Associated Viruses Vectors (AAVs)
7.1.3. Lentiviral Vectors (LVs)
- (i)
- sustained gene transfer through stable integration of the vector into the host genome.
- (ii)
- ability to infect both dividing and non-dividing cells.
- (iii)
- broad tissue and cell orientation.
- (iv)
- no expression of viral proteins in the host cells after vector transduction.
- (v)
- ability to deliver complex gene elements.
- (vi)
- a more secure integrated site profile.
- (vii)
- relatively easy vector manipulation and production.
7.2. Non-Viral Vectors
7.3. Direct Delivery of Epigenetically Acting Fusion Proteins
8. Conclusions
- Opens the gene to context-dependent, situation-appropriate stimuli
- Potentially heritable (at least mitotically) effects in tissues
- Mechanistic studies in epigenetics (environmental epigenetics, immunotoxicology)
- Vector-free methods will allow studying causality of immunoregulatory genes
- Forms a new class of ‘biologic’ drugs
- Not every gene can likely be targeted
- Gene-specificity/off-target effects
- Non-viral delivery challenges
- Cell-specific delivery
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ao, C.; Gao, L.; Yu, L. Research Progress in Predicting DNA Methylation Modifications and the Relation with Human Diseases. Curr. Med. Chem. 2022, 29, 822–836. [Google Scholar] [CrossRef] [PubMed]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Ghoshal, K.; Majumder, S.; Li, Z.; Dong, X.; Jacob, S.T. Suppression of Metallothionein Gene Expression in a Rat Hepatoma Because of Promoter-specific DNA Methylation. J. Biol. Chem. 2000, 275, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Jaenisch, R.; Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003, 33, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Meissner, A.; Mikkelsen, T.S.; Gu, H.; Wernig, M.; Hanna, J.; Sivachenko, A.; Zhang, X.; Bernstein, B.E.; Nusbaum, C.; Jaffe, D.B.; et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008, 454, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.; Li, Y.; Robertson, K.D. DNA methylation: Superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Onabote, O.; Hassan, H.M.; Isovic, M.; Torchia, J. The Role of Thymine DNA Glycosylase in Transcription, Active DNA Demethylation, and Cancer. Cancers 2022, 14, 765. [Google Scholar] [CrossRef]
- Ren, W.; Gao, L.; Song, J. Structural Basis of DNMT1 and DNMT3A-Mediated DNA Methylation. Genes 2018, 9, 620. [Google Scholar] [CrossRef]
- Guo, F.; Li, X.; Liang, D.; Li, T.; Zhu, P.; Guo, H.; Wu, X.; Wen, L.; Gu, T.-P.; Hu, B.; et al. Active and Passive Demethylation of Male and Female Pronuclear DNA in the Mammalian Zygote. Cell Stem Cell 2014, 15, 447–459. [Google Scholar] [CrossRef]
- Suelves, M.; Carrió, E.; Núñez-Álvarez, Y.; Peinado, M.A. DNA methylation dynamics in cellular commitment and differentiation. Briefings Funct. Genom. 2016, 15, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, G.P.; Steigerwald, S.D.; Grünwald, S. The DNA methylation system in proliferating and differentiated cells. Cell Biochem. Biophys. 1989, 15, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Wilks, A.; Seldran, M.; Jost, J.-P. An estrogen-dependent demethylation at the 5′ end of the chicken vitellogenin gene is independent of DNA synthesis. Nucleic Acids Res. 1984, 12, 1163–1177. [Google Scholar] [CrossRef] [PubMed]
- Niehrs, C. Active DNA demethylation and DNA repair. Differentiation 2009, 77, 1–11. [Google Scholar] [CrossRef]
- Szyf, M.; Eliasson, L.; Mann, V.; Klein, G.; Razin, A. Cellular and viral DNA hypomethylation associated with induction of Epstein-Barr virus lytic cycle. Proc. Natl. Acad. Sci. USA 1985, 82, 8090–8094. [Google Scholar] [CrossRef]
- Mayer, W.; Niveleau, A.; Walter, J.; Fundele, R.; Haaf, T. Demethylation of the zygotic paternal genome. Nature 2000, 403, 501–502. [Google Scholar] [CrossRef]
- Ou, J.-N.; Torrisani, J.; Unterberger, A.; Provençal, N.; Shikimi, K.; Karimi, M.; Ekström, T.J.; Szyf, M. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem. Pharmacol. 2007, 73, 1297–1307. [Google Scholar] [CrossRef]
- Gehring, M.; Huh, J.H.; Hsieh, T.-F.; Penterman, J.; Choi, Y.; Harada, J.J.; Goldberg, R.B.; Fischer, R.L. DEMETER DNA Glycosylase Establishes MEDEA Polycomb Gene Self-Imprinting by Allele-Specific Demethylation. Cell 2006, 124, 495–506. [Google Scholar] [CrossRef]
- Barreto, G.; Schäfer, A.; Marhold, J.; Stach, D.; Swaminathan, S.K.; Handa, V.; Döderlein, G.; Maltry, N.; Wu, W.; Lyko, F.; et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature 2007, 445, 671–675. [Google Scholar] [CrossRef]
- Razin, A.; Szyf, M.; Kafri, T.; Roll, M.; Giloh, H.; Scarpa, S.; Carotti, D.; Cantoni, G.L. Replacement of 5-methylcytosine by cytosine: A possible mechanism for transient DNA demethylation during differentiation. Proc. Natl. Acad. Sci. USA 1986, 83, 2827–2831. [Google Scholar] [CrossRef]
- Weaver, I.C.; Cervoni, N.; Champagne, F.A.; D’Alessio, A.C.; Sharma, S.; Seckl, J.R.; Dymov, S.; Szyf, M.; Meaney, M.J. Epi-genetic programming by maternal behavior. Nat. Neurosci. 2004, 7, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.T.; Fields, P.E.; Flavell, R.A. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proc. Natl. Acad. Sci. USA 2007, 104, 17052–17057. [Google Scholar] [CrossRef] [PubMed]
- Kangaspeska, S.; Stride, B.; Métivier, R.; Polycarpou-Schwarz, M.; Ibberson, D.; Carmouche, R.P.; Benes, V.; Gannon, F.; Reid, G. Transient cyclical methylation of promoter DNA. Nature 2008, 452, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-G.; Zhan, W.; Yan, L.; Qin, R.-Y.; Yan, Y.-P.; Yang, Z.-J.; Liu, G.-C.; Li, G.-Q.; Wang, H.-F.; Li, X.-L.; et al. TET1 partially mediates HDAC inhibitor-induced suppression of breast cancer invasion. Mol. Med. Rep. 2014, 10, 2595–2600. [Google Scholar] [CrossRef]
- Bayraktar, G.; Kreutz, M.R. The Role of Activity-Dependent DNA Demethylation in the Adult Brain and in Neurological Disorders. Front. Mol. Neurosci. 2018, 11, 169. [Google Scholar] [CrossRef]
- Sleutels, F.; Barlow, D.P. The origins of genomic imprinting in mammals. Adv. Genet. 2002, 46, 119–163. [Google Scholar] [CrossRef]
- Jeziorska, D.M.; Murray, R.J.S.; De Gobbi, M.; Gaentzsch, R.; Garrick, D.; Ayyub, H.; Chen, T.; Li, E.; Telenius, J.; Lynch, M.; et al. DNA methylation of intragenic CpG islands depends on their transcriptional activity during differentiation and disease. Proc. Natl. Acad. Sci. USA 2017, 114, E7526–E7535. [Google Scholar] [CrossRef]
- Tang, L.; Ye, H.; Hong, Q.; Wang, L.; Wang, Q.; Wang, H.; Xu, L.; Bu, S.; Zhang, L.; Cheng, J.; et al. Elevated CpG island methylation of GCK gene predicts the risk of type 2 diabetes in Chinese males. Gene 2014, 547, 329–333. [Google Scholar] [CrossRef]
- de la Rocha, C.; Zaina, S.; Lund, G. Is Any Cardiovascular Disease-Specific DNA Methylation Biomarker Within Reach? Curr. Atheroscler. Rep. 2020, 22, 62. [Google Scholar] [CrossRef]
- Kitamoto, T.; Kitamoto, A.; Ogawa, Y.; Honda, Y.; Imajo, K.; Saito, S.; Yoneda, M.; Nakamura, T.; Nakajima, A.; Hotta, K. Targeted-bisulfite sequence analysis of the methylation of CpG islands in genes encoding PNPLA3, SAMM50, and PARVB of patients with non-alcoholic fatty liver disease. J. Hepatol. 2015, 63, 494–502. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yang, Y.; Tian, H.; Quan, H.; Liu, S.; Zhang, L.; Yang, L.; Zhu, H.; Wu, H.; Gao, Y.Q. Computational characterization of domain-segregated 3D chromatin structure and segmented DNA methylation status in carcinogenesis. Mol. Oncol. 2021, 16, 699–716. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-T.; Lin, W.-D.; Liao, W.-L.; Lin, Y.-J.; Chang, J.-G.; Tsai, F.-J. PTPRD silencing by DNA hypermethylation decreases insulin receptor signaling and leads to type 2 diabetes. Oncotarget 2015, 6, 12997–13005. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, Y.; Yamada, H.; Munetsuna, E.; Fujii, R.; Yamazaki, M.; Ando, Y.; Mizuno, G.; Ishikawa, H.; Ohashi, K.; Hashimoto, S.; et al. Global DNA hypermethylation in peripheral blood mononuclear cells and cardiovascular disease risk: A population-based propensity score-matched cohort study. J. Epidemiol. Community Health 2021, 75, 890–895. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.J.; McKay, G.J.; Maxwell, A.P.; McKnight, A.J. DNA hypermethylation and DNA hypomethylation is present at different loci in chronic kidney disease. Epigenetics 2013, 9, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Dinardo, A.R.; Rajapakshe, K.; Nishiguchi, T.; Grimm, S.L.; Mtetwa, G.; Dlamini, Q.; Kahari, J.; Mahapatra, S.; Kay, A.W.; Maphalala, G.; et al. DNA hypermethylation during tuberculosis dampens host immune responsiveness. J. Clin. Investig. 2020, 130, 3113–3123. [Google Scholar] [CrossRef]
