Dissecting the Molecular Role of ADIPOQ SNPs in Saudi Women Diagnosed with Gestational Diabetes Mellitus
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of GDM Women
2.2. Data Collection from Pregnant Women
2.3. EDTA Blood Analysis
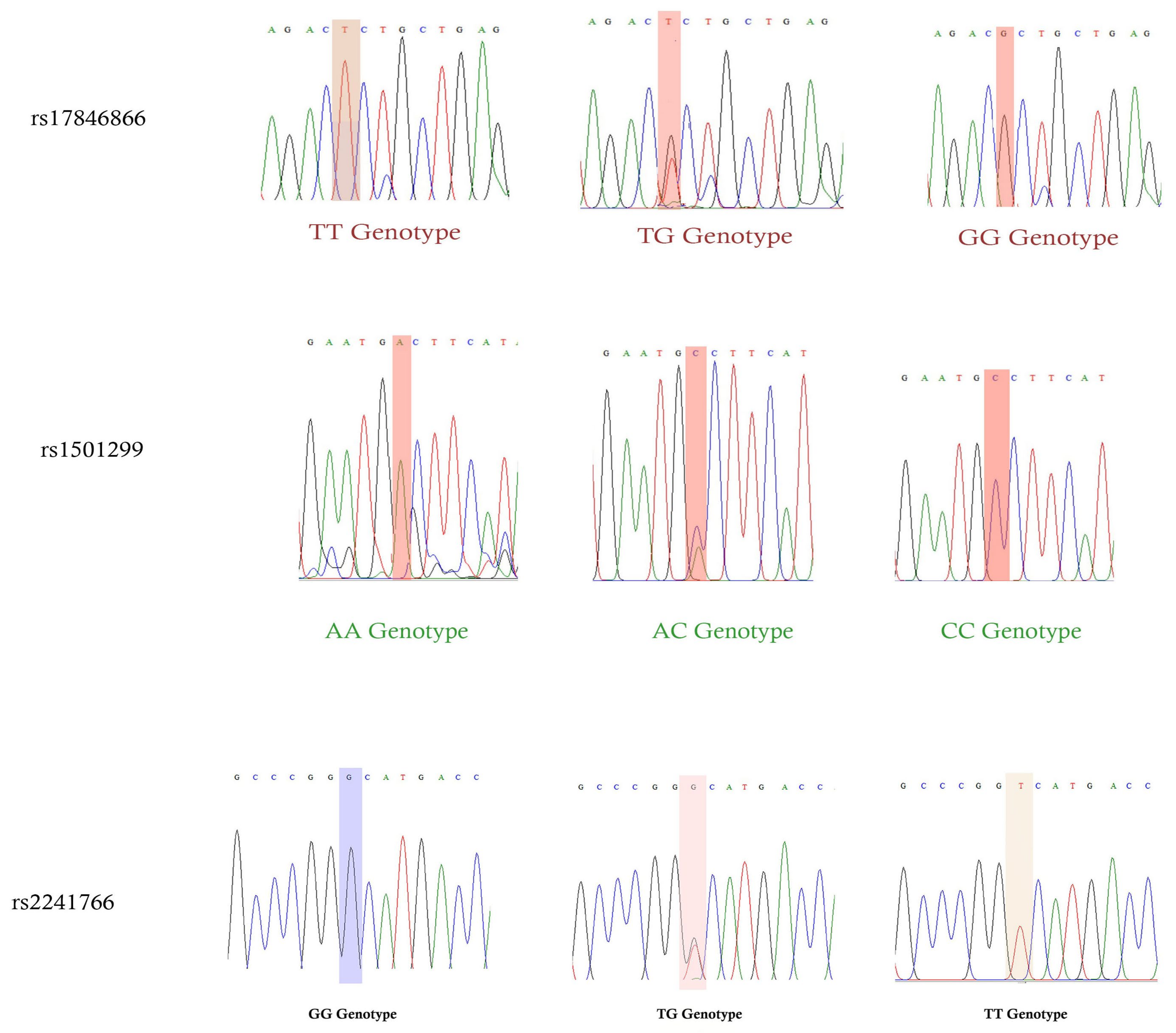
2.4. Sanger Sequencing Analysis
2.5. Statistical Analysis
3. Results
3.1. Details from Questionnaire for Patients with GDM and Control Patients
3.2. Hardy–Weinberg Equilibrium
3.3. Calculation of Genetic Frequencies
3.4. Calculation of Allelic Frequencies
3.5. Logistic Regression Analysis Studied in GDM Women
3.6. One-Way ANOVA
3.7. Analysis in ADIPOQ Variants
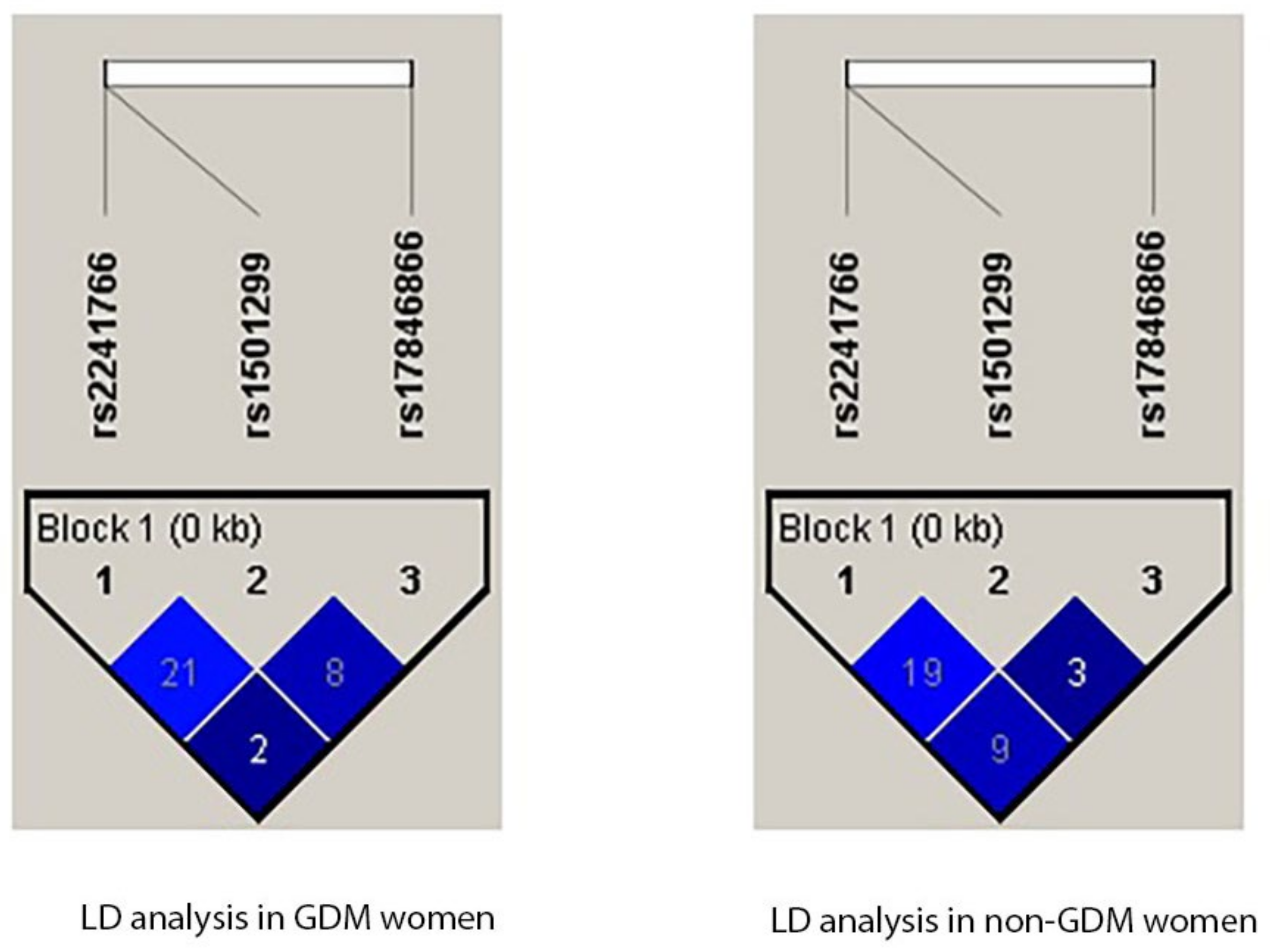
3.8. Linkage Disequilibrium Analysis
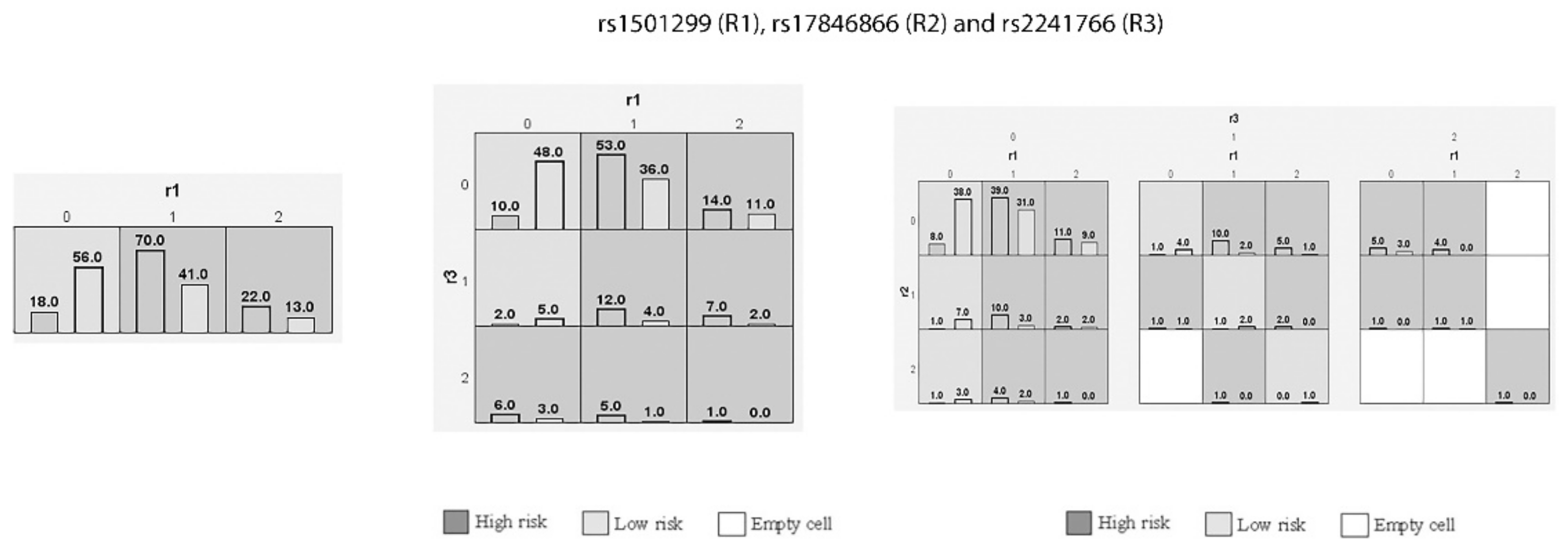
3.9. Interaction of Genetic Variants in GDM Women through MDR and GMDR Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Egan, A.M.; Dunne, F.P. Diagnosis of gestational diabetes mellitus: The debate continues. Nat. Rev. Endocrinol. 2022, 18, 723–724. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K.A.; Chang, L.; Cheung, J.; Aouizerat, B.E.; Jelliffe-Pawlowski, L.L.; McLemore, M.R.; Piening, B.; Rand, L.; Ryckman, K.K.; Flowers, E. Systematic review of transcriptome and microRNAome associations with gestational diabetes mellitus. Front. Endocrinol. 2023, 13, 3544. [Google Scholar] [CrossRef] [PubMed]
- Davidsen, E.; Maindal, H.T.; Rod, M.H.; Olesen, K.; Byrne, M.; Damm, P.; Nielsen, K.K. The stigma associated with gestational diabetes mellitus: A scoping review. eClinicalMedicine 2022, 52, 101614. [Google Scholar] [CrossRef] [PubMed]
- Sweeting, A.; Wong, J.; Murphy, H.R.; Ross, G.P. A clinical update on gestational diabetes mellitus. Endocr. Rev. 2022, 43, 763–793. [Google Scholar] [CrossRef]
- Al-Rifai, R.H.; Abdo, N.M.; Paulo, M.S.; Saha, S.; Ahmed, L.A. Prevalence of gestational diabetes mellitus in the middle east and North Africa, 2000–2019: A Systematic Review, Meta-Analysis, and Meta-Regression. Front. Endocrinol. 2021, 12, 668447. [Google Scholar] [CrossRef]
- Khan, R.S.; Malik, H. Diagnostic Biomarkers for Gestational Diabetes Mellitus Using Spectroscopy Techniques: A Systematic Review. Diseases 2023, 11, 16. [Google Scholar] [CrossRef]
- Blanco, J.A.; Lambert, C.; Fernandez-Sanjurjo, M.; Morales-Sanchez, P.; Pujante, P.; Pinto-Hernandez, P.; Iglesias-Gutierrez, E.; Torre, E.M.; Delgado, E. miR-24-3p and Body Mass Index as Type 2 Diabetes Risk Factors in Spanish Women 15 Years after Gestational Diabetes Mellitus Diagnosis. Int. J. Mol. Sci. 2023, 24, 1152. [Google Scholar] [CrossRef] [PubMed]
- Perämäki, R.; Gissler, M.; Ollila, M.-M.; Hukkanen, J.; Vääräsmäki, M.; Uotila, J.; Metso, S.; Hakkarainen, H.; Rintamäki, R.; Kaaja, R. The risk of developing type 2 diabetes after gestational diabetes: A registry study from Finland. Diabetes Epidemiol. Manag. 2023, 10, 100124. [Google Scholar] [CrossRef]
- Wu, L.; Cui, L.; Tam, W.H.; Ma, R.C.; Wang, C.C. Genetic variants associated with gestational diabetes mellitus: A meta-analysis and subgroup analysis. Sci. Rep. 2016, 6, 30539. [Google Scholar] [CrossRef]
- Naeh, A.; Maor-Sagie, E.; Hallak, M.; Gabbay-Benziv, R. Early Identification of the Maternal, Placental and Fetal Dialog in Gestational Diabetes and Its Prevention. Reprod. Med. 2022, 3, 1–14. [Google Scholar] [CrossRef]
- Zakaria, H.; Abusanana, S.; Mussa, B.M.; Al Dhaheri, A.S.; Stojanovska, L.; Mohamad, M.N.; Saleh, S.T.; Ali, H.I.; Ismail, L.C. The Role of Lifestyle Interventions in the Prevention and Treatment of Gestational Diabetes Mellitus. Medicina 2023, 59, 287. [Google Scholar] [CrossRef] [PubMed]
