Exploring the Role of Hypoxia-Inducible Carbonic Anhydrase IX (CAIX) in Circulating Tumor Cells (CTCs) of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Anoikis Screening of Suspension Cultured Cell Lines
2.3. Detection of Breast Cancer Stem Cell Populations
2.4. Immunoblotting
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results
3.1. Non-Adherent Suspension Condition Enriches for a Circulating Cancer Stem Cell Population
3.2. Identification of a Molecular Signature of Circulating Breast Cancer Stem Cells
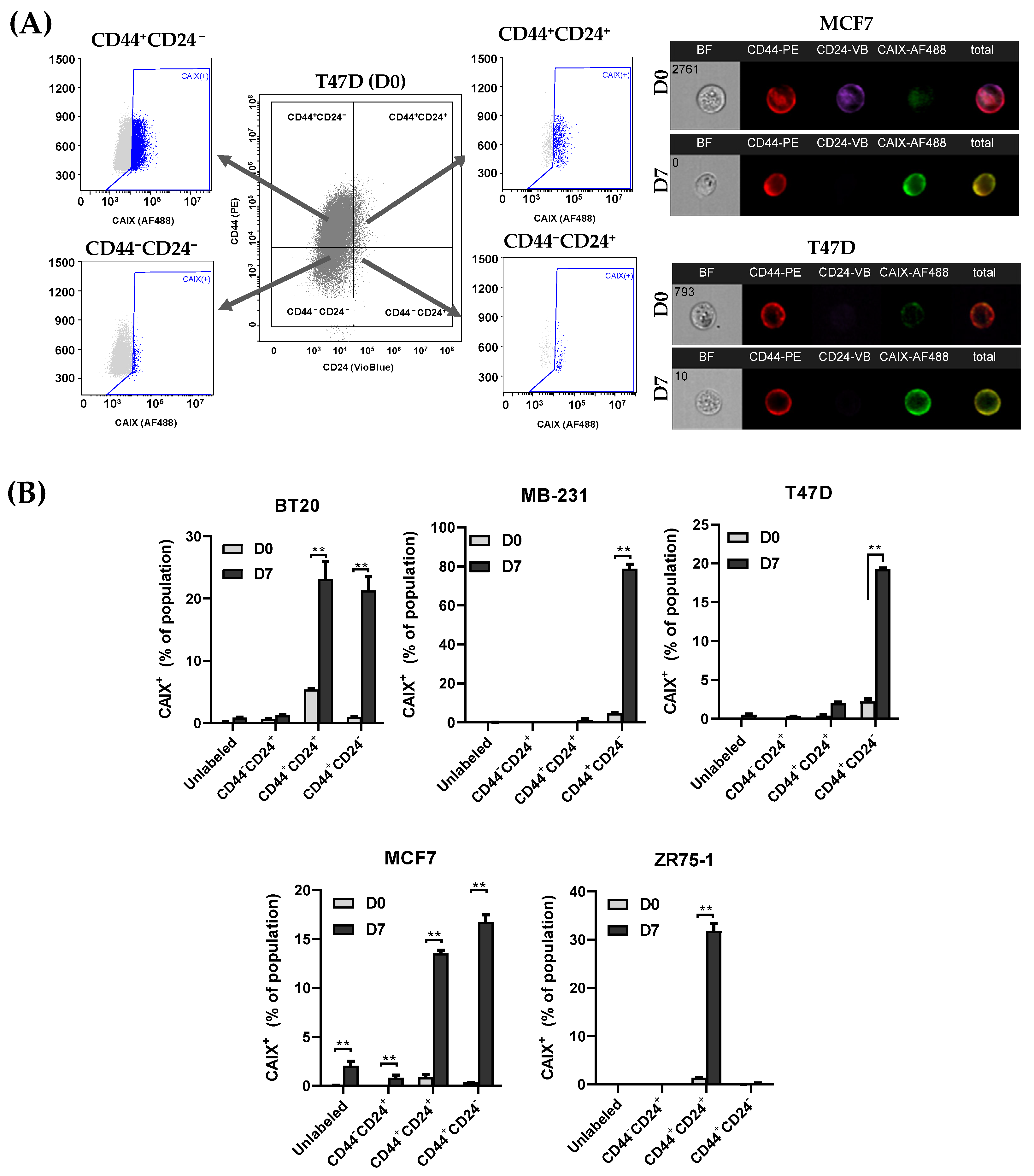
3.3. CAIX Expression Is Upregulated in the CD44+ Population, and the Inhibition of CAIX Reduces the cCSC Population
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Velasco-Velázquez, M.A.; Popov, V.M.; Lisanti, M.P.; Pestell, R.G. The role of breast cancer stem cells in metastasis and therapeutic implications. Am. J. Pathol. 2011, 179, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Maly, V.; Maly, O.; Kolostova, K.; Bobek, V. Circulating Tumor Cells in Diagnosis and Treatment of Lung Cancer. In Vivo 2019, 33, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-H.; Hsieh, J.C.-H.; Wu, T.M.-H.; Yeh, T.-S.; Wang, H.-M.; Lin, Y.-C.; Chen, J.-S.; Lee, C.-L.; Huang, W.-K.; Hung, T.-M.; et al. Baseline circulating stem-like cells predict survival in patients with metastatic breast Cancer. BMC Cancer 2019, 19, 1167. [Google Scholar] [CrossRef]
- Yan, W.T.; Cui, X.; Chen, Q.; Li, Y.F.; Cui, Y.H.; Wang, Y.; Jiang, J. Circulating tumor cell status monitors the treatment responses in breast cancer patients: A meta-analysis. Sci. Rep. 2017, 7, 43464. [Google Scholar] [CrossRef]
- Toss, A.; Mu, Z.; Fernandez, S.; Cristofanilli, M. CTC enumeration and characterization: Moving toward personalized medicine. Ann. Transl. Med. 2014, 2, 108. [Google Scholar] [CrossRef]
- Raimondi, C.; Nicolazzo, C.; Gradilone, A. Circulating tumor cells isolation: The “post-EpCAM era”. Chin. J. Cancer Res. 2015, 27, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.E.; Ring, A.; Porras, T.; Kaur, P.; Forte, V.A.; Mineyev, N.; Tripathy, D.; Press, M.F.; Campo, D. RNA-Seq of Circulating Tumor Cells in Stage II-III Breast Cancer. Ann. Surg. Oncol. 2018, 25, 2261–2270. [Google Scholar] [CrossRef] [PubMed]