- Chen, H.-C.; Chen, Y.-Z.; Wang, C.-H.; Lin, F.-J. The nonalcoholic fatty liver disease-like phenotype and lowered serum VLDL are associated with decreased expression and DNA hypermethylation of hepatic ApoB in male offspring of ApoE deficient mothers fed a with Western diet. J. Nutr. Biochem. 2019, 77, 108319. [Google Scholar] [CrossRef]
- Xiang, T.; Li, L.; Yin, X.; Yuan, C.; Tan, C.; Su, X.; Xiong, L.; Putti, T.C.; Oberst, M.; Kelly, K.; et al. The Ubiquitin Peptidase UCHL1 Induces G0/G1 Cell Cycle Arrest and Apoptosis Through Stabilizing p53 and Is Frequently Silenced in Breast Cancer. PLoS ONE 2012, 7, e29783. [Google Scholar] [CrossRef]
- Saavedra, K.; Valbuena, J.; Olivares, W.; Marchant, M.J.; Rodríguez, A.; Torres-Estay, V.; Carrasco-Aviño, G.; Guzmán, L.; Aguayo, F.; Roa, J.C.; et al. Loss of Expression of Reprimo, a p53-induced Cell Cycle Arrest Gene, Correlates with Invasive Stage of Tumor Progression and p73 Expression in Gastric Cancer. PLoS ONE 2015, 10, e0125834. [Google Scholar] [CrossRef]
- Xu, G.; Fan, L.; Zhao, S.; OuYang, C. Neuronal pentraxin II (NPTX2) hypermethylation promotes cell proliferation but inhibits cell cycle arrest and apoptosis in gastric cancer cells by suppressing the p53 signaling pathway. Bioengineered 2021, 12, 1311–1323. [Google Scholar] [CrossRef]
- Keller, J.A.; Erson-Bensan, A.E.; Petty, E.M. Connections between CHFR, the cell cycle, and chemosensitivity: Are they critical in cancer? Cancer Biol. Ther. 2010, 10, 942–944. [Google Scholar] [CrossRef]
- Johnson, B.E. Emerging gene mutation targets in lung cancer. Clin. Adv. Hematol. Oncol. 2015, 13, 812–814. [Google Scholar] [PubMed]
- Takeshita, M.; Koga, T.; Takayama, K.; Yano, T.; Maehara, Y.; Nakanishi, Y.; Sueishi, K. Alternative efficacy-predicting markers for paclitaxel instead of CHFR in non-small-cell lung cancer. Cancer Biol. Ther. 2010, 10, 933–941. [Google Scholar] [CrossRef]
- Toyota, M.; Sasaki, Y.; Satoh, A.; Ogi, K.; Kikuchi, T.; Suzuki, H.; Mita, H.; Tanaka, N.; Itoh, F.; Issa, J.-P.J.; et al. Epigenetic inactivation of CHFR in human tumors. Proc. Natl. Acad. Sci. USA 2003, 100, 7818–7823. [Google Scholar] [CrossRef] [PubMed]
- Scolnick, D.M.; Halazonetis, T.D. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature 2000, 406, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.-D.; Hu, S.-L.; Sun, Y.-B.; Xu, W.-P.; Shen, G.; Kong, X.-Y. Promoter methylation of CHFR gene in gastric carcinoma tissues detected using two methods. Chin. J. Cancer 2010, 29, 163–166. [Google Scholar] [CrossRef]
- Castiel, A.; Danieli, M.M.; David, A.; Moshkovitz, S.; Aplan, P.D.; Kirsch, I.R.; Brandeis, M.; Krämer, A.; Izraeli, S. The Stil protein regulates centrosome integrity and mitosis through suppression of Chfr. J. Cell Sci. 2011, 124, 532–539. [Google Scholar] [CrossRef]
- Erson, A.E.; Petty, E.M. CHFR-associated early G2/M checkpoint defects in breast cancer cells. Mol. Carcinog. 2003, 39, 26–33. [Google Scholar] [CrossRef]
- Yanokura, M.; Banno, K.; Kawaguchi, M.; Hirao, N.; Hirasawa, A.; Susumu, N.; Tsukazaki, K.; Aoki, D. Relationship of aberrant DNA hypermethylation of CHFR with sensitivity to taxanes in endometrial cancer. Oncol. Rep. 2007, 17, 41–48. [Google Scholar] [CrossRef]
- Helling, B.A.; Gerber, N.A.; Kadiyala, V.; Sasse, S.K.; Pedersen, B.S.; Sparks, L.; Nakano, Y.; Okamoto, T.; Evans, C.M.; Yang, I.V.; et al. Regulation of MUC5B expression in idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2017, 57, 91–99. [Google Scholar] [CrossRef]
- Wang, H.; Maurano, M.T.; Qu, H.; Varley, K.E.; Gertz, J.; Pauli, F.; Lee, K.; Canfield, T.; Weaver, M.; Sandstrom, R.; et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012, 22, 1680–1688. [Google Scholar] [CrossRef] [PubMed]
- Murayama, A.; Sakura, K.; Nakama, M.; Yasuzawa-Tanaka, K.; Fujita, E.; Tateishi, Y.; Wang, Y.; Ushijima, T.; Baba, T.; Shibuya, K.; et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006, 25, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Green, B.B.; Marsit, C.J. Select Prenatal Environmental Exposures and Subsequent Alterations of Gene-Specific and Repetitive Element DNA Methylation in Fetal Tissues. Curr. Environ. Health Rep. 2015, 2, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Bollati, V. Epigenetics and environmental chemicals. Curr. Opin. Pediatr. 2009, 21, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Mund, C.; Hackanson, B.; Stresemann, C.; Lübbert, M.; Lyko, F. Characterization of DNA Demethylation Effects Induced by 5-Aza-2′-Deoxycytidine in Patients with Myelodysplastic Syndrome. Cancer Res. 2005, 65, 7086–7090. [Google Scholar] [CrossRef]
- Patra, S.K.; Bettuzzi, S. Epigenetic DNA-(cytosine-5-carbon) modifications: 5-aza-2′-deoxycytidine and DNA-demethylation. Biochemistry 2009, 74, 613–619. [Google Scholar] [CrossRef]
- Singh, K.P.; Treas, J.; Tyagi, T.; Gao, W. DNA demethylation by 5-aza-2-deoxycytidine treatment abrogates 17 beta-estradiol-induced cell growth and restores expression of DNA repair genes in human breast cancer cells. Cancer Lett. 2012, 316, 62–69. [Google Scholar] [CrossRef]
- Gnyszka, A.; Jastrzebski, Z.; Flis, S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013, 33, 2989–2996. [Google Scholar]
- Stewart, D.J.; Issa, J.-P.; Kurzrock, R.; Nunez, M.I.; Jelinek, J.; Hong, D.; Oki, Y.; Guo, Z.; Gupta, S.; Wistuba, I.I. Decitabine Effect on Tumor Global DNA Methylation and Other Parameters in a Phase I Trial in Refractory Solid Tumors and Lymphomas. Clin. Cancer Res. 2009, 15, 3881–3888. [Google Scholar] [CrossRef]
- Schuermann, D.; Weber, A.R.; Schär, P. Active DNA demethylation by DNA repair: Facts and uncertainties. DNA Repair. 2016, 44, 92–102. [Google Scholar] [CrossRef]
- De Groote, M.L.; Verschure, P.J.; Rots, M.G. Epigenetic Editing: Targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucleic Acids Res. 2012, 40, 10596–10613. [Google Scholar] [CrossRef] [PubMed]
- Gjerset, R.A.; Martin, D.W., Jr. Presence of a DNA demethylating activity in the nucleus of murine erythroleukemic cells. J. Biol. Chem. 1982, 257, 8581–8583. [Google Scholar] [CrossRef] [PubMed]
- Kavoosi, S.; Sudhamalla, B.; Dey, D.; Shriver, K.; Arora, S.; Sappa, S.; Islam, K. Site- and degree-specific C–H oxidation on 5-methylcytosine homologues for probing active DNA demethylation. Chem. Sci. 2019, 10, 10550–10555. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev. 2011, 25, 2436–2452. [Google Scholar] [CrossRef]
- Ito, S.; D’alessio, A.C.; Taranova, O.V.; Hong, K.; Sowers, L.C.; Zhang, Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010, 466, 1129–1133. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- DeNizio, J.E.; Dow, B.J.; Serrano, J.C.; Ghanty, U.; Drohat, A.C.; Kohli, R.M. TET-TDG Active DNA Demethylation at CpG and Non-CpG Sites. J. Mol. Biol. 2021, 433, 166877. [Google Scholar] [CrossRef]
- Lorsbach, R.B.; Moore, J.; Mathew, S.; Raimondi, S.C.; Mukatira, S.T.; Downing, J.R. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia 2003, 17, 637–641. [Google Scholar] [CrossRef]
- Guo, J.U.; Su, Y.; Zhong, C.; Ming, G.-L.; Song, H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell 2011, 145, 423–434. [Google Scholar] [CrossRef]
- Seiler, C.L.; Fernandez, J.; Koerperich, Z.; Andersen, M.P.; Kotandeniya, D.; Nguyen, M.E.; Sham, Y.; Tretyakova, N.Y. Maintenance DNA Methyltransferase Activity in the Presence of Oxidized Forms of 5-Methylcytosine: Structural Basis for Ten Eleven Translocation-Mediated DNA Demethylation. Biochemistry 2018, 57, 6061–6069. [Google Scholar] [CrossRef]
- Ross, S.E.; Bogdanovic, O. TET enzymes, DNA demethylation and pluripotency. Biochem. Soc. Trans. 2019, 47, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Fuks, F. TET proteins: On the frenetic hunt for new cytosine modifications. Briefings Funct. Genom. 2013, 12, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Seethy, A.; Pethusamy, K.; Chattopadhyay, I.; Sah, R.; Chopra, A.; Dhar, R.; Karmakar, S. TETology: Epigenetic Mastermind in Action. Appl. Biochem. Biotechnol. 2021, 193, 1701–1726. [Google Scholar] [CrossRef] [PubMed]