- Karami, M.; Mousavi, S.H.; Rafiee, M.; Heidari, R.; Shahrokhi, S.Z. Biochemical and molecular biomarkers: Unraveling their role in gestational diabetes mellitus. Diabetol. Metab. Syndr. 2023, 15, 5. [Google Scholar] [CrossRef] [PubMed]
- Abualhamael, S.; Mosli, H.; Baig, M.; Noor, A.M.; Alshehri, F.M. Prevalence and associated risk factors of gestational diabetes mellitus at a university hospital in Saudi Arabia. Pak. J. Med. Sci. 2019, 35, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Wahabi, H.A.; Fayed, A.; Esmaeil, S.A. Maternal and perinatal outcomes of pregnancies complicated with pre-gestational and gestational diabetes mellitus in Saudi Arabia. J. Diabetes Metab. 2014, 5, 2. [Google Scholar] [CrossRef]
- Lee, K.W.; Ching, S.M.; Ramachandran, V.; Yee, A.; Hoo, F.K.; Chia, Y.C.; Sulaiman, W.A.W.; Suppiah, S.; Mohamed, M.H.; Veettil, S.K. Prevalence and risk factors of gestational diabetes mellitus in Asia: A systematic review and meta-analysis. BMC Pregnancy Childbirth 2018, 18, 494. [Google Scholar] [CrossRef]
- Badakhsh, M.; Daneshi, F.; Abavisani, M.; Rafiemanesh, H.; Bouya, S.; Sheyback, M.; Keikhaie, K.R.; Balouchi, A. Prevalence of gestational diabetes mellitus in Eastern Mediterranean region: A systematic review and meta-analysis. Endocrine 2019, 65, 505–514. [Google Scholar] [CrossRef]
- World Health Organization. Noncommunicable Diseases Country Profiles 2018; World Health Organization: Geneva, Switzerland, 2018. [Google Scholar]
- Fallatah, A.M.; AlNoury, A.; Fallatah, E.M.; Nassibi, K.M.; Babatin, H.; Alghamdi, O.A.; Tarabaih, B.Y.; Abduljabbar, H.S. Obesity among pregnant women in Saudi Arabia: A retrospective single-center medical record review. Cureus 2021, 13, e13454. [Google Scholar] [CrossRef] [PubMed]
- Alba-Linares, J.J.; Pérez, R.F.; Tejedor, J.R.; Bastante-Rodríguez, D.; Ponce, F.; Carbonell, N.G.; Zafra, R.G.; Fernández, A.F.; Fraga, M.F.; Lurbe, E. Maternal obesity and gestational diabetes reprogram the methylome of offspring beyond birth by inducing epigenetic signatures in metabolic and developmental pathways. Cardiovasc. Diabetol. 2023, 22, 44. [Google Scholar] [CrossRef]
- Catalano, P. The impact of gestational diabetes and maternal obesity on the mother and her offspring. J. Dev. Orig. Health Dis. 2010, 1, 208–215. [Google Scholar] [CrossRef]
- Chiefari, E.; Arcidiacono, B.; Foti, D.; Brunetti, A. Gestational diabetes mellitus: An updated overview. J. Endocrinol. Investig. 2017, 40, 899–909. [Google Scholar] [CrossRef]
- Ramachandrayya, S.A.; D’Cunha, P.; Rebeiro, C. Maternal circulating levels of Adipocytokines and insulin resistance as predictors of gestational diabetes mellitus: Preliminary findings of a longitudinal descriptive study. J. Diabetes Metab. Disord. 2020, 19, 1447–1452. [Google Scholar] [CrossRef] [PubMed]
- Kampmann, U.; Knorr, S.; Fuglsang, J.; Ovesen, P. Determinants of maternal insulin resistance during pregnancy: An updated overview. J. Diabetes Res. 2019, 2019, 5320156. [Google Scholar] [CrossRef] [PubMed]
- Beltcheva, O.; Boyadzhieva, M.; Angelova, O.; Mitev, V.; Kaneva, R.; Atanasova, I. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch. Gynecol. Obstet. 2014, 289, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Tang, L.; Li, L. The roles of ADIPOQ rs266729 and MTNR1B rs10830963 polymorphisms in patients with gestational diabetes mellitus: A meta-analysis. Gene 2020, 730, 144302. [Google Scholar] [CrossRef]