- Tayoun, T.; Faugeroux, V.; Oulhen, M.; Aberlenc, A.; Pawlikowska, P.; Farace, F. CTC-Derived Models: A Window into the Seeding Capacity of Circulating Tumor Cells (CTCs). Cells 2019, 8, 1145. [Google Scholar] [CrossRef]
- Lock, F.E.; McDonald, P.C.; Lou, Y.; Serrano, I.; Chafe, S.C.; Ostlund, C.; Aparicio, S.; Winum, J.Y.; Supuran, C.T.; Dedhar, S. Targeting carbonic anhydrase IX depletes breast cancer stem cells within the hypoxic niche. Oncogene 2012, 32, 5210. [Google Scholar] [CrossRef]
- Rota, L.M.; Lazzarino, D.A.; Ziegler, A.N.; LeRoith, D.; Wood, T.L. Determining mammosphere-forming potential: Application of the limiting dilution analysis. J. Mammary Gland. Biol. Neoplasia 2012, 17, 119–123. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, D.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014, 2, 78–91. [Google Scholar] [CrossRef]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [PubMed]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef]
- Nakamura, K.; Terai, Y.; Tanabe, A.; Ono, Y.J.; Hayashi, M.; Maeda, K.; Fujiwara, S.; Ashihara, K.; Nakamura, M.; Tanaka, Y.; et al. CD24 expression is a marker for predicting clinical outcome and regulates the epithelial-mesenchymal transition in ovarian cancer via both the Akt and ERK pathways. Oncol. Rep. 2017, 37, 3189–3200. [Google Scholar] [CrossRef] [PubMed]
- Lubeseder-Martellato, C.; Hidalgo-Sastre, A.; Hartmann, C.; Alexandrow, K.; Kamyabi-Moghaddam, Z.; Sipos, B.; Wirth, M.; Neff, F.; Reichert, M.; Heid, I.; et al. Membranous CD24 drives the epithelial phenotype of pancreatic cancer. Oncotarget 2016, 7, 49156. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Twomey, J.D.; Zhang, B. Circulating Tumor Cells Develop Resistance to TRAIL-Induced Apoptosis Through Autophagic Removal of Death Receptor 5: Evidence from an In Vitro Model. Cancers 2019, 11, 94. [Google Scholar] [CrossRef]
- Twomey, J.D.; Zhao, L.; Luo, S.; Xu, Q.; Zhang, B. Tubulin couples death receptor 5 to regulate apoptosis. Oncotarget 2018, 9, 36804–36815. [Google Scholar] [CrossRef] [PubMed]