- Akahori, H.; Guindon, S.; Yoshizaki, S.; Muto, Y. Molecular Evolution of the TET Gene Family in Mammals. Int. J. Mol. Sci. 2015, 16, 28472–28485. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Guo, B.; Wu, H.; Tan, L.; Lu, Q. TET Family of Dioxygenases: Crucial Roles and Underlying Mechanisms. Cytogenet. Genome Res. 2015, 146, 171–180. [Google Scholar] [CrossRef]
- Fromme, J.; Verdine, G.L. Base Excision Repair. Cold Spring Harb. Perspect. Biol. 2004, 69, a012583. [Google Scholar] [CrossRef]
- Mol, C.D.; Hosfield, D.J.; A Tainer, J. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: The 3′ ends justify the means. Mutat. Res. Repair 2000, 460, 211–229. [Google Scholar] [CrossRef]
- Ronemus, M.J.; Galbiati, M.; Ticknor, C.; Chen, J.; Dellaporta, S.L. Demethylation-Induced Developmental Pleiotropy in Arabidopsis. Science 1996, 273, 654–657. [Google Scholar] [CrossRef]
- Zhu, B.; Zheng, Y.; Hess, D.; Angliker, H.; Schwarz, S.; Siegmann, M.; Thiry, S.; Jost, J.-P. 5-Methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc. Natl. Acad. Sci. USA 2000, 97, 5135–5139. [Google Scholar] [CrossRef]
- Hashimoto, H.; Hong, S.; Bhagwat, A.S.; Zhang, X.; Cheng, X. Excision of 5-hydroxymethyluracil and 5-carboxylcytosine by the thymine DNA glycosylase domain: Its structural basis and implications for active DNA demethylation. Nucleic Acids Res. 2012, 40, 10203–10214. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, X.; Lu, J.; Liang, H.; Dai, Q.; Xu, G.-L.; Luo, C.; Jiang, H.; He, C. Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA. Nat. Chem. Biol. 2012, 8, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Zhang, X.; Cheng, X. Selective Excision of 5-Carboxylcytosine by a Thymine DNA Glycosylase Mutant. J. Mol. Biol. 2013, 425, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.T.; Rodgers, M.T.; Hebert, A.S.; Ruslander, L.E.; Eisele, L.; Drohat, A.C. Specificity of Human Thymine DNA Glycosylase Depends on N-Glycosidic Bond Stability. J. Am. Chem. Soc. 2006, 128, 12510–12519. [Google Scholar] [CrossRef] [PubMed]
- Onodera, A.; González-Avalos, E.; Lio, C.-W.J.; Georges, R.O.; Bellacosa, A.; Nakayama, T.; Rao, A. Roles of TET and TDG in DNA demethylation in proliferating and non-proliferating immune cells. Genome Biol. 2021, 22, 186. [Google Scholar] [CrossRef] [PubMed]
- Gregory, D.J.; Mikhaylova, L.; Fedulov, A.V. Selective DNA demethylation by fusion of TDG with a sequence-specific DNA-binding domain. Epigenetics 2012, 7, 344–349. [Google Scholar] [CrossRef]
- Gregory, D.J.; Zhang, Y.; Kobzik, L.; Fedulov, A.V. Specific transcriptional enhancement of inducible nitric oxide synthase by targeted promoter demethylation. Epigenetics 2013, 8, 1205–1212. [Google Scholar] [CrossRef]
- Hassan, H.M.; Kolendowski, B.; Isovic, M.; Bose, K.; Dranse, H.J.; Sampaio, A.V.; Underhill, T.M.; Torchia, J. Regulation of Active DNA Demethylation through RAR-Mediated Recruitment of a TET/TDG Complex. Cell Rep. 2017, 19, 1685–1697. [Google Scholar] [CrossRef]
- Weber, A.R.; Krawczyk, C.; Robertson, A.B.; Kuśnierczyk, A.; Vågbø, C.B.; Schuermann, D.; Klungland, A.; Schär, P. Biochemical reconstitution of TET1–TDG–BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat. Commun. 2016, 7, 10806. [Google Scholar] [CrossRef]
- Cortellino, S.; Xu, J.; Sannai, M.; Moore, R.; Caretti, E.; Cigliano, A.; Le Coz, M.; Devarajan, K.; Wessels, A.; Soprano, D.; et al. Thymine DNA Glycosylase Is Essential for Active DNA Demethylation by Linked Deamination-Base Excision Repair. Cell 2011, 146, 67–79. [Google Scholar] [CrossRef]
- Niehrs, C.; Schäfer, A. Active DNA demethylation by Gadd45 and DNA repair. Trends Cell Biol. 2012, 22, 220–227. [Google Scholar] [CrossRef]
- Rai, K.; Huggins, I.J.; James, S.R.; Karpf, A.R.; Jones, D.A.; Cairns, B.R. DNA Demethylation in Zebrafish Involves the Coupling of a Deaminase, a Glycosylase, and Gadd45. Cell 2008, 135, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.-G.; Guo, C.; Pfeifer, G.P. GADD45A Does Not Promote DNA Demethylation. PLOS Genet. 2008, 4, e1000013. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.; Yang, K.; Kuraguchi, M.; Werling, U.; Avdievich, E.; Fan, K.; Fazzari, M.; Jin, B.; Brown, A.M.C.; Lipkin, M.; et al. Mbd4 inactivation increases C→T transition mutations and promotes gastrointestinal tumor formation. Proc. Natl. Acad. Sci. USA 2002, 99, 14937–14942. [Google Scholar] [CrossRef] [PubMed]
- Hackett, J.A.; Sengupta, R.; Zylicz, J.J.; Murakami, K.; Lee, C.; Down, T.A.; Surani, M.A. Germline DNA Demethylation Dynamics and Imprint Erasure Through 5-Hydroxymethylcytosine. Science 2013, 339, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Hong, K.; Liu, R.; Inoue, A.; Shen, L.; Zhang, K.; Zhang, Y. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res. 2013, 23, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Wang, K.-Y.; Shen, C.-K.J. DNA 5-Methylcytosine Demethylation Activities of the Mammalian DNA Methyltransferases. J. Biol. Chem. 2013, 288, 9084–9091. [Google Scholar] [CrossRef]
- Waheed, S.O.; Ramanan, R.; Chaturvedi, S.S.; Lehnert, N.; Schofield, C.J.; Christov, C.Z.; Karabencheva-Christova, T.G. Role of Structural Dynamics in Selectivity and Mechanism of Non-heme Fe(II) and 2-Oxoglutarate-Dependent Oxygenases Involved in DNA Repair. ACS Central Sci. 2020, 6, 795–814. [Google Scholar] [CrossRef]
- Bian, K.; Lenz, S.A.P.; Tang, Q.; Chen, F.; Qi, R.; Jost, M.; Drennan, C.L.; Essigmann, J.M.; Wetmore, S.D.; Li, D. DNA repair enzymes ALKBH2, ALKBH3, and AlkB oxidize 5-methylcytosine to 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in vitro. Nucleic Acids Res. 2019, 47, 5522–5529. [Google Scholar] [CrossRef]
- Liutkevičiūtė, Z.; Kriukienė, E.; Ličytė, J.; Rudytė, M.; Urbanavičiūtė, G.; Klimašauskas, S. Direct Decarboxylation of 5-Carboxylcytosine by DNA C5- Methyltransferases. J. Am. Chem. Soc. 2014, 136, 5884–5887. [Google Scholar] [CrossRef]
- Tsai, W.C.; Strieter, R.M.; A Zisman, D.; Wilkowski, J.M.; A Bucknell, K.; Chen, G.H.; Standiford, T.J. Nitric oxide is required for effective innate immunity against Klebsiella pneumoniae. Infect. Immun. 1997, 65, 1870–1875. [Google Scholar] [CrossRef]
- Richardson, A.R.; Libby, S.J.; Fang, F.C. A Nitric Oxide–Inducible Lactate Dehydrogenase Enables Staphylococcus aureus to Resist Innate Immunity. Science 2008, 319, 1672–1676. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.R.; Payne, E.C.; Younger, N.; Karlinsey, J.E.; Thomas, V.C.; Becker, L.A.; Navarre, W.W.; Castor, M.E.; Libby, S.J.; Fang, F.C. Multiple Targets of Nitric Oxide in the Tricarboxylic Acid Cycle of Salmonella enterica Serovar Typhimurium. Cell Host Microbe 2011, 10, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Li, Z. Regulatory role of nitric oxide in lipopolysaccharides-triggered plant innate immunity. Plant Signal. Behav. 2013, 8, e22554. [Google Scholar] [CrossRef] [PubMed]
- Förstermann, U.; Li, H. Therapeutic effect of enhancing endothelial nitric oxide synthase (eNOS) expression and preventing eNOS uncoupling. Br. J. Pharmacol. 2011, 164, 213–223. [Google Scholar] [CrossRef]
- Kraan, C.M.; E Godler, D.; Amor, D.J. Epigenetics of fragile X syndrome and fragile X-related disorders. Dev. Med. Child Neurol. 2018, 61, 121–127. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992. [Google Scholar] [CrossRef]
- Weisenberger, D.J.; Liang, G.; Lenz, H.-J. DNA methylation aberrancies delineate clinically distinct subsets of colorectal cancer and provide novel targets for epigenetic therapies. Oncogene 2017, 37, 566–577. [Google Scholar] [CrossRef]
- Mann, J.; Oakley, F.; Akiboye, F.; Elsharkawy, A.; Thorne, A.W.; A Mann, D. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: Implications for wound healing and fibrogenesis. Cell Death Differ. 2006, 14, 275–285. [Google Scholar] [CrossRef]
- Mathison, M.; Sanagasetti, D.; Singh, V.P.; Pugazenthi, A.; Pinnamaneni, J.P.; Ryan, C.T.; Yang, J.; Rosengart, T.K. Fibroblast transition to an endothelial “trans” state improves cell reprogramming efficiency. Sci. Rep. 2021, 11, 22605. [Google Scholar] [CrossRef]
- Li, X. Epigenetics and cell cycle regulation in cystogenesis. Cell Signal. 2019, 68, 109509. [Google Scholar] [CrossRef]
- Dong, Y.; Zhao, H.; Li, H.; Li, X.; Yang, S. DNA methylation as an early diagnostic marker of cancer (Review). Biomed. Rep. 2014, 2, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhang, L.; Liu, Y.; Sun, H.; Onwuka, J.U.; Zhao, Z.; Tian, W.; Xu, J.; Zhao, Y.; Xu, H. Specific DNA methylation markers in the diagnosis and prognosis of esophageal cancer. Aging 2019, 11, 11640–11658. [Google Scholar] [CrossRef]