- Dias, S.; Adam, S.; Rheeder, P.; Pheiffer, C. No association between ADIPOQ or MTHFR polymorphisms and gestational diabetes mellitus in South African women. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 791–800. [Google Scholar] [CrossRef]
- Li, Z.-P.; Zhang, M.; Gao, J.; Zhou, G.-Y.; Li, S.-Q.; An, Z.-M. Relation between ADIPOQ gene polymorphisms and type 2 diabetes. Genes 2015, 6, 512–519. [Google Scholar] [CrossRef]
- Li, P.; Jiang, R.; Li, L.; Liu, C.; Yang, F.; Qiu, Y. Correlation of serum adiponectin and adiponectin gene polymorphism with metabolic syndrome in Chinese adolescents. Eur. J. Clin. Nutr. 2015, 69, 62–67. [Google Scholar] [CrossRef]
- Elghazy, A.M.; Elsaeid, A.M.; Refaat, M.; Youssef, M.M. Biochemical studies of adiponectin gene polymorphism in patients with obesity in Egyptians. Arch. Physiol. Biochem. 2022, 128, 43–50. [Google Scholar] [CrossRef]
- Pawlik, A.; Teler, J.; Maciejewska, A.; Sawczuk, M.; Safranow, K.; Dziedziejko, V. Adiponectin and leptin gene polymorphisms in women with gestational diabetes mellitus. J. Assist. Reprod. Genet. 2017, 34, 511–516. [Google Scholar] [CrossRef]
- Demirci, H.; Yilmaz, M.; Ali Ergun, M.; Yurtcu, E.; Bukan, N.; Ayvaz, G. Frequency of adiponectin gene polymorphisms in polycystic ovary syndrome and the association with serum adiponectin, androgen levels, insulin resistance and clinical parameters. Gynecol. Endocrinol. 2010, 26, 348–355. [Google Scholar] [CrossRef]
- Pheiffer, C.; Dias, S.; Jack, B.; Malaza, N.; Adam, S. Adiponectin as a potential biomarker for pregnancy disorders. Int. J. Mol. Sci. 2021, 22, 1326. [Google Scholar] [CrossRef] [PubMed]
- Al-Daghri, N.M.; Al-Attas, O.S.; Alokail, M.S.; Alkharfy, K.M.; Hussain, T. Adiponectin gene variants and the risk of coronary artery disease in patients with type 2 diabetes. Mol. Biol. Rep. 2011, 38, 3703–3708. [Google Scholar] [CrossRef] [PubMed]
- Al-Daghri, N.M.; Al-Attas, O.S.; Alokail, M.S.; Alkharfy, K.M.; Hussain, T.; Yakout, S.; Vinodson, B.; Sabico, S. Adiponectin gene polymorphisms (T45G and G276T), adiponectin levels and risk for metabolic diseases in an Arab population. Gene 2012, 493, 142–147. [Google Scholar] [CrossRef] [PubMed]
- Ezzidi, I.; Mtiraoui, N.; Mohmmed Ali, M.E.; Al Masoudi, A.; Abu Duhier, F. Adiponectin (ADIPOQ) gene variants and haplotypes in Saudi Arabian women with polycystic ovary syndrome (PCOS): A case-control study. Gynecol. Endocrinol. 2020, 36, 66–71. [Google Scholar] [CrossRef]
- Mackawy, A.M. Association of the +45T>G adiponectin gene polymorphism with insulin resistance in non-diabetic Saudi women. Gene 2013, 530, 158–163. [Google Scholar] [CrossRef]
- Al-Nbaheen, M.S. Effect of Genetic Variations in the ADIPOQ Gene on Susceptibility to Type 2 Diabetes Mellitus. Diabetes Metab. Syndr. Obes. Targets Ther. 2022, 15, 2753–2761. [Google Scholar] [CrossRef] [PubMed]
- Alharbi, K.K.; Khan, I.A.; Abotalib, Z.; Al-Hakeem, M.M. Insulin receptor substrate-1 (IRS-1) Gly927Arg: Correlation with gestational diabetes mellitus in Saudi women. BioMed Res. Int. 2014, 2014, 146495. [Google Scholar] [CrossRef]
- Al-Harithy, R.N.; Al-Zahrani, M.H. The adiponectin gene, ADIPOQ, and genetic susceptibility to colon cancer. Oncol. Lett. 2012, 3, 176–180. [Google Scholar] [CrossRef]
- Gu, H.F. Biomarkers of adiponectin: Plasma protein variation and genomic DNA polymorphisms. Biomark. Insights 2009, 4, 123–133. [Google Scholar] [CrossRef]