- Eslami-S, Z.; Cortés-Hernández, L.E.; Thomas, F.; Pantel, K.; Alix-Panabières, C. Functional analysis of circulating tumour cells: The KEY to understand the biology of the metastatic cascade. Br. J. Cancer 2022, 127, 800–810. [Google Scholar] [CrossRef]
- Gencoglu, M.F.; Barney, L.E.; Hall, C.L.; Brooks, E.A.; Schwartz, A.D.; Corbett, D.C.; Stevens, K.R.; Peyton, S.R. Comparative Study of Multicellular Tumor Spheroid Formation Methods and Implications for Drug Screening. ACS Biomater. Sci. Eng. 2018, 4, 410–420. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. ASSAY Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S.A. Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Hugo, H.J.; Gunasinghe, N.; Hollier, B.G.; Tanaka, T.; Blick, T.; Toh, A.; Hill, P.; Gilles, C.; Waltham, M.; Thompson, E.W. Epithelial requirement for in vitro proliferation and xenograft growth and metastasis of MDA-MB-468 human breast cancer cells: Oncogenic rather than tumor-suppressive role of E-cadherin. Breast Cancer Res. 2017, 19, 86. [Google Scholar] [CrossRef] [PubMed]
- McSweeney, K.M.; Bozza, W.P.; Alterovitz, W.-L.; Zhang, B. Transcriptomic profiling reveals p53 as a key regulator of doxorubicin-induced cardiotoxicity. Cell Death Discov. 2019, 5, 102. [Google Scholar] [CrossRef]
- Ciccone, V.; Terzuoli, E.; Donnini, S.; Giachetti, A.; Morbidelli, L.; Ziche, M. Stemness marker ALDH1A1 promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF signalling in MCF-7 breast cancer cells. J. Exp. Clin. Cancer Res. 2018, 37, 311. [Google Scholar] [CrossRef]
- Alves, W.E.F.M.; Bonatelli, M.; Dufloth, R.; Kerr, L.M.; Carrara, G.F.A.; da Costa, R.F.A.; Scapulatempo-Neto, C.; Tiezzi, D.; da Costa Vieira, R.A.; Pinheiro, C. CAIX is a predictor of pathological complete response and is associated with higher survival in locally advanced breast cancer submitted to neoadjuvant chemotherapy. BMC Cancer 2019, 19, 1173. [Google Scholar] [CrossRef] [PubMed]
- Güttler, A.; Theuerkorn, K.; Riemann, A.; Wichmann, H.; Kessler, J.; Thews, O.; Bache, M.; Vordermark, D. Cellular and radiobiological effects of carbonic anhydrase IX in human breast cancer cells. Oncol. Rep. 2019, 41, 2585–2594. [Google Scholar] [CrossRef] [PubMed]