- Tserpeli, V.; Stergiopoulou, D.; Londra, D.; Giannopoulou, L.; Buderath, P.; Balgkouranidou, I.; Xenidis, N.; Grech, C.; Obermayr, E.; Zeillinger, R.; et al. Prognostic Significance of SLFN11 Methylation in Plasma Cell-Free DNA in Advanced High-Grade Serous Ovarian Cancer. Cancers 2021, 14, 4. [Google Scholar] [CrossRef] [PubMed]
- Pangeni, R.P.; Zhang, Z.; Alvarez, A.A.; Wan, X.; Sastry, N.; Lu, S.; Shi, T.; Huang, T.; Lei, C.X.; James, C.D.; et al. Genome-wide methylomic and transcriptomic analyses identify subtype-specific epigenetic signatures commonly dysregulated in glioma stem cells and glioblastoma. Epigenetics 2018, 13, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, M.; Song, C.; Xu, Q.; Huo, R.; Shen, L.; Xing, Q.; Cui, D.; Li, W.; Zhao, J.; et al. Combined study of genetic and epigenetic biomarker risperidone treatment efficacy in Chinese Han schizophrenia patients. Transl. Psychiatry 2017, 7, e1170. [Google Scholar] [CrossRef] [PubMed]
- Falahi, F.; Sgro, A.; Blancafort, P. Epigenome Engineering in Cancer: Fairytale or a Realistic Path to the Clinic? Front. Oncol. 2015, 5, 22. [Google Scholar] [CrossRef]
- Roubroeks, J.A.; Smith, A.R.; Smith, R.G.; Pishva, E.; Ibrahim, Z.; Sattlecker, M.; Hannon, E.J.; Kłoszewska, I.; Mecocci, P.; Soininen, H.; et al. An epigenome-wide association study of Alzheimer’s disease blood highlights robust DNA hypermethylation in the HOXB6 gene. Neurobiol. Aging 2020, 95, 26–45. [Google Scholar] [CrossRef] [PubMed]
- Semick, S.A.; Bharadwaj, R.A.; Collado-Torres, L.; Tao, R.; Shin, J.H.; Deep-Soboslay, A.; Weiss, J.R.; Weinberger, D.R.; Hyde, T.M.; Kleinman, J.E.; et al. Integrated DNA methylation and gene expression profiling across multiple brain regions implicate novel genes in Alzheimer’s disease. Acta Neuropathol. 2019, 137, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Vallerga, C.L.; Zhang, F.; Fowdar, J.; McRae, A.F.; Qi, T.; Nabais, M.F.; Zhang, Q.; Kassam, I.; Henders, A.K.; Wallace, L.; et al. Analysis of DNA methylation associates the cystine–glutamate antiporter SLC7A11 with risk of Parkinson’s disease. Nat. Commun. 2020, 11, 1238. [Google Scholar] [CrossRef]
- Liu, L.; Hu, Y.; Lu, Y.; Hu, L.; Gao, C.; Nie, S. Sex-dependent DNA hypermethylation of SLC6A4 in patients with schizophrenia. Neurosci. Lett. 2021, 769, 136394. [Google Scholar] [CrossRef]
- Thomas, M.; Knoblich, N.; Wallisch, A.; Glowacz, K.; Becker-Sadzio, J.; Gundel, F.; Brückmann, C.; Nieratschker, V. Increased BDNF methylation in saliva, but not blood, of patients with borderline personality disorder. Clin. Epigenetics 2018, 10, 109. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wang, H.; Liu, B.; Jiang, M.; Gu, Y.; Yan, S.; Han, X.; Hou, A.Y.; Tang, C.; Jiang, Z.; et al. Differential DNA Methylation Profiles in Patients with Temporal Lobe Epilepsy and Hippocampal Sclerosis ILAE Type I. J. Mol. Neurosci. 2021, 71, 1951–1966. [Google Scholar] [CrossRef] [PubMed]
- Dereix, A.E.; Ledyard, R.; Redhunt, A.M.; Bloomquist, T.R.; Brennan, K.J.; A Baccarelli, A.; Hacker, M.R.; Burris, H.H. Maternal anxiety and depression in pregnancy and DNA methylation of the NR3C1 glucocorticoid receptor gene. Epigenomics 2021, 13, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Starnawska, A.; Bukowski, L.; Chernomorchenko, A.; Elfving, B.; Müller, H.K.; Oord, E.V.D.; Aberg, K.; Guintivano, J.; Grove, J.; Mors, O.; et al. DNA methylation of the KLK8 gene in depression symptomatology. Clin. Epigenetics 2021, 13, 200. [Google Scholar] [CrossRef]
- Gao, B. Identification of Feature Autophagy-Related Genes and DNA Methylation Profiles in Systemic Lupus Erythematosus Patients. Experiment 2021, 27, e933425. [Google Scholar] [CrossRef]
- Miao, C.; Chang, J.; Dou, J.; Xiong, Y.; Zhou, G. DNA hypermethylation of SFRP2 influences the pathology of rheumatoid arthritis through the canonical Wnt signaling in model rats. Autoimmunity 2018, 51, 319–332. [Google Scholar] [CrossRef]
- Zhang, Y.; Pötter, S.; Chen, C.-W.; Liang, R.; Gelse, K.; Ludolph, I.; Horch, R.E.; Distler, O.; Schett, G.; Distler, J.H.W.; et al. Poly(ADP-ribose) polymerase-1 regulates fibroblast activation in systemic sclerosis. Ann. Rheum. Dis. 2018, 77, 744–751. [Google Scholar] [CrossRef]
- Sepúlveda, D.; Barrera, M.-J.; Castro, I.; Aguilera, S.; Carvajal, P.; Lagos, C.; González, S.; Albornoz, N.; Bahamondes, V.; Quest, A.F.G.; et al. Impaired IRE1α/XBP-1 pathway associated to DNA methylation might contribute to salivary gland dysfunction in Sjögren’s syndrome patients. Rheumatology 2018, 57, 1021–1032. [Google Scholar] [CrossRef]
- Cai, T.; Qin, Q.; Song, R.; Zhao, J.; Wang, G.; Zhang, J. Identifying and Validating Differentially Methylated Regions in Newly Diagnosed Patients with Graves’ Disease. DNA Cell Biol. 2021, 40, 482–490. [Google Scholar] [CrossRef]
- Ye, J.; Stefan-Lifshitz, M.; Tomer, Y. Genetic and environmental factors regulate the type 1 diabetes gene CTSH via differential DNA methylation. J. Biol. Chem. 2021, 296, 100774. [Google Scholar] [CrossRef]
- Wu, H.; Liu, H.; Liu, H.; Chen, Y.; Liu, T.; Shen, X.; Liu, L. Genome-wide DNA methylation profiling in differentiating Crohn’s disease from intestinal tuberculosis. Genes Genom. 2022, 44, 603–615. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, X.; Wei, A.; Chen, F.; Gao, Q.; Lu, K.; Jiang, Q.; Cao, W. Nrf2 epigenetic derepression induced by running exercise protects against osteoporosis. Bone Res. 2021, 9, 15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Peng, H.; Gao, W.; Cao, W.; Lv, J.; Yu, C.; Huang, T.; Sun, D.; Wang, B.; Liao, C.; et al. Blood DNA methylation markers associated with type 2 diabetes, fasting glucose, and HbA1c levels: An epigenome-wide association study in 316 adult twin pairs. Genomics 2021, 113, 4206–4213. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Yamada, H.; Munetsuna, E.; Yamazaki, M.; Kageyama, I.; Teshigawara, A.; Nouchi, Y.; Fujii, R.; Mizuno, G.; Sadamoto, N.; et al. Maternal high-fructose corn syrup consumption causes insulin resistance and hyperlipidemia in offspring via DNA methylation of the Pparα promoter region. J. Nutr. Biochem. 2022, 103, 108951. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Tarazón, E.; Gil-Cayuela, C.; Martínez-Dolz, L.; Lago, F.; González-Juanatey, J.R.; Sandoval, J.; Portolés, M.; Roselló-Lletí, E.; Rivera, M. ASB1 differential methylation in ischaemic cardiomyopathy: Relationship with left ventricular performance in end-stage heart failure patients. ESC Heart Fail. 2018, 5, 732–737. [Google Scholar] [CrossRef]
- Sun, Q.; Zhang, J.; Zhou, N.; Liu, X.; Shen, Y. DNA Methylation in Cosmc Promoter Region and Aberrantly Glycosylated IgA1 Associated with Pediatric IgA Nephropathy. PLoS ONE 2015, 10, e0112305. [Google Scholar] [CrossRef]
- Li, Y.; Ren, D.; Shen, Y.; Zheng, X.; Xu, G. Altered DNA methylation of TRIM13 in diabetic nephropathy suppresses mesangial collagen synthesis by promoting ubiquitination of CHOP. Ebiomedicine 2020, 51, 102582. [Google Scholar] [CrossRef]
- Wei, A.; Gao, Q.; Chen, F.; Zhu, X.; Chen, X.; Zhang, L.; Su, X.; Dai, J.; Shi, Y.; Cao, W. Inhibition of DNA methylation de-represses peroxisome proliferator-activated receptor-γ and attenuates pulmonary fibrosis. Br. J. Pharmacol. 2022, 179, 1304–1318. [Google Scholar] [CrossRef]
- Vucic, E.A.; Chari, R.; Thu, K.L.; Wilson, I.M.; Cotton, A.M.; Kennett, J.Y.; Zhang, M.; Lonergan, K.M.; Steiling, K.; Brown, C.; et al. DNA Methylation Is Globally Disrupted and Associated with Expression Changes in Chronic Obstructive Pulmonary Disease Small Airways. Am. J. Respir. Cell Mol. Biol. 2014, 50, 912–922. [Google Scholar] [CrossRef]
- Devi, P.; Ota, S.; Punga, T.; Bergqvist, A. Hepatitis C Virus Core Protein Down-Regulates Expression of Src-Homology 2 Domain Containing Protein Tyrosine Phosphatase by Modulating Promoter DNA Methylation. Viruses 2021, 13, 2514. [Google Scholar] [CrossRef]
- Chen, H.; Kazemier, H.G.; De Groote, M.L.; Ruiters, M.H.J.; Xu, G.-L.; Rots, M.G. Induced DNA demethylation by targeting Ten-Eleven Translocation 2 to the human ICAM-1 promoter. Nucleic Acids Res. 2013, 42, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Angstman, J.F.; Richardson, M.E.; Linder, S.J.; Cascio, V.M.; Tsai, S.Q.; Ho, Q.H.; Sander, J.D.; Reyon, D.; Bernstein, B.E.; et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013, 31, 1137–1142. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Noguchi, H.; Horii, T.; Nakabayashi, K.; Kimura, M.; Okamura, K.; Sakai, A.; Nakashima, H.; Hata, K.N.K.; Nakashima, K.; et al. Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv–TET1 catalytic domain fusions. Nat. Biotechnol. 2016, 34, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Horii, T.; Hatada, I. Editing of DNA Methylation Using dCas9-Peptide Repeat and scFv-TET1 Catalytic Domain Fusions. Epigenome Ed. Methods Protoc. 2018, 1767, 419–428. [Google Scholar] [CrossRef]