- Hussain, M.K.; Deli, F.A.; Algenabi, A.H.A.; Abdul-Rudha, K.H. Adiponectin gene polymorphisms as a predictor for development of type 2 diabetes mellitus in Iraqi population. Gene 2018, 662, 118–122. [Google Scholar] [CrossRef]
- Vimaleswaran, K.S.; Radha, V.; Ramya, K.; Babu, H.N.S.; Savitha, N.; Roopa, V.; Monalisa, D.; Deepa, R.; Ghosh, S.; Majumder, P.P. A novel association of a polymorphism in the first intron of adiponectin gene with type 2 diabetes, obesity and hypoadiponectinemia in Asian Indians. Hum. Genet. 2008, 123, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Boutin, P.; Mori, Y.; Tobe, K.; Dina, C.; Yasuda, K.; Yamauchi, T.; Otabe, S.; Okada, T.; Eto, K. Genetic variation in the gene encoding adiponectin is associated with an increased risk of type 2 diabetes in the Japanese population. Diabetes 2002, 51, 536–540. [Google Scholar] [CrossRef]
- Howlader, M.; Sultana, M.I.; Akter, F.; Hossain, M.M. Adiponectin gene polymorphisms associated with diabetes mellitus: A descriptive review. Heliyon 2021, 7, e07851. [Google Scholar] [CrossRef] [PubMed]
- Al-Goblan, A.S.; Al-Alfi, M.A.; Khan, M.Z. Mechanism linking diabetes mellitus and obesity. Diabetes Metab. Syndr. Obes. Targets Ther. 2014, 7, 587–591. [Google Scholar] [CrossRef] [PubMed]
- AlShahrani, M.S. Prevalence of obesity and overweight among type 2 diabetic patients in Bisha, Saudi Arabia. J. Fam. Med. Prim. Care 2021, 10, 143. [Google Scholar] [CrossRef]
- Jatoi, N.-A.; Al-Qassab, R.M.; Al Salem, F.H.; Al Muzayan, F.M.; AlShammari, R.Z. Prevalence of Obesity and Cardiovascular Risk Factors Among Type 2 Diabetes Mellitus Patients in Al-Khobar, Saudi Arabia. Cureus 2022, 14, e30539. [Google Scholar] [CrossRef]
- Aljulifi, M.Z. Prevalence and reasons of increased type 2 diabetes in Gulf Cooperation Council Countries. Saudi Med. J. 2021, 42, 481. [Google Scholar] [CrossRef]
- Al Mansour, M.A. The prevalence and risk factors of type 2 diabetes mellitus (DMT2) in a semi-urban Saudi population. Int. J. Environ. Res. Public Health 2020, 17, 7. [Google Scholar] [CrossRef]
- Zayani, N.; Omezzine, A.; Boumaiza, I.; Achour, O.; Rebhi, L.; Rejeb, J.; Ben Rejeb, N.; Ben Abdelaziz, A.; Bouslama, A. Association of ADIPOQ, leptin, LEPR, and resistin polymorphisms with obesity parameters in Hammam Sousse Sahloul Heart Study. J. Clin. Lab. Anal. 2017, 31, e22148. [Google Scholar] [CrossRef]
- Phillips, C.M. Nutrigenetics and metabolic disease: Current status and implications for personalised nutrition. Nutrients 2013, 5, 32–57. [Google Scholar] [CrossRef]
- Low, C.F.; Mohd Tohit, E.R.; Chong, P.P.; Idris, F. Adiponectin SNP45TG is associated with gestational diabetes mellitus. Arch. Gynecol. Obstet. 2011, 283, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Daher, S.; Torloni, M.R.; Gueuvoghlanian-Silva, B.Y.; Moron, A.F.; Mattar, R. Inflammatory mediator gene polymorphisms and gestational diabetes: A review of the literature. J. Reprod. Immunol. 2011, 90, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Du, J.; Du, J. Relationship between diponectin gene polymorphism and gestational diabetes mellitus. Xiandai Fuchanke Jinzhan 2011, 20, 718–721. [Google Scholar]
- Han, Y.; Zheng, Y.; Liu, M.; Fan, Y. Association of adiponectin gene single nucleotide polymorphism with gestational diabetes mellitus and pregnancy outcomes. Shiyong Fuchanke Zazhi 2012, 22, 743–746. [Google Scholar]
- Li, G.; Kong, L.; Zhang, L.; Zhang, W. Association of adiponectin gene polymorphisms+ 45T/G with gestational diabetes mellitus and neonate birth weight. Zhonghua Yi Xue Za Zhi 2013, 93, 3770–3772. [Google Scholar] [PubMed]