- Peiró, C.H.F.; Perez, M.M.; de Aquino, G.S.A.; Encinas, J.F.A.; Sousa, L.V.d.A.; da Veiga, G.L.; del Giglio, A.; Fonseca, F.L.A.; da Costa Aguiar Alves, B. Diagnostic potential of hypoxia-induced genes in liquid biopsies of breast cancer patients. Sci. Rep. 2021, 11, 8724. [Google Scholar] [CrossRef]
- Meehan, J.; Ward, C.; Turnbull, A.; Bukowski-Wills, J.; Finch, A.J.; Jarman, E.J.; Xintaropoulou, C.; Martinez-Perez, C.; Gray, M.; Pearson, M.; et al. Inhibition of pH regulation as a therapeutic strategy in hypoxic human breast cancer cells. Oncotarget 2017, 8, 42857–42875. [Google Scholar] [CrossRef]
- Chen, Z.; Ai, L.; Mboge, M.Y.; Tu, C.; McKenna, R.; Brown, K.D.; Heldermon, C.D.; Frost, S.C. Differential expression and function of CAIX and CAXII in breast cancer: A comparison between tumorgraft models and cells. PLoS ONE 2018, 13, e0199476. [Google Scholar] [CrossRef]
- Tafreshi, N.K.; Lloyd, M.C.; Proemsey, J.B.; Bui, M.M.; Kim, J.; Gillies, R.J.; Morse, D.L. Evaluation of CAIX and CAXII Expression in Breast Cancer at Varied O2 Levels: CAIX is the Superior Surrogate Imaging Biomarker of Tumor Hypoxia. Mol. Imaging Biol. 2016, 18, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Emami Nejad, A.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Haghjooy Javanmard, S.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef] [PubMed]
- McDonald, P.C.; Chia, S.; Bedard, P.L.; Chu, Q.; Lyle, M.; Tang, L.; Singh, M.; Zhang, Z.; Supuran, C.T.; Renouf, D.J.; et al. A Phase 1 Study of SLC-0111, a Novel Inhibitor of Carbonic Anhydrase IX, in Patients With Advanced Solid Tumors. Am. J. Clin. Oncol. 2020, 43, 484–490. [Google Scholar] [CrossRef]
- Peppicelli, S.; Andreucci, E.; Ruzzolini, J.; Bianchini, F.; Nediani, C.; Supuran, C.T.; Calorini, L. The Carbonic Anhydrase IX inhibitor SLC-0111 as emerging agent against the mesenchymal stem cell-derived pro-survival effects on melanoma cells. J. Enzym. Inhib. Med. Chem. 2020, 35, 1185–1193. [Google Scholar] [CrossRef]