- Morita, S.; Horii, T.; Kimura, M.; Hatada, I. Synergistic Upregulation of Target Genes by TET1 and VP64 in the dCas9–SunTag Platform. Int. J. Mol. Sci. 2020, 21, 1574. [Google Scholar] [CrossRef]
- Taghbalout, A.; Du, M.; Jillette, N.; Rosikiewicz, W.; Rath, A.; Heinen, C.D.; Li, S.; Cheng, A.W. Enhanced CRISPR-based DNA demethylation by Casilio-ME-mediated RNA-guided coupling of methylcytosine oxidation and DNA repair pathways. Nat. Commun. 2019, 10, 4296. [Google Scholar] [CrossRef]
- Chan, W.F.; Coughlan, H.D.; Chen, Y.; Keenan, C.R.; Smyth, G.K.; Perkins, A.C.; Johanson, T.M.; Allan, R.S. Activation of stably silenced genes by recruitment of a synthetic de-methylating module. Nat. Commun. 2022, 13, 5582. [Google Scholar] [CrossRef]
- Wilk, C.; Effenberg, L.; Abberger, H.; Steenpass, L.; Hansen, W.; Zeschnigk, M.; Kirschning, C.; Buer, J.; Kehrmann, J. CRISPR/Cas9-mediated demethylation of FOXP3-TSDR toward Treg-characteristic programming of Jurkat T cells. Cell Immunol. 2021, 371, 104471. [Google Scholar] [CrossRef]
- Okada, M.; Kanamori, M.; Someya, K.; Nakatsukasa, H.; Yoshimura, A. Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin 2017, 10, 24. [Google Scholar] [CrossRef]
- Xu, C.; Jiang, S.; Ma, X.; Jiang, Z.; Pan, Y.; Li, X.; Zhang, L.; Zhou, H.; Chen, S.; Xing, X.; et al. CRISPR-based DNA methylation editing of NNT rescues the cisplatin resistance of lung cancer cells by reducing autophagy. Arch. Toxicol. 2022, 97, 441–456. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, H.; Yang, J.; Zhao, J.; Liang, Z.; Kang, X.; Wang, R. The identification and validation of EphA7 hypermethylation, a novel biomarker, in cervical cancer. BMC Cancer 2022, 22, 636. [Google Scholar] [CrossRef]
- Lindström, A.K.; Ekman, K.; Stendahl, U.; Tot, T.; Henriksson, R.; Hedman, H.; Hellberg, D. LRIG1 and squamous epithelial uterine cervical cancer: Correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int. J. Gynecol. Cancer 2008, 18, 312–317. [Google Scholar] [CrossRef] [PubMed]
- Umeh-Garcia, M.; O’geen, H.; Simion, C.; Gephart, M.H.; Segal, D.J.; Sweeney, C.A. Aberrant promoter methylation contributes to LRIG1 silencing in basal/triple-negative breast cancer. Br. J. Cancer 2022, 127, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zhang, C.; Xu, Z.; Wang, S.; Li, X.; Stringer-Reasor, E.; Bae, S.; Zeng, L.; Zhao, D.; Liu, R.; et al. Dual CRISPR interference and activation for targeted reactivation of X-linked endogenous FOXP3 in human breast cancer cells. Mol. Cancer 2022, 21, 38. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, S. TRIM Family Proteins: Roles in Autophagy, Immunity, and Carcinogenesis. Trends Biochem. Sci. 2017, 42, 297–311. [Google Scholar] [CrossRef]
- Gan, Y.; Cao, C.; Li, A.; Song, H.; Kuang, G.; Ma, B.; Zhang, Q. Silencing of the TRIM58 Gene by Aberrant Promoter Methylation is Associated with a Poor Patient Outcome and Promotes Cell Proliferation and Migration in Clear Cell Renal Cell Carcinoma. Front. Mol. Biosci. 2021, 8, 655126. [Google Scholar] [CrossRef]
- Iżykowska, K.; Rassek, K.; Żurawek, M.; Nowicka, K.; Paczkowska, J.; Ziółkowska-Suchanek, I.; Podralska, M.; Dzikiewicz-Krawczyk, A.; Joks, M.; Olek-Hrab, K.; et al. Hypomethylation of the promoter region drives ectopic expression of TMEM244 in Sézary cells. J. Cell. Mol. Med. 2020, 24, 10970–10977. [Google Scholar] [CrossRef]
- Josipović, G.; Tadić, V.; Klasić, M.; Zanki, V.; Bečeheli, I.; Chung, F.; Ghantous, A.; Keser, T.; Madunić, J.; Bošković, M.; et al. Antagonistic and synergistic epigenetic modulation using orthologous CRISPR/dCas9-based modular system. Nucleic Acids Res. 2019, 47, 9637–9657. [Google Scholar] [CrossRef]
- Le Berre, G.; Hossard, V.; Riou, J.-F.; Guieysse-Peugeot, A.-L. Repression of TERRA Expression by Subtelomeric DNA Methylation Is Dependent on NRF1 Binding. Int. J. Mol. Sci. 2019, 20, 2791. [Google Scholar] [CrossRef]
- Fang, S.; Cui, D.; Hong, T.; Guo, L.; Lee, Y.-T.; Lee, M.; Isgandarova, S.; Martinez-Moczygemba, M.; Zhou, Y.; Li, J.; et al. Ten-Eleven Translocation Ablation Impairs Cardiac Differentiation of Mouse Embryonic Stem Cells. Stem Cells 2022, 40, 260–272. [Google Scholar] [CrossRef]
- Halmai, J.A.N.M.; Deng, P.; E Gonzalez, C.; Coggins, N.B.; Cameron, D.; Carter, J.L.; Buchanan, F.K.B.; Waldo, J.J.; Lock, S.R.; Anderson, J.D.; et al. Artificial escape from XCI by DNA methylation editing of the CDKL5 gene. Nucleic Acids Res. 2020, 48, 2372–2387. [Google Scholar] [CrossRef]
- Kyle, S.M.; Vashi, N.; Justice, M.J. Rett syndrome: A neurological disorder with metabolic components. Open Biol. 2018, 8, 170216. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Guan, X.; Xie, B.; Xu, C.; Niu, J.; Tang, X.; Li, C.H.; Colecraft, H.M.; Jaenisch, R.; Liu, X.S. Multiplex epigenome editing of MECP2 to rescue Rett syndrome neurons. Sci. Transl. Med. 2023, 15, eadd4666. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Park, J.S.; Ko, J.-H.; Kim, Y.-S. Regulation of gene expression by altered promoter methylation using a CRISPR/Cas9-mediated epigenetic editing system. Sci. Rep. 2019, 9, 11960. [Google Scholar] [CrossRef] [PubMed]
- Marx, N.; Grünwald-Gruber, C.; Bydlinski, N.; Dhiman, H.; Nguyen, L.N.; Klanert, G.; Borth, N. CRISPR-Based Targeted Epigenetic Editing Enables Gene Expression Modulation of the Silenced Beta-Galactoside Alpha-2,6-Sialyltransferase 1 in CHO Cells. Biotechnol. J. 2018, 13, e1700217. [Google Scholar] [CrossRef] [PubMed]
- Hanzawa, N.; Hashimoto, K.; Yuan, X.; Kawahori, K.; Tsujimoto, K.; Hamaguchi, M.; Tanaka, T.; Nagaoka, Y.; Nishina, H.; Morita, S.; et al. Targeted DNA demethylation of the Fgf21 promoter by CRISPR/dCas9-mediated epigenome editing. Sci. Rep. 2020, 10, 5181. [Google Scholar] [CrossRef]
- Horii, T.; Morita, S.; Hino, S.; Kimura, M.; Hino, Y.; Kogo, H.; Nakao, M.; Hatada, I. Successful generation of epigenetic disease model mice by targeted demethylation of the epigenome. Genome Biol. 2020, 21, 77. [Google Scholar] [CrossRef]
- Noack, F.; Pataskar, A.; Schneider, M.; Buchholz, F.; Tiwari, V.K.; Calegari, F. Assessment and site-specific manipulation of DNA (hydroxy-)methylation during mouse corticogenesis. Life Sci. Alliance 2019, 2, e201900331. [Google Scholar] [CrossRef]
- Liu, R.; Lang, Z. The mechanism and function of active DNA demethylation in plants. J. Integr. Plant Biol. 2020, 62, 148–159, Erratum in: J. Integr. Plant Biol. 2020, 62, 1638–1639. [Google Scholar] [CrossRef]
- Parrilla-Doblas, J.T.; Ariza, R.R.; Roldán-Arjona, T. Targeted DNA demethylation in human cells by fusion of a plant 5-methylcytosine DNA glycosylase to a sequence-specific DNA binding domain. Epigenetics 2017, 12, 296–303. [Google Scholar] [CrossRef]
- Hilton, I.B.; D’ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.-S.; Kang, J.G.; Lee, J.-H.; Lee, J.-J.; Jeon, S.K.; Ko, J.-H.; Kim, D.-S.; Park, K.-H.; Kim, Y.-S.; Kim, N.-S. Direct regulation of E-cadherin by targeted histone methylation of TALE-SET fusion protein in cancer cells. Oncotarget 2015, 6, 23837–23844. [Google Scholar] [CrossRef] [PubMed]
- Honarmand, A.; Mayall, R.; George, I.; Oberding, L.; Dastidar, H.; Fegan, J.; Chaudhuri, S.; Dole, J.; Feng, S.; Hoang, D.; et al. A Multiplexed Transcription Activator-like Effector System for Detecting Specific DNA Sequences. ACS Synth. Biol. 2014, 3, 953–955. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef]
- Garcia-Bloj, B.; Moses, C.; Sgro, A.; Plani-Lam, J.; Arooj, M.; Duffy, C.; Thiruvengadam, S.; Sorolla, A.; Rashwan, R.; Mancera, R.L.; et al. Waking up dormant tumor suppressor genes with zinc fingers, TALEs and the CRISPR/dCas9 system. Oncotarget 2016, 7, 60535–60554. [Google Scholar] [CrossRef]
- Chou, C.-C.; Lou, Y.-C.; Tang, T.K.; Chen, C. Structure and DNA binding characteristics of the three-Cys2His2 domain of mouse testis zinc finger protein. Proteins Struct. Funct. Bioinform. 2010, 78, 2202–2212. [Google Scholar] [CrossRef]
- Huber, P.W.; Morii, T.; Mei, H.Y.; Barton, J.K. Structural polymorphism in the major groove of a 5S RNA gene complements the zinc finger domains of transcription factor IIIA. Proc. Natl. Acad. Sci. USA 1991, 88, 10801–10805. [Google Scholar] [CrossRef]