- Zhang, C.; Liang, X. The relationship between human adiponectin gene polymorphism of Guangxi Zhuang ethnic group and gestational diabetes mellitus. J. Chin. Physician 2014, 12, 1221–1223. [Google Scholar]
- Luan, Y.; Guo, X.; Yang, J. Study on the correlation between the damage of adiponectin gene polymorphism with gestational impaired glucose. Zhongguo Shiyanzhenduanxue Zazhi 2015, 19, 1093–1096. [Google Scholar]
- Takhshid, M.A.; Haem, Z.; Aboualizadeh, F. The association of circulating adiponectin and +45 T/G polymorphism of adiponectin gene with gestational diabetes mellitus in Iranian population. J. Diabetes Metab. Disord. 2015, 14, 30. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Zhou, G.; Pu, X. Study of the correlation between adiponectin gene polymorphism and gestational diabetes mellitus. Zhongguo Fuyoubaojian Zazhi 2016, 31, 2546–2549. [Google Scholar]
- Gao, Y.; Wang, W.; Wang, S. Association of Adiponectin Gene Single Nucleotide Polymorphism with Gestational Diabetes Mellitus. Jilin Yixue 2016, 6, 1301–1302. [Google Scholar]
- Li, J.; Ma, S.; Zhao, J.; Duan, L.; Sun, H. The correlation between single nucleotide polymorphism of adiponectin gene and gestational diabetes and its effect on pregnancy outcome. Zhongguo Fuyoubaojian Zazhi 2017, 22, 5674–5677. [Google Scholar]
- Feng, Y.; Jiang, C.-D.; Chang, A.-M.; Shi, Y.; Gao, J.; Zhu, L.; Zhang, Z. Interactions among insulin resistance, inflammation factors, obesity-related gene polymorphisms, environmental risk factors, and diet in the development of gestational diabetes mellitus. J. Matern. Fetal Neonatal Med. 2019, 32, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhang, S.; Wang, T.; Diao, J.; Li, J.; Li, Y.; Zhao, L.; Chen, L.; Ye, Z.; Huang, P. Associations of maternal diabetes mellitus and adiponectin gene polymorphisms with congenital heart disease in offspring: A case-control study. Medicine 2021, 100, e24672. [Google Scholar] [CrossRef] [PubMed]
- Tangjittipokin, W.; Thanatummatis, B.; Wardati, F.; Narkdontri, T.; Teerawattanapong, N.; Boriboonhirunsarn, D. The genetic polymorphisms and levels of adipokines and adipocytokines that influence the risk of developing gestational diabetes mellitus in Thai pregnant women. Gene 2023, 860, 147228. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Li, P. Novel Single Nucleotide Polymorphisms in Gestational Diabetes Mellitus. Clin. Chim. Acta 2022, 538, 60–64. [Google Scholar] [CrossRef]
- Huang, L.-T.; Wu, S.-L.; Liao, X.; Ma, S.-J.; Tan, H.-Z. Adiponectin gene polymorphisms and risk of gestational diabetes mellitus: A meta-analysis. World J. Clin. Cases 2019, 7, 572. [Google Scholar] [CrossRef]
- Dong, Y.; Huang, G.; Wang, X.; Chu, Z.; Miao, J.; Zhou, H. Meta-analysis of the association between adiponectin SNP 45, SNP 276, and type 2 diabetes mellitus. PLoS ONE 2020, 15, e0241078. [Google Scholar] [CrossRef]
- Timasheva, Y.; Balkhiyarova, Z.; Avzaletdinova, D.; Rassoleeva, I.; Morugova, T.V.; Korytina, G.; Prokopenko, I.; Kochetova, O. Integrating Common Risk Factors with Polygenic Scores Improves the Prediction of Type 2 Diabetes. Int. J. Mol. Sci. 2023, 24, 984. [Google Scholar] [CrossRef]



| Gene | ADIPOQ | ADIPOQ | ADIPOQ |
|---|---|---|---|
| rs Number | rs1501299 | rs17846866 | rs2241766 |
| SNPs | A-C/(c.276G > C) | T-G/(+10211T > G) | T-G/(+45T > G) |
| Location | Intron 2 | Intron 1 | Exon 2 |
| Forward Primer | GGCCTCTTTCATCACAGACC | GCTAAGTATTACAGATTTCAGGGCAG | GAAGTAGACTCTGCTTGAGATGG |
| Reverse Primer | AGATGCAGCAAAGCCAAAGT | CAGCAACAGCATCCTGAGC | TATCAGTGTAGGAGGTCTGTGATG |