- Tellez-Gabriel, M.; Cochonneau, D.; Cadé, M.; Jubellin, C.; Heymann, M.F.; Heymann, D. Circulating Tumor Cell-Derived Pre-Clinical Models for Personalized Medicine. Cancers 2018, 11, 19. [Google Scholar] [CrossRef]
- Zhang, L.; Ridgway, L.D.; Wetzel, M.D.; Ngo, J.; Yin, W.; Kumar, D.; Goodman, J.C.; Groves, M.D.; Marchetti, D. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci. Transl. Med. 2013, 5, 180ra148. [Google Scholar] [CrossRef] [PubMed]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bäuerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef]
- Cayrefourcq, L.; Mazard, T.; Joosse, S.; Solassol, J.; Ramos, J.; Assenat, E.; Schumacher, U.; Costes, V.; Maudelonde, T.; Pantel, K.; et al. Establishment and characterization of a cell line from human circulating colon cancer cells. Cancer Res. 2015, 75, 892–901. [Google Scholar] [CrossRef]
- Bade, R.M.; Schehr, J.L.; Emamekhoo, H.; Gibbs, B.K.; Rodems, T.S.; Mannino, M.C.; Desotelle, J.A.; Heninger, E.; Stahlfeld, C.N.; Sperger, J.M.; et al. Development and initial clinical testing of a multiplexed circulating tumor cell assay in patients with clear cell renal cell carcinoma. Mol. Oncol. 2021, 15, 2330–2344. [Google Scholar] [CrossRef]
- Hamilton, G.; Rath, B. Role of circulating tumor cell spheroids in drug resistance. Cancer Drug Resist. 2019, 2, 762–772. [Google Scholar] [CrossRef] [PubMed]
- Fabisiewicz, A.; Grzybowska, E. CTC clusters in cancer progression and metastasis. Med. Oncol. 2017, 34, 12. [Google Scholar] [CrossRef]
- Krol, I.; Schwab, F.D.; Carbone, R.; Ritter, M.; Picocci, S.; De Marni, M.L.; Stepien, G.; Franchi, G.M.; Zanardi, A.; Rissoglio, M.D.; et al. Detection of clustered circulating tumour cells in early breast cancer. Br. J. Cancer 2021, 125, 23–27. [Google Scholar] [CrossRef]
- Franco, S.; Szczesna, K.; Iliou, M.S.; Al-Qahtani, M.; Mobasheri, A.; Kobolák, J.; Dinnyés, A. In vitro models of cancer stem cells and clinical applications. BMC Cancer 2016, 16, 738. [Google Scholar] [CrossRef]
- Potter, C.; Harris, A.L. Hypoxia inducible carbonic anhydrase IX, marker of tumour hypoxia, survival pathway and therapy target. Cell Cycle 2004, 3, 164–167. [Google Scholar] [CrossRef]