- Mandell, J.G.; Barbas, C.F. Zinc Finger Tools: Custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006, 34, W516–W523. [Google Scholar] [CrossRef]
- Urnov, F.D.; Miller, J.C.; Lee, Y.-L.; Beausejour, C.M.; Rock, J.M.; Augustus, S.; Jamieson, A.C.; Porteus, M.H.; Gregory, P.D.; Holmes, M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 2005, 435, 646–651. [Google Scholar] [CrossRef]
- Lim, W.F.; Burdach, J.; Funnell, A.P.; Pearson, R.C.; Quinlan, K.G.; Crossley, M. Directing an artificial zinc finger protein to new targets by fusion to a non-DNA-binding domain. Nucleic Acids Res. 2015, 44, 3118–3130. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, B.; Kuang, H.; Korakavi, G.; Lu, L.-Y.; Yu, X. Zinc Finger Protein 618 Regulates the Function of UHRF2 (Ubiquitin-like with PHD and Ring Finger Domains 2) as a Specific 5-Hydroxymethylcytosine Reader. J. Biol. Chem. 2016, 291, 13679–13688. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Maeder, M.L.; Reyon, D.; Voytas, D.F.; Joung, J.K.; Dobbs, D. ZiFiT (Zinc Finger Targeter): An updated zinc finger engineering tool. Nucleic Acids Res. 2010, 38, W462–W468. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Thibodeau-Beganny, S.; Sander, J.D.; Voytas, D.F.; Joung, J.K. Oligomerized pool engineering (OPEN): An ‘open-source’ protocol for making customized zinc-finger arrays. Nat. Protoc. 2009, 4, 1471–1501. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Dahlborg, E.J.; Goodwin, M.J.; Cade, L.; Zhang, F.; Cifuentes, D.; Curtin, S.J.; Blackburn, J.; Thibodeau-Beganny, S.; Qi, Y.; et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat. Methods 2010, 8, 67–69. [Google Scholar] [CrossRef]
- Yee, J.-K. Off-target effects of engineered nucleases. FEBS J. 2016, 283, 3239–3248. [Google Scholar] [CrossRef]
- Rogers, J.M.; Barrera, L.A.; Reyon, D.; Sander, J.D.; Kellis, M.; Joung, J.K.; Bulyk, M.L. Context influences on TALE–DNA binding revealed by quantitative profiling. Nat. Commun. 2015, 6, 7440. [Google Scholar] [CrossRef]
- Nitsch, S.; Mussolino, C. Generation of TALE-Based Designer Epigenome Modifiers. Epigenome Ed. Methods Protoc. 2018, 1767, 89–109. [Google Scholar] [CrossRef]
- Waryah, C.B.; Moses, C.; Arooj, M.; Blancafort, P. Zinc Fingers, TALEs, and CRISPR Systems: A Comparison of Tools for Epigenome Editing. Epigenome Ed. Methods Protoc. 2018, 1767, 19–63. [Google Scholar] [CrossRef]
- Sung, C.K.; Yim, H. CRISPR-mediated promoter de/methylation technologies for gene regulation. Arch. Pharmacal Res. 2020, 43, 705–713. [Google Scholar] [CrossRef]
- Nguyen, T.V.; Lister, R. Genomic targeting of TET activity for targeted demethylation using CRISPR/Cas9. Methods Mol. Biol. 2021, 2272, 181–194. [Google Scholar] [CrossRef]
- Sapozhnikov, D.M.; Szyf, M. Unraveling the functional role of DNA demethylation at specific promoters by targeted steric blockage of DNA methyltransferase with CRISPR/dCas9. Nat. Commun. 2021, 12, 5711. [Google Scholar] [CrossRef]
- Xu, X.; Tao, Y.; Gao, X.; Zhang, L.; Li, X.; Zou, W.; Ruan, K.; Wang, F.; Xu, G.-L.; Hu, R. A CRISPR-based approach for targeted DNA demethylation. Cell Discov. 2016, 2, 16009. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Jillette, N.; Lee, P.; Plaskon, D.; Fujiwara, Y.; Wang, W.; Taghbalout, A.; Wang, H. Casilio: A versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labeling. Cell Res. 2016, 26, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gaj, T.; Barbas, C.F., 3rd. Directed Evolution of an Enhanced and Highly Efficient FokI Cleavage Domain for Zinc Finger Nucleases. J. Mol. Biol. 2010, 400, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Z.; Huang, J.; Luo, H.; Zhu, S.; Yi, H.; Zheng, L.; Hu, B.; Yu, L.; Li, L.; et al. Specific zinc finger-induced methylation of PD-L1 promoter inhibits its expression. FEBS Open Bio 2018, 9, 1063–1070. [Google Scholar] [CrossRef]
- Bennett, C. Are Zinc Finger Nucleases Making a Comeback? 30 November 2017 Genetic Engineering & Biotechnology News. Available online: https://www.genengnews.com/insights/are-zinc-finger-nucleases-making-a-comeback/ (accessed on 8 February 2023).
- Lanio, T.; Jeltsch, A.; Pingoud, A. Towards the design of rare cutting restriction endonucleases: Using directed evolution to generate variants of EcoRV differing in their substrate specificity by two orders of magnitude. J. Mol. Biol. 1998, 283, 59–69. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, X.; Su, J.; Jeong, M.; Gundry, M.C.; Huang, Y.-H.; Zhou, Y.; Li, W.; Goodell, M.A. Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat. Commun. 2017, 8, 16026. [Google Scholar] [CrossRef]
- Li, R.; Meng, Q.; Qi, J.; Hu, L.; Huang, J.; Zhang, Y.; Yang, J.; Sun, J. Microinjection-based CRISPR/Cas9 mutagenesis in the decapoda crustaceans Neocaridina heteropoda and Eriocheir sinensis. J. Exp. Biol. 2022, 225, jeb243702. [Google Scholar] [CrossRef]
- Laustsen, A.; Bak, R.O. Electroporation-Based CRISPR/Cas9 Gene Editing Using Cas9 Protein and Chemically Modified sgRNAs. CRISPR Gene Ed. Methods Protoc. 2019, 1961, 127–134. [Google Scholar] [CrossRef]
- Danthinne, X.; Imperiale, M.J. Production of first generation adenovirus vectors: A review. Gene Ther. 2000, 7, 1707–1714. [Google Scholar] [CrossRef]
- Rice, S.A.; Klessig, D.F.; Williams, J. Multiple effects of the 72-kDa, adenovirus-specified DNA binding protein on the efficiency of cellular transformation. Virology 1987, 156, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Schneider, R.J. Adenovirus inhibition of cell translation facilitates release of virus particles and enhances degradation of the cytokeratin network. J. Virol. 1994, 68, 2544–2555. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ertl, H.C.; Wilson, J.M. MHC class I-cestricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity 1994, 1, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Jerome, S. Adenovirus vectors deleted for genes essential for viral DNA replication. Front. Biosci. 2005, 10, 1146–1155. [Google Scholar] [CrossRef]
- Cannon, P.; June, C.H. Chemokine receptor 5 knockout strategies. Curr. Opin. HIV AIDS 2011, 6, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Holkers, M.; de Vries, A.A.F.; Gonçalves, M.A.F.V. Nonspaced inverted DNA repeats are preferential targets for homology-directed gene repair in mammalian cells. Nucleic Acids Res. 2012, 40, 1984–1999. [Google Scholar] [CrossRef]
- Hu, Z.; Ding, W.; Zhu, D.; Yu, L.; Jiang, X.; Wang, X.; Zhang, C.; Wang, L.; Ji, T.; Liu, D.; et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPV-related cervical malignancy. J. Clin. Investig. 2014, 125, 425–436. [Google Scholar] [CrossRef]
- Krasnykh, V.; Dmitriev, I.; Navarro, J.G.; Belousova, N.; Kashentseva, E.; Xiang, J.; Douglas, J.T.; Curiel, D.T. Advanced generation adenoviral vectors possess augmented gene transfer efficiency based upon coxsackie adenovirus receptor-independent cellular entry capacity. Cancer Res. 2000, 60, 6784–6787. [Google Scholar]
- Hensen, L.C.; Hoeben, R.C.; Bots, S.T. Adenovirus Receptor Expression in Cancer and Its Multifaceted Role in Oncolytic Adenovirus Therapy. Int. J. Mol. Sci. 2020, 21, 6828. [Google Scholar] [CrossRef]
- Lowenstein, P.R.; Mandel, R.J.; Xiong, W.-D.; Kroeger, K.; Castro, M.G. Immune responses to adenovirus and adeno-associated vectors used for gene therapy of brain diseases: The role of immunological synapses in understanding the cell biology of neuroimmune interactions. Curr. Gene Ther. 2007, 7, 347–360. [Google Scholar] [CrossRef]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral vectors: A look back and ahead on gene transfer technology. New Microbiol. 2013, 36, 1–22. [Google Scholar] [PubMed]
- Anguela, X.M.; Sharma, R.; Doyon, Y.; Miller, J.C.; Li, H.; Haurigot, V.A.; Rohde, M.E.; Wong, S.Y.; Davidson, R.J.; Zhou, S.; et al. Robust ZFN-mediated genome editing in adult hemophilic mice. Blood 2013, 122, 3283–3287. [Google Scholar] [CrossRef]
- Händel, E.-M.; Gellhaus, K.; Khan, K.; Bednarski, C.; Cornu, T.I.; Müller-Lerch, F.; Kotin, R.M.; Heilbronn, R.; Cathomen, T. Versatile and Efficient Genome Editing in Human Cells by Combining Zinc-Finger Nucleases With Adeno-Associated Viral Vectors. Hum. Gene Ther. 2012, 23, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Rahman, S.H.; Bobis-Wozowicz, S.; Chatterjee, D.; Gellhaus, K.; Pars, K.; Heilbronn, R.; Jacobs, R.; Cathomen, T. The Nontoxic Cell Cycle Modulator Indirubin Augments Transduction of Adeno-Associated Viral Vectors and Zinc-Finger Nuclease-Mediated Gene Targeting. Hum. Gene Ther. 2013, 24, 67–77. [Google Scholar] [CrossRef]