| PCR Size | 196bp | 222bp | 372bp |
| Annealing Temperature | 64 °C | 66 °C | 62 °C |
| Restriction Enzyme | BsmI (GAATGCC↑) | HgaI (GAATGCC↑) | SmaI (GGG↑CCC) |
| Digested Products | A-196bp; C-146/50bp | T-222bp; G-115/107bp | T-372bp; G-219/153bp |
| Covariates | Controls (n = 110) | GDM (n = 110) | p-Value |
|---|---|---|---|
| Age (year) | 29.46 ± 6.16 | 33.02 ± 5.87 | 0.0001 |
| Gender (male: female) | 0 (0%):110 (100%) | 0 (0%):110 (100%) | 1.00 |
| Weight (kilograms) | 73.14 ± 11.64 | 79.25 ± 12.32 | 0.0002 |
| Height (centimeters) | 157.92 ± 5.01 | 157.91 ± 5.49 | 0.98 |
| BMI (kg/m2) | 29.29 ± 4.07 | 31.80 ± 4.42 | 0.0001 |
| SBP (mmHg) | 110.42 ± 11.86 | 120.15 ± 10.09 | <0.0001 |
| DBP (mmHg) | 64. 37 ± 3.01 | 74.40 ± 3.31 | <0.0001 |
| FBG (mmol/L) | 4.31 ± 0.98 | 5.93 ± 1.18 | <0.0001 |
| PPBG (mmol/L) | 4.96 ± 12.32 | 9.31 ± 15.73 | <0.0001 |
| GCT (mmol/L) | 6.21 ± 1.58 | 9.33 ± 1.04 | <0.0001 |
| OGTT (F) (mmol/L) | 4.92 ± 1.08 | 6.59 ± 2.14 | <0.0001 |
| OGTT (1) (mmol/L) | 7.23 ± 1.65 | 10.61 ± 1.76 | <0.0001 |
| OGTT (2) (mmol/L) | 6.32 ± 1.47 | 9.09 ± 1.74 | <0.0001 |
| OGTT (3) (mmol/L) | 4.13 ± 1.27 | 6.16 ± 1.79 | <0.0001 |
| Hb1Ac (%) | 4.78 ± 0.28 | 5.41 ± 0.34 | <0.0001 |
| TC (mmol/L) | 5.06 ± 1.14 | 5.75 ± 1.27 | 0.0003 |
| TG (mmol/L) | 1.71 ± 1.23 | 2.33 ± 1.95 | 0.005 |
| Hdlc (mmol/L) | 0.71 ± 0.27 | 0.95 ± 0.41 | 0.001 |
| Ldlc (mmol/L) | 3.71 ± 0.94 | 3.85 ± 0.97 | 0.27 |
| Medication (Insulin) | NA | 08 (7.3%) | NA |
| Family History of T2DM | 28 (25.5%) | 110 (100%) | <0.0001 |
| Family History of GDM | 10 (9.1%) | 37 (33.7%) | <0.0001 |
| SNPs | Minor Allele | Genotype Frequencies for Controls (n = 110) | ꭓ2 |
|---|---|---|---|
| rs1501299 | C | AA-50.9%, AC-37.3% and CC-11.8% | 1.58 |
| rs17846866 | G | TT-80%, TG-14.6% and GG-5.5% | 13.1 |
| rs2241766 | G | TT-86.4%, TG-10% and GG-3.6% | 14.7 |
| Gene (rs Number) | Genotypes | GDM (n = 110) | Non-GDM (n = 110) | OR (95%CI) and p-Value |
|---|---|---|---|---|
| ADIPOQ (rs1501299) | AA | 18 (16.4%) | 56 (50.9%) | 1.00 |
| AC | 70 (63.6%) | 41 (37.3%) | OR-5.31 (2.76–10.24); p < 0.0001 | |
| CC | 22 (20%) | 13 (11.8%) | OR-5.26 (2.21–12.53); p < 0.0001 | |
| AC + CC vs. AA | 92 (83.6%) | 44 (49.1%) | OR-5.30 (2.83–9.94); p < 0.0001 | |
| CC + AA vs. AC | 40 (36.4%) | 69 (71.7%) | OR-1.87 (0.89–3.92); p = 0.09 | |
| AA + AC vs. CC | 98 (80%) | 97 (88.2%) | OR-2.95 (1.70–5.09); p < 0.0001 | |
| ADIPOQ (rs17846866) | TT | 83 (75.5%) | 88 (80%) | 1.00 |
| TG | 19 (17.3%) | 16 (14.5%) | OR-1.26 (0.61–2.61); p = 0.71 | |
| GG | 8 (7.3%) | 6 (5.5%) | OR-1.41 (0.47–4.25); p = 0.73 | |
| TG + GG vs. TT | 27 (24.6%) | 22 (20%) | OR-1.30 (0.69–2.46); p = 0.42 | |
| TT + GG vs. TG | 91 (82.8%) | 94 (85.5%) | OR-1.36 (0.46–4.06); p = 0.58 | |
| TT + TG vs. GG | 102 (92.7%) | 104 94.5%) | OR-1.23 (0.59–2.53); p = 0.58 | |
| ADIPOQ (rs2241766) | TT | 77 (70%) | 95 (86.4%) | 1.00 |
| TG | 21 (19.1%) | 11 (10%) | OR-2.36 (1.07–5.18); p = 0.009 | |
| GG | 12 (10.9%) | 4 (3.6%) | OR-3.70 (1.15–11.94); p = 0.02 | |
| TG + GG vs. TT | 33 (30%) | 15 (13.6%) | OR-2.71 (1.37–5.36); p = 0.003 | |
| TT + GG vs. TG | 89 (80.9%) | 99 (90%) | OR-3.24 (1.01–10.40); p = 0.03 | |
| TT + TG vs. GG | 98 (89.1%) | 106 (96.4%) | OR-2.12 (0.97–4.65); p = 0.054 |
| Gene (rs Number) | Genotypes | GDM (n = 110) | Control (n = 110) | OR (95%CI) and p-Value |
|---|---|---|---|---|
| ADIPOQ (rs1501299) | A | 106 (48.2%) | 153 (69.5%) | Reference |
| C | 114 (51.8%) | 67 (30.5%) | OR-2.45 (1.66–3.63); p < 0.0001 | |