- İlie, M.; Mazure, N.M.; Hofman, V.; Ammadi, R.E.; Ortholan, C.; Bonnetaud, C.; Havet, K.; Venissac, N.; Mograbi, B.; Mouroux, J.; et al. High levels of carbonic anhydrase IX in tumour tissue and plasma are biomarkers of poor prognostic in patients with non-small cell lung cancer. Br. J. Cancer 2010, 102, 1627–1635. [Google Scholar] [CrossRef]
- Ong, C.H.C.; Lee, D.Y.; Lee, B.; Li, H.; Lim, J.C.T.; Lim, J.X.; Yeong, J.P.S.; Lau, H.Y.; Thike, A.A.; Tan, P.H.; et al. Hypoxia-regulated carbonic anhydrase IX (CAIX) protein is an independent prognostic indicator in triple negative breast cancer. Breast Cancer Res. 2022, 24, 38. [Google Scholar] [CrossRef]
- Kallergi, G.; Markomanolaki, H.; Giannoukaraki, V.; Papadaki, M.A.; Strati, A.; Lianidou, E.S.; Georgoulias, V.; Mavroudis, D.; Agelaki, S. Hypoxia-inducible factor-1alpha and vascular endothelial growth factor expression in circulating tumor cells of breast cancer patients. Breast Cancer Res. 2009, 11, R84. [Google Scholar] [CrossRef]
- Ward, C.; Meehan, J.; Gray, M.; Kunkler, I.H.; Langdon, S.P.; Argyle, D.J. Carbonic Anhydrase IX (CAIX), Cancer, and Radiation Responsiveness. Metabolites 2018, 8, 13. [Google Scholar] [CrossRef]
- Chamie, K.; Donin, N.M.; Klöpfer, P.; Bevan, P.; Fall, B.; Wilhelm, O.; Störkel, S.; Said, J.; Gambla, M.; Hawkins, R.E.; et al. Adjuvant Weekly Girentuximab Following Nephrectomy for High-Risk Renal Cell Carcinoma: The ARISER Randomized Clinical Trial. JAMA Oncol. 2017, 3, 913–920. [Google Scholar] [CrossRef]
- Petrul, H.M.; Schatz, C.A.; Kopitz, C.C.; Adnane, L.; McCabe, T.J.; Trail, P.; Ha, S.; Chang, Y.S.; Voznesensky, A.; Ranges, G.; et al. Therapeutic mechanism and efficacy of the antibody-drug conjugate BAY 79-4620 targeting human carbonic anhydrase 9. Mol. Cancer Ther. 2012, 11, 340–349. [Google Scholar] [CrossRef]
- Chafe, S.C.; McDonald, P.C.; Saberi, S.; Nemirovsky, O.; Venkateswaran, G.; Burugu, S.; Gao, D.; Delaidelli, A.; Kyle, A.H.; Baker, J.H.E.; et al. Targeting Hypoxia-Induced Carbonic Anhydrase IX Enhances Immune-Checkpoint Blockade Locally and Systemically. Cancer Immunol. Res. 2019, 7, 1064–1078. [Google Scholar] [CrossRef]