- Swiech, L.; Heidenreich, M.; Banerjee, A.; Habib, N.; Li, Y.; Trombetta, J.J.; Sur, M.; Zhang, F. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol. 2014, 33, 102–106. [Google Scholar] [CrossRef] [PubMed]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Zabaleta, N.; Dai, W.; Bhatt, U.; Hérate, C.; Maisonnasse, P.; Chichester, J.A.; Sanmiguel, J.; Estelien, R.; Michalson, K.T.; Diop, C.; et al. An AAV-based, room-temperature-stable, single-dose COVID-19 vaccine provides durable immunogenicity and protection in non-human primates. Cell Host Microbe 2021, 29, 1437–1453. [Google Scholar] [CrossRef]
- Zhao, S.; Ke, J.; Yang, B.; Tan, F.; Yang, J.; Lin, C.-P.; Wang, H.; Zhong, G. A protective AAV vaccine for SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 310. [Google Scholar] [CrossRef]
- Narayan, O.; Zink, M.; Huso, D.; Sheffer, D.; Crane, S.; Kennedy-Stoskopf, S.; Jolly, P.; Clements, J. Lentiviruses of animals are biological models of the human immunodeficiency viruses. Microb. Pathog. 1988, 5, 149–157. [Google Scholar] [CrossRef]
- Sakuma, T.; Barry, M.A.; Ikeda, Y. Lentiviral vectors: Basic to translational. Biochem. J. 2012, 443, 603–618. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.E.; Ehrhardt, A.; Kay, M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003, 4, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Mátrai, J.; Chuah, M.K.; Vandendriessche, T. Recent Advances in Lentiviral Vector Development and Applications. Mol. Ther. 2010, 18, 477–490. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, A.L.; Genovese, P.; Beausejour, C.M.; Colleoni, S.; Lee, Y.-L.; A Kim, K.; Ando, D.; Urnov, F.D.; Galli, C.; Gregory, P.; et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007, 25, 1298–1306. [Google Scholar] [CrossRef] [PubMed]
- Apolonia, L.; Waddington, S.; Fernandes, C.; Ward, N.J.; Bouma, G.; Blundell, M.; Thrasher, A.J.; Collins, M.K.; Philpott, N.J. Stable Gene Transfer to Muscle Using Non-integrating Lentiviral Vectors. Mol. Ther. 2007, 15, 1947–1954. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Herzog, R.W. Progress and prospects: Immune responses to viral vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-S.; Liu, D.-P.; Liang, C.-C. Challenges and strategies: The immune responses in gene therapy. Med. Res. Rev. 2004, 24, 748–761. [Google Scholar] [CrossRef] [PubMed]
- Nair, V. Retrovirus-induced oncogenesis and safety of retroviral vectors. Curr. Opin. Mol. Ther. 2008, 10, 431–438. [Google Scholar]
- Baum, C.; Kustikova, O.; Modlich, U.; Li, Z.; Fehse, B. Mutagenesis and Oncogenesis by Chromosomal Insertion of Gene Transfer Vectors. Hum. Gene Ther. 2006, 17, 253–263. [Google Scholar] [CrossRef]
- Themis, M.; Waddington, S.N.; Schmidt, M.; von Kalle, C.; Wang, Y.; Al-Allaf, F.; Gregory, L.G.; Nivsarkar, M.; Themis, M.; Holder, M.V.; et al. Oncogenesis Following Delivery of a Nonprimate Lentiviral Gene Therapy Vector to Fetal and Neonatal Mice. Mol. Ther. 2005, 12, 763–771. [Google Scholar] [CrossRef]
- Baum, C.; Düllmann, J.; Li, Z.; Fehse, B.; Meyer, J.; Williams, D.A.; Von Kalle, C. Side effects of retroviral gene transfer into hematopoietic stem cells. Blood 2003, 101, 2099–2113. [Google Scholar] [CrossRef] [PubMed]
- Modlich, U.; Kustikova, O.S.; Schmidt, M.; Rudolph, C.; Meyer, J.; Li, Z.; Kamino, K.; Von Neuhoff, N.; Schlegelberger, B.; Kuehlcke, K.; et al. Leukemias following retroviral transfer of multidrug resistance 1 (MDR1) are driven by combinatorial insertional mutagenesis. Blood 2005, 105, 4235–4246. [Google Scholar] [CrossRef] [PubMed]
- Nowrouzi, A.; Glimm, H.; Von Kalle, C.; Schmidt, M. Retroviral Vectors: Post Entry Events and Genomic Alterations. Viruses 2011, 3, 429–455. [Google Scholar] [CrossRef] [PubMed]
- Baum, C.; von Kalle, C.; Staal, F.J.; Li, Z.; Fehse, B.; Schmidt, M.; Weerkamp, F.; Karlsson, S.; Wagemaker, G.; A Williams, D. Chance or necessity? Insertional Mutagenesis in Gene Therapy and Its Consequences. Mol. Ther. 2004, 9, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, L.B.; Calvin, S.A.; Lobenhofer, E.K. Transcriptional effects of transfection: The potential for misinterpretation of gene expression data generated from transiently transfected cells. Biotechniques 2009, 47, 617–624. [Google Scholar] [CrossRef]
- Capretto, L.; Carugo, D.; Mazzitelli, S.; Nastruzzi, C.; Zhang, X. Microfluidic and lab-on-a-chip preparation routes for organic nanoparticles and vesicular systems for nanomedicine applications. Adv. Drug Deliv. Rev. 2013, 65, 1496–1532. [Google Scholar] [CrossRef]
- Rawal, S.; Patel, M.M. Threatening cancer with nanoparticle aided combination oncotherapy. J. Control. Release 2019, 301, 76–109. [Google Scholar] [CrossRef]
- Shoari, A.; Tooyserkani, R.; Tahmasebi, M.; Löwik, D.W.P.M. Delivery of Various Cargos into Cancer Cells and Tissues via Cell-Penetrating Peptides: A Review of the Last Decade. Pharmaceutics 2021, 13, 1391. [Google Scholar] [CrossRef]
- de Ilarduya, C.T.; Sun, Y.; Düzgüneş, N. Gene delivery by lipoplexes and polyplexes. Eur. J. Pharm. Sci. 2010, 40, 159–170. [Google Scholar] [CrossRef]
- Zu, H.; Gao, D. Non-viral Vectors in Gene Therapy: Recent Development, Challenges, and Prospects. AAPS J. 2021, 23, 78. [Google Scholar] [CrossRef]
- Obika, S.; Yu, W.; Shimoyama, A.; Uneda, T.; Minami, T.; Miyashita, K.; Doi, T.; Imanishi, T. Properties of Cationic Liposomes Composed of Cationic Lipid YKS-220 Having an Ester Linkage: Adequate Stability, High Transfection Efficiency, and Low Cytotoxicity. Biol. Pharm. Bull. 1999, 22, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Almofti, M.R.; Harashima, H.; Shinohara, Y.; Almofti, A.; Baba, Y.; Kiwada, H. Cationic liposome-mediated gene delivery: Biophysical study and mechanism of internalization. Arch. Biochem. Biophys. 2003, 410, 246–253. [Google Scholar] [CrossRef]
- Conde, J.; Bao, C.; Tan, Y.; Cui, D.; Edelman, E.R.; Azevedo, H.S.; Byrne, H.J.; Artzi, N.; Tian, F. Dual Targeted Immunotherapy via In Vivo Delivery of Biohybrid RNAi-Peptide Nanoparticles to Tumor-Associated Macrophages and Cancer Cells. Adv. Funct. Mater. 2015, 25, 4183–4194. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Xu, L.; Dai, S.; Wang, M.; Wang, H. A facile supramolecular approach to fabricate multifunctional upconversion nanoparticles as a versatile platform for drug loading, in vivo delivery and tumor imaging. J. Mater. Chem. B 2017, 5, 2425–2435. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Sun, R.; Bao, Y.; Zhang, C.; Wu, Y.; Gong, Y. In Vivo Delivery of siRNAs Targeting EGFR and BRD4 Expression by Peptide-Modified Redox Responsive PEG–PEI Nanoparticles for the Treatment of Triple-Negative Breast Cancer. Mol. Pharm. 2021, 18, 3990–3998. [Google Scholar] [CrossRef] [PubMed]
- Han, J.P.; Kim, M.; Choi, B.S.; Lee, J.H.; Lee, G.S.; Jeong, M.; Lee, Y.; Kim, E.-A.; Oh, H.-K.; Go, N.; et al. In vivo delivery of CRISPR-Cas9 using lipid nanoparticles enables antithrombin gene editing for sustainable hemophilia A and B therapy. Sci. Adv. 2022, 8, eabj6901. [Google Scholar] [CrossRef]
- Lokugamage, M.P.; Vanover, D.; Beyersdorf, J.; Hatit, M.Z.C.; Rotolo, L.; Echeverri, E.S.; Peck, H.E.; Ni, H.; Yoon, J.-K.; Kim, Y.; et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 2021, 5, 1059–1068. [Google Scholar] [CrossRef]
- Kim, J.; Jozic, A.; Lin, Y.; Eygeris, Y.; Bloom, E.; Tan, X.; Acosta, C.; MacDonald, K.D.; Welsher, K.D.; Sahay, G. Engineering Lipid Nanoparticles for Enhanced Intracellular Delivery of mRNA through Inhalation. ACS Nano 2022, 16, 14792–14806. [Google Scholar] [CrossRef]
- Rotolo, L.; Vanover, D.; Bruno, N.C.; Peck, H.E.; Zurla, C.; Murray, J.; Noel, R.K.; O’farrell, L.; Araínga, M.; Orr-Burks, N.; et al. Species-agnostic polymeric formulations for inhalable messenger RNA delivery to the lung. Nat. Mater. 2022, 22, 369–379. [Google Scholar] [CrossRef]
- Pei, Y.; Bao, Y.; Sacchetti, C.; Brady, J.; Gillard, K.; Yu, H.; Roberts, S.; Rajappan, K.; Tanis, S.P.; Perez-Garcia, C.G.; et al. Synthesis and bioactivity of readily hydrolysable novel cationic lipids for potential lung delivery application of mRNAs. Chem. Phys. Lipids 2022, 243, 105178. [Google Scholar] [CrossRef]