| ADIPOQ (rs17846866) | T | 185 (84.1%) | 192 (87.3%) | Reference |
| G | 35 (15.9%) | 28 (12.7%) | OR-1.29 (0.75–2.23); p = 0.41 | |
| ADIPOQ (rs2241766) | T | 175 (79.5%) | 210 (91.4%) | Reference |
| G | 45 (20.5%) | 19 (8.6%) | OR-2.71 (1.54–4.90); p < 0.0006 |
| Covariates | R-Value a | Adjusted R Square Value | Standardized β-Coefficient for rs1501299 | Standardized β-Coefficient for rs17846866 | Standardized β-Coefficient for rs2241766 | F | p Value b |
|---|---|---|---|---|---|---|---|
| Age | 0.130 | −0.011 | 0.018 | −0.120 | 0.052 | 0.609 | 0.610 |
| Weight | 0.067 | −0.024 | −0.030 | −0.376 | −0.051 | 0.159 | 0.924 |
| BMI | 0.102 | −0.018 | −0.025 | −0.098 | −0.003 | 0.371 | 0.774 |
| SBP | 0.046 | −0.026 | 0.012 | −0.045 | 0.004 | 0.075 | 0.974 |
| DBP | 0.128 | −0.012 | −0.068 | 0.111 | −0.007 | 0.585 | 0.626 |
| FBG | 0.125 | −0.012 | −0.036 | 0.052 | −0.113 | 0.559 | 0.643 |
| PPBG | 0.119 | −0.014 | −0.021 | 0.104 | −0.057 | 0.506 | 0.679 |
| GCT | 0.192 | 0.010 | 0.168 | −0.064 | −0.055 | 1.359 | 0.259 |
| OGTT (F) | 0.152 | −0.005 | 0.345 | 0.341 | 0.306 | 0.833 | 0.479 |
| OGTT (1) | 0.125 | −0.012 | 0.285 | 0.282 | 0.253 | 0.565 | 0.639 |
| OGTT (2) | 0.105 | −0.017 | 0.282 | 0.279 | 0.250 | 0.390 | 0.760 |
| OGTT (3) | 0.207 | 0.016 | 1.173 | 1.163 | 1.043 | 1.582 | 0.198 |
| Hb1Ac | 0.165 | 0.000 | 0.120 | −0.054 | 0.122 | 0.993 | 0.399 |
| TC | 0.135 | −0.010 | −0.122 | 0.065 | −0.027 | 0.657 | 0.580 |
| TG | 0.098 | −0.018 | 0.019 | −0.078 | −0.055 | 0.343 | 0.794 |
| HDLc | 0.077 | −0.022 | 0.051 | 0.047 | −0.024 | 0.213 | 0.888 |
| LDLc | 0.241 | 0.032 | 0.171 | −0.036 | 0.195 | 2.188 | 0.094 |
| ADIPOQ (rs1501299) | ADIPOQ (rs17846866) | ADIPOQ (rs2241766) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AA (n = 18) | AC (n = 70) | CC (n = 22) | p-Value | GG (n = 08) | GT (n = 19) | TT (n = 83) | p-value | GG (n = 12) | GT (n = 21) | TT (n = 77) | p-Value | |
| Age | 33.50 ± 6.89 | 32.75 ± 5.83 | 33.5 ± 4.64 | 0.24 | 33.25 ± 5.42 | 30.42 ± 5.77 | 33.60 ± 5.7 | 0.98 | 34.08 ± 5.30 | 32.85 ± 5.40 | 32.90 ± 5.98 | 0.78 |
| Weight | 81.08 ± 10.71 | 78.58 ± 13.15 | 79.85 ± 10.17 | 0.28 | 82.07 ± 14.59 | 75.27 ± 11.83 | 79.88 ± 11.91 | 0.74 | 80.4 ± 9.50 | 74.47 ± 14.17 | 79.98 ± 12.43 | 0.37 |
| BMI | 32.72 ± 3.99 | 31.45 ± 4.7 | 32.17 ± 3.51 | 0.25 | 31.91 ± 4.71 | 30.19 ± 4.43 | 32.16 ± 4.2 | 0.89 | 32.70 ± 2.70 | 30.77 ± 4.68 | 31.94 ± 4.49 | 0.15 |
| SBP | 120.38 ± 11.66 | 119.94 ± 9.05 | 120.6 ± 11.47 | 0.22 | 119.37 ± 9.87 | 119.26 ± 8.35 | 120.43 ± 10.39 | 0.53 | 120.0 ± 10.6 | 120.4 ± 9.91 | 120.1 ± 9.98 | 0.95 |
| DBP | 74.44 ± 2.83 | 74.58 ± 3.14 | 73.81 ± 3.99 | 0.25 | 75.5 ± 3.35 | 74.73 ± 3.41 | 74.22 ± 3.24 | 0.95 | 74.20 ± 3.20 | 74.61 ± 3.84 | 74.37 ± 3.14 | 0.49 |
| FBG | 5.83 ± 1.14 | 6.0 ± 1.25 | 5.79 ± 0.91 | 0.25 | 5.9 ± 0.88 | 6.17 ± 1.08 | 5.88 ± 1.21 | 0.51 | 5.50 ± 1.1 | 5.88 ± 0.93 | 6.0 ± 1.23 | 0.31 |
| PPBG | 7.87 ± 1.55 | 10.17 ± 19.54 | 7.75 ± 1.2 | 0.01 * | 7.17 ± 0.81 | 16.76 ± 36.61 | 7.81 ± 1.58 | 0.01 * | 7.4 ± 1.5 | 8.43 ± 1.62 | 9.85 ± 18.65 | 0.01 * |
| GCT | 8.95 ± 0.77 | 9.37 ± 1.08 | 9.58 ± 1.05 | 0.26 | 9.1 ± 1.12 | 9.39 ± 0.98 | 9.35 ± 1.04 | 0.90 | 9 ± 0.9 | 9.34 ± 0.61 | 9.5 ± 7.14 | 0.01 * |
| OGTT (F) | 7.27 ± 2.85 | 6.5 ± 1.96 | 6.19 ± 1.8 | 0.06 | 7.53 ± 2.23 | 5.61 ± 1.17 | 6.72 ± 2.21 | 0.01 * | 6 ± 1.9 | 7.2 ± 2.04 | 6.49 ± 2.14 | 0.91 |
| OGTT (1) | 10.02 ± 1.47 | 10.69 ± 1.93 | 10.70 ± 1.23 | 0.04 * | 10.68 ± 0.95 | 10.50 ± 1.69 | 10.61 ± 1.83 | 0.14 | 10.3 ± 1.8 | 10.90 ± 1.24 | 10.55 ± 1.85 | 0.11 |