- Juhász, P.; Hasulyó, D.; Bedekovics, J.; Beke, L.; Kacsala, N.; Török, M.; Méhes, G. Carbonic Anhydrase IX (CAIX) Expressing Hypoxic Micro-environment Hampers CD8+ Immune Cell Infiltrate in Breast Carcinoma. Appl. Immunohistochem. Mol. Morphol. 2023, 31, 26–32. [Google Scholar] [CrossRef]

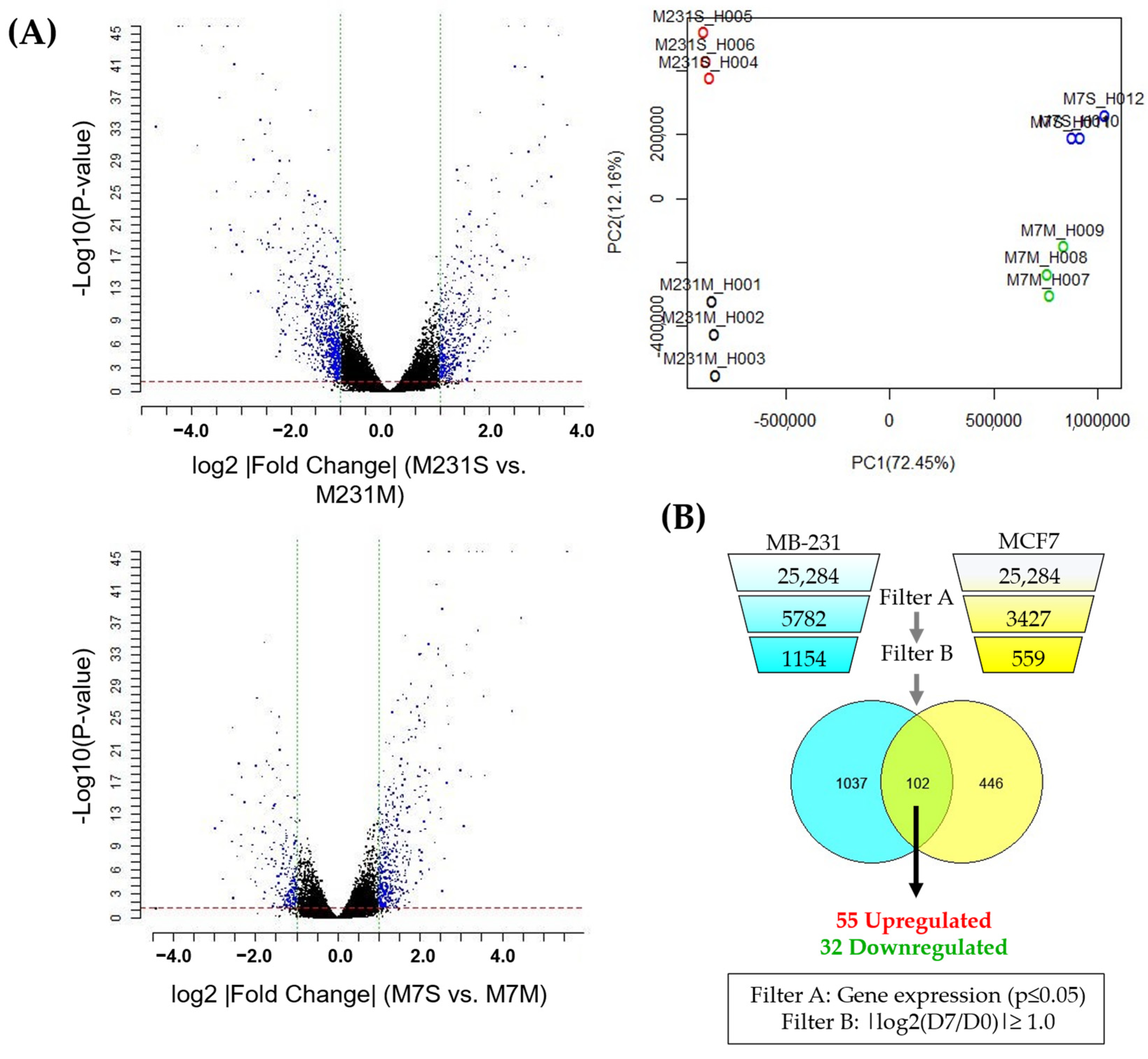



| Gene Symbol | Description | log2 (Ratio) (D7/D0) | Uniprot Subcellular Localization | |
|---|---|---|---|---|
| MB-231 | MCF7 | |||
| CA9 | Carbonic anhydrase IX | 1.380 | 5.581 |
|
| DTNA | Dystrobrevin, alpha | 1.453 | 1.696 |
|
| ENO2 | Enolase 2 (gamma, neuronal) | 1.481 | 2.485 |
|
| GJA1 | Gap junction protein, alpha 1, 43 kDa; Connexin-43 | 1.470 | 1.044 |
|
| IL1RAP | Interleukin 1 receptor accessory protein | 1.176 | 1.060 |
|
| RGS2 | Regulator of G-protein signaling 2, 24 kDa | 1.268 | 3.077 |
|
| RND1 | Rho family GTPase 1 | 1.073 | 1.080 |
|
| SLC2A1/GLUT1 | Solute carrier family 2 (facilitated glucose transporter), member 1 | 1.163 | 2.025 |
|
| SLC6A6 | Solute carrier family 6 (neurotransmitter transporter, taurine), member 6 | 1.493 | 1.696 |
|
| UBE2C | Ubiquitin-conjugating enzyme E2C | 1.486 | 1.274 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Twomey, J.D.; Zhang, B. Exploring the Role of Hypoxia-Inducible Carbonic Anhydrase IX (CAIX) in Circulating Tumor Cells (CTCs) of Breast Cancer. Biomedicines 2023, 11, 934. https://doi.org/10.3390/biomedicines11030934
Twomey JD, Zhang B. Exploring the Role of Hypoxia-Inducible Carbonic Anhydrase IX (CAIX) in Circulating Tumor Cells (CTCs) of Breast Cancer. Biomedicines. 2023; 11(3):934. https://doi.org/10.3390/biomedicines11030934
Chicago/Turabian StyleTwomey, Julianne D., and Baolin Zhang. 2023. "Exploring the Role of Hypoxia-Inducible Carbonic Anhydrase IX (CAIX) in Circulating Tumor Cells (CTCs) of Breast Cancer" Biomedicines 11, no. 3: 934. https://doi.org/10.3390/biomedicines11030934
APA StyleTwomey, J. D., & Zhang, B. (2023). Exploring the Role of Hypoxia-Inducible Carbonic Anhydrase IX (CAIX) in Circulating Tumor Cells (CTCs) of Breast Cancer. Biomedicines, 11(3), 934. https://doi.org/10.3390/biomedicines11030934