- Tam, A.; Kulkarni, J.; An, K.; Li, L.; Dorscheid; Singhera, G.; Bernatchez, P.; Reid, G.; Chan, K.; Witzigmann, D.; et al. Lipid nanoparticle formulations for optimal RNA-based topical delivery to murine airways. Eur. J. Pharm. Sci. 2022, 176, 106234. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Jeong, M.; Lee, G.; Kim, M.; Yoo, Y.; Kim, H.J.; Cho, J.; Lee, Y.; Lee, H. Engineered Lipid Nanoparticles for the Treatment of Pulmonary Fibrosis by Regulating Epithelial-Mesenchymal Transition in the Lungs. Adv. Funct. Mater. 2022, 33, 2209432. [Google Scholar] [CrossRef]
- Wadhwa, A.; Aljabbari, A.; Lokras, A.; Foged, C.; Thakur, A. Opportunities and Challenges in the Delivery of mRNA-Based Vaccines. Pharmaceutics 2020, 12, 102. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Guo, J.; Kato, Y.; Sirk, S.J.; Barbas, C.F. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat. Methods 2012, 9, 805–807. [Google Scholar] [CrossRef] [PubMed]
- Rádis-Baptista, G.; Campelo, I.S.; Morlighem, J.R.; Melo, L.M.; Freitas, V.J. Cell-penetrating peptides (CPPs): From delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J. Biotechnol. 2017, 252, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Gagat, M.; Zielińska, W.; Grzanka, A. Cell-penetrating peptides and their utility in genome function modifications (Review). Int. J. Mol. Med. 2017, 40, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, J.; Kadungure, T.; Beyene, J.; Zhang, H.; Lu, Q. ARMMs as a versatile platform for intracellular delivery of macromolecules. Nat. Commun. 2018, 9, 960. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat. Commun. 2017, 8, 709. [Google Scholar] [CrossRef]
- Ramakrishna, S.; Kim, Y.-H.; Kim, H. Stability of Zinc Finger Nuclease Protein Is Enhanced by the Proteasome Inhibitor MG132. PLoS ONE 2013, 8, e54282. [Google Scholar] [CrossRef]
- Lo, C.-L.; Choudhury, S.R.; Irudayaraj, J.; Zhou, F.C. Epigenetic Editing of Ascl1 Gene in Neural Stem Cells by Optogenetics. Sci. Rep. 2017, 7, srep42047. [Google Scholar] [CrossRef]
- Huisman, C.; van der Wijst, M.; Schokker, M.; Blancafort, P.; Terpstra, M.M.; Kok, K.; van der Zee, A.G.J.; Schuuring, E.; A Wisman, G.B.; Rots, M.G. Re-expression of Selected Epigenetically Silenced Candidate Tumor Suppressor Genes in Cervical Cancer by TET2-directed Demethylation. Mol. Ther. 2016, 24, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Gallego-Bartolome, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.-C.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Cui, Y.; Lubecka, K.; Stefanska, B.; Irudayaraj, J. CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 2016, 7, 46545–46556. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; He, L.; Chen, B.; Wang, Y.; Wang, L.; Zhou, W.; Zhang, T.; Cao, L.; Zhang, P.; Xie, L.; et al. Transgenerationally Transmitted DNA Demethylation of a Spontaneous Epialleles Using CRISPR/dCas9-TET1cd Targeted Epigenetic Editing in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 10492. [Google Scholar] [CrossRef]
| Category | Disease | Gene | Year | References |
|---|---|---|---|---|
| nervous | Alzheimer’s disease | HOXB6 a, ANKRD30B b, etc. | 2019, 2020 | [117,118] |
| Parkinson’s disease | Solute Carrier Family 7A11 | 2020 | [119] | |
| schizophrenia | Solute Carrier Family 6A4 | 2022 | [120] | |
| borderline personality disorder | BDNF c | 2018 | [121] | |
| epilepsy | PABPC1 d, ARGLU1 e, etc. | 2021 | [122] | |
| depression | KLK8 f, NR3C1 g | 2021 | [123,124] | |
| fragile X syndrome | FMR1 h | 2019 | [105] | |
| immunological | systemic lupus erythematosus | NCR3 i | 2021 | [125] |
| rheumatoid arthritis | SFRP2 j | 2018 | [126] | |
| systemic sclerosis | PARP-1 k | 2018 | [127] | |
| Sjogren’s syndrome | IRE1α l, XBP-1 m, GRP78 n | 2018 | [128] | |
| Graves’ disease | CRHR1 o, B3GNT2 p | 2021 | [129] | |
| type 1 diabetes | Cathepsin H | 2021 | [130] | |
| Crohn’s disease | KCNJ15 q | 2022 | [131] | |
| endocrine/metabolic | osteoporosis | Nrf2 r | 2021 | [132] |
| type 2 diabetes | TXNIP s | 2021 | [133] | |
| hyperlipidemia | PPARα t | 2022 | [134] | |
| cardiovascular | cardiomyopathy | ASB1 u | 2018 | [135] |
| renal | IgA nephropathy | Cosmc v | 2015 | [136] |
| diabetic nephropathy | TRIM13 w | 2020 | [137] | |
| pulmonary | idiopathic pulmonary fibrosis | PPARγ | 2022 | [138] |
| chronic obstructive pulmonary disease | CYP4F11 x, SNRPN y, etc. | 2014 | [139] | |
| hepatic | hepatitis C | SHP-1 z | 2021 | [140] |
| ZFA | TALE | CRISPR/dCas9 | |
|---|---|---|---|
| Components | Zinc finger domain | TALE DNA-binding domain | crRNA/tracrRNA or sgRNA, dCas9 |
| Size | ~1 kb + demethylase | ~2 kb + demethylase | 4.2 kb (dCas9) + 0.1 kb (sgRNA) |
| Working mechanism | DNA/protein recognition | DNA/protein recognition | DNA/RNA recognition |
| Length of the target sequence | 9–36 bp | 30–40 bp per TALE pair | 20–22 bp |
| Target recognition efficiency | High | High | Very High |
| Off target effects | Low for long arrays | Unknown | Lowest |
| Multiplexing | Possible | Difficult | Possible |
| Cloning | Essential | Essential | Not essential |
| Advantages | Small protein size (<1 kDa) facilitates packaging and purification | High specificity with each module recognizing 1bp; no need to engineer linkage between repeats | Enable multiplexing |
| When administered as a naked protein, spontaneously internalize into cells and translocate into nuclei, delivering the fused enzymatic payload | Only a replacement of sgRNA is required to retarget new genes | ||
| Limitations | Re-designing the entire protein is required to re-target | Re-designing is required to target new genes | Large protein size (~150 kDa) makes packaging into vectors or nanovesicles difficult |
| Cloning methods that require additional linker sequence to fuse modules together add variability | Repetitive sequences may induce undesirable recombination events | When administered as a naked protein, additional measures for cellular internalization and nuclear translocation are required | |
| Has difficulty binding methylated DNA | sgRNA may be unstable under in vivo and in vitro biological conditions, may require packaging/protection |
| AdVs * | AAVs ** | LVs *** | |
|---|---|---|---|
| Cell affinity | Inefficient for some types of cells | Dependent on viral serotypes | Broad |
| Infection into non-dividing cells | + | + | + |
| Transient/Stable expression | Stable expression by genome integration | Transient, episomal | Transient, episomal |
| Maximum titer | Very high | High | High |
| In vivo immunogenicity | High | Very low | Low |
| DBDs | Enzymes | Targeted Gene | Research Materials | Publication Year | Ref |
|---|---|---|---|---|---|
| TALE | TET1 | RHOXF2 a | HeLa cells | 2013 | [142] |
| TALE | TET1 | Ascl1 b | Neural stem cells | 2017 | [261] |
| ZFA | TDG | Nos2 c | NIH-3T3 cells | 2013 | [86] |
| ZFA | TET2 | C13ORF18 d, CCNA1 e, TFPI2 f, SERPINB5 g | HeLa cells, SiHa cells, CaSki cells, C33a cells | 2016 | [262] |
| ZFA | TET1 | FWA h | Arabidopsis | 2016 | [263] |
| CRISPR | TET1 | RANKL i, MAGEB2 j, MMP2 k | HEK293FT cells | 2016 | [192] |
| CRISPR | TET1 | H19 l, RHOXF2, CARD9 m, SH3BP2 n, CNKSR1 o, GFAP p | ESC, cancer cell lines, primary neural precursor cells, mouse fetuses | 2016 | [143] |
| CRISPR | TET1 | BRCA1 q | HeLa cells, MCF7 cells | 2016 | [264] |
| CRISPR | TET1 | ST6GAL1 r | CHO cells | 2018 | [165] |
| CRISPR | TET1 | Oct4 s | NIH-3T3 cells | 2022 | [164] |
| CRISPR | TET1 | FOXP3 t | HCC202 cells, HEK293T cells | 2022 | [154] |
| CRISPR | TET1 | PPH u | Arabidopsis | 2022 | [265] |
| Rel-homology domain (RHD) of NFκB | TDG | Nos2 | NIH-3T3 cells | 2012 | [85] |
| DNA-binding domain of yeast GAL4 | ROS1 5mC DNA glycosylase | Targeted reporter gene | HEK293 cells | 2013 | [170] |
| Proteins coded by the genes | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yano, N.; Fedulov, A.V. Targeted DNA Demethylation: Vectors, Effectors and Perspectives. Biomedicines 2023, 11, 1334. https://doi.org/10.3390/biomedicines11051334
Yano N, Fedulov AV. Targeted DNA Demethylation: Vectors, Effectors and Perspectives. Biomedicines. 2023; 11(5):1334. https://doi.org/10.3390/biomedicines11051334
Chicago/Turabian StyleYano, Naohiro, and Alexey V. Fedulov. 2023. "Targeted DNA Demethylation: Vectors, Effectors and Perspectives" Biomedicines 11, no. 5: 1334. https://doi.org/10.3390/biomedicines11051334
APA StyleYano, N., & Fedulov, A. V. (2023). Targeted DNA Demethylation: Vectors, Effectors and Perspectives. Biomedicines, 11(5), 1334. https://doi.org/10.3390/biomedicines11051334