| OGTT (2) | 9.11 ± 0.96 | 9.05 ± 1.79 | 9.21 ± 1.99 | 0.01 * | 9.50 ± 1.05 | 8.23 ± 1.69 | 9.25 ± 1.73 | 0.30 | 9.1 ± 1.2 | 9.3 ± 2.06 | 9.02 ± 1.69 | 0.16 |
| OGTT (3) | 5.9 ± 1.5 | 7.96 ± 7.03 | 10.58 ± 10.08 | 0.02 * | 5.62 ± 1.18 | 9.20 ± 0.45 | 8.16 ± 7.49 | 0.01 * | 8.1 ± 7.3 | 9.10 ± 7.96 | 7.9 ± 7.16 | 0.85 |
| Hb1Ac | 5.30 ± 0.35 | 5.40 ± 0.29 | 5.48 ± 0.45 | 0.52 | 5.27 ± 0.3 | 5.46 ± 0.70 | 5.41 ± 0.30 | 0.01 * | 5.5 ± 0.3 | 5.4 ± 0.43 | 5.39 ± 0.3 | 0.09 |
| TC | 6.27 ± 1.44 | 5.62 ± 1.23 | 5.73 ± 1.11 | 0.01 * | 5.76 ± 1.99 | 6.0 ± 0.36 | 5.69 ± 1.21 | 0.01 * | 5.7 ± 1.3 | 5.71 ± 0.87 | 5.76 ± 1.34 | <0.03 * |
| TG | 2.28 ± 0.95 | 2.32 ± 2.16 | 2.43 ± 1.80 | 0.96 | 2.25 ± 1.15 | 1.89 ± 0.83 | 2.44 ± 2.17 | 0.01 * | 2.0 ± 0.6 | 2.3 ± 1.4 | 2.3 ± 2.18 | 0.01 * |
| Hdlc | 1.02 ± 0.41 | 0.89 ± 0.39 | 1.08 ± 0.4 | 0.01 * | 1.01 ± 0.54 | 0.97 ± 0.36 | 0.94 ± 0.40 | 0.39 | 0.9 ± 0.3 | 0.95 ± 0.41 | 0.96 ± 0.41 | 0.45 |
| Ldlc | 3.58 ± 1.01 | 3.86 ± 0.98 | 4.03 ± 0.8 | 0.50 | 3.93 ± 0.91 | 3.69 ± 0.83 | 3.88 ± 0.99 | 0.64 | 4.1 ± 0.9 | 4.08 ± 1.09 | 3.74 ± 0.90 | 0.53 |
| S. No | rs1501299 | rs17846866 | rs2241766 | Freq | OR (95% CI) | p-Value |
|---|---|---|---|---|---|---|
| 1 | A | T | T | 0.4275 | 1.00 | - |
| 2 | C | T | T | 0.3087 | 3.01 (1.73–5.25) | p = 0.004 |
| 3 | A | T | G | 0.0852 | 2.26 (1.14–4.48) | p = 0.02 |
| 4 | A | G | T | 0.0718 | 1.34 (0.56–3.17) | p = 0.51 |
| 5 | C | G | T | 0.0466 | 4.06 (1.24–13.31) | p = 0.02 |
| 6 | C | T | G | 0.0354 | 16.66 (2.13–130.42) | p = 0.008 |
| 7 | C | G | G | 0.0207 | 2.57 (0.66–10.01) | p = 0.18 |
| Subjects | L1 | L2 | D′ | r^2 |
|---|---|---|---|---|
| GDM cases | rs2241766 | rs1501299 | 0.212 | 0.012 |
| GDM cases | rs2241766 | rs17846866 | 0.028 | 0.0 |
| GDM cases | rs1501299 | rs17846866 | 0.087 | 0.001 |
| non-GDM | rs2241766 | rs1501299 | 0.19 | 0.001 |
| non-GDM | rs2241766 | rs17846866 | 0.094 | 0.006 |
| Model No | Genes Included in Best Combination in Each Model | Training Accuracy | Testing Accuracy | CVC | p-Value |
|---|---|---|---|---|---|
| 1 | rs1501299 (R1) | 0.6727 | 0.6727 | 10/10 | <0.001 |
| 2 | rs1501299, rs2241766 (R1, R3) | 0.6869 | 0.6682 | 10/10 | <0.001 |
| 3 | rs1501299, rs17846866, rs2241766 (R1, R2, R3) | 0.698 | 0.65 | 10/10 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammary, A.F.; Ansar, S.; Farzan, R.; Alsobaie, S.F.; Alageel, A.A.; Al-Hakeem, M.M.; Ali Khan, I. Dissecting the Molecular Role of ADIPOQ SNPs in Saudi Women Diagnosed with Gestational Diabetes Mellitus. Biomedicines 2023, 11, 1289. https://doi.org/10.3390/biomedicines11051289
Alshammary AF, Ansar S, Farzan R, Alsobaie SF, Alageel AA, Al-Hakeem MM, Ali Khan I. Dissecting the Molecular Role of ADIPOQ SNPs in Saudi Women Diagnosed with Gestational Diabetes Mellitus. Biomedicines. 2023; 11(5):1289. https://doi.org/10.3390/biomedicines11051289
Chicago/Turabian StyleAlshammary, Amal F., Sabah Ansar, Raed Farzan, Sarah F. Alsobaie, Arwa A. Alageel, Malak Mohammed Al-Hakeem, and Imran Ali Khan. 2023. "Dissecting the Molecular Role of ADIPOQ SNPs in Saudi Women Diagnosed with Gestational Diabetes Mellitus" Biomedicines 11, no. 5: 1289. https://doi.org/10.3390/biomedicines11051289
APA StyleAlshammary, A. F., Ansar, S., Farzan, R., Alsobaie, S. F., Alageel, A. A., Al-Hakeem, M. M., & Ali Khan, I. (2023). Dissecting the Molecular Role of ADIPOQ SNPs in Saudi Women Diagnosed with Gestational Diabetes Mellitus. Biomedicines, 11(5), 1289. https://doi.org/10.3390/biomedicines11051289