Abstract
Hearing loss is the most frequent sensorineural disorder, affecting approximately 1:1000 newborns. Hereditary forms (HHL) represent 50–60% of cases, highlighting the relevance of genetic testing in deaf patients. HHL is classified as non-syndromic (NSHL—70% of cases) or syndromic (SHL—30% of cases). In this study, a multistep and integrative approach aimed at identifying the molecular cause of HHL in 102 patients, whose GJB2 analysis already showed a negative result, is described. In NSHL patients, multiplex ligation probe amplification and long-range PCR analyses of the STRC gene solved 13 cases, while whole exome sequencing (WES) identified the genetic diagnosis in 26 additional ones, with a total detection rate of 47.6%. Concerning SHL, WES detected the molecular cause in 55% of cases. Peculiar findings are represented by the identification of four subjects displaying a dual molecular diagnosis and eight affected by non-syndromic mimics, five of them presenting Usher syndrome type 2. Overall, this study provides a detailed characterisation of the genetic causes of HHL in the Italian population. Furthermore, we highlighted the frequency of Usher syndrome type 2 carriers in the Italian population to pave the way for a more effective implementation of diagnostic and follow-up strategies for this disease.
1. Introduction
The primary aim of medical genetics is the identification of the molecular cause of rare diseases, which, despite being singularly rare, may affect up to 8% of newborns [1]. However, recognising the specific genetic condition that affects a patient is anything but easy, since the number of rare disorders is exceptionally high and many of them are characterised by a significant clinical and genetic heterogeneity, as in hereditary hearing loss (HHL).
Congenital hearing loss is the most frequent sensorineural disorder, with a prevalence of approximately 1:1000 live births [2]. In developed countries, 50% to 60% of all cases are due to genetic causes (https://www.cdc.gov/ncbddd/hearingloss/genetics.html, accessed on 25 January 2023) and are referred to as HHL. HHL can be classified as non-syndromic (NSHL), when deafness is the only present sign, or syndromic (SHL), when hearing loss (HL) is accompanied by other clinical features [3]. NSHL accounts for approximately 70% of HHL cases and, so far, more than 120 genes have been associated with this condition. Among them, 51 are causative of autosomal dominant (AD) forms, 77 of autosomal recessive (AR) forms, and five of X-linked (XL) forms; notably, some genes are responsible for both AD and AR NSHL, additionally highlighting the complexity of inheritance patterns associated with this condition (Hereditary Hearing Loss Homepage; https://hereditaryhearingloss.org/, last accessed on 25 January 2023). According to the literature, the major players involved in NSHL are the GJB2 gene, which accounts for approximately 50% of all AR cases, and STRC, which is increasingly recognized as the second-most significant contributor to AR NSHL [4]. On the other hand, SHL represents 30% of cases of HHL and about 400 syndromes that include this phenotype have been reported in the literature [5]. Some of them account for a substantial fraction of cases, such as Pendred, Usher, Alport, and Waardenburg syndromes [6], whereas others are very rare, such as Perrault and Barakat syndromes [7,8].
In this entangled genetic landscape, defining the molecular cause of HHL poses a real challenge to diagnosticians, since there are further peculiar circumstances to take into consideration. For instance, it has recently been demonstrated that the presence of a dual molecular diagnosis in HHL patients is a particularly frequent event that needs to be suspected whenever signs and symptoms do not fit a known syndromic pattern or whenever there is a high intrafamilial clinical variability [9]. Another occurrence that complicates HHL diagnosis is represented by non-syndromic mimics: they are defined as syndromic forms of HL that masquerade as NSHL, since additional clinical features either are extremely subtle or develop later in life [3,10].
In this light, the optimisation of high-throughput sequencing technologies, such as whole exome sequencing (WES), together with complementary genetic diagnostic approaches, such as multiplex ligation probe amplification(MLPA) and long-range PCR (LR-PCR) have enormously improved molecular and clinical geneticists’ ability to define the precise molecular diagnosis of HHL patients. Indeed, the possibility to simultaneously analyse thousands of genes with integrated approaches is the cornerstone to face HHL’s extensive genetic and clinical heterogeneity.
In this context, we hereby report the results of our study aimed at the identification of the genetic cause of HHL in 102 patients recruited in the last 18 months and analysed through a multistep and integrative approach. The final goal of this study is to provide a detailed characterisation of the genetic causes of HHL other than GJB2 mutations in the Italian population, with particular attention to the identification of peculiar scenarios such as the presence of dual molecular diagnoses or non-syndromic mimics.
2. Materials and Methods
2.1. Ethical Statement
All the analyses were performed following the relevant guidelines and regulations. Written informed consent was obtained from all participants or, in the case of underage patients, their legal guardians. The study was conducted in accordance with the tenets of the Helsinki Declaration and was approved by the Ethics Committee of the I.R.C.C.S. “Burlo Garofolo” of Trieste.
2.2. Study Design
In the last 18 months, 102 patients affected by NSHL and SHL and negative on GJB2, GJB6, and MT-RNR1 first-tier screenings were referred to the Medical Genetics Unit of the I.R.C.C.S. “Burlo Garofolo” (Trieste, Italy), the Medical Genetics Unit of the “AUSL della Romagna” (Cesena, Italy), the Medical Genetics Unit of the “Azienda USL—I.R.C.C.S. di Reggio Emilia” (Reggio Emilia, Italy), the Medical Genetics Unit of the I.R.C.C.S. Azienda Ospedaliero-Universitaria “Policlinico di Sant’Orsola” (Bologna, Italy), or the Medical Genetics Unit of the “Sant’Anna” University Hospital (Ferrara, Italy).
The enrolled subjects were analysed through a multistep approach that comprises the following steps: (1) a detailed clinical evaluation to distinguish NSHL patients from SHL ones; (2) MLPA analysis of STRC-CATSPER2 and OTOA in NSHL patients; (3) LR-PCR and STRC sequencing in patients carrying a heterozygous deletion of STRC and an audiometric pattern suggestive of Deafness, autosomal recessive 16; (4) WES analysis in patients negative to steps 2 and 3 and in SHL subjects.
Regarding the clinical evaluation, all participants were deeply characterised through a detailed anamnesis, a dysmorphological examination, an audiological evaluation, and further investigations. In particular, a complete familial anamnesis was collected in order to identify possible other affected family members and a personal medical history was obtained to highlight potential confounding factors (e.g., infections, trauma, or other non-genetic causes of HL). A careful physical examination was carried out to identify possible dysmorphic features, with particular attention to the overall appearance and to facial, ectodermal, skeletal, and genital features. All patients underwent pure tone audiometry testing (PTA) or auditory brainstem response (ABR), according to the proband’s age, to define the degree of HL [11]. Ancillary clinical tests included brain magnetic resonance imaging (MRI) and computed tomography (CT), thyroid function assessment, abdominal ultrasound, and neurological, ophthalmological, and cardiological evaluations.
2.3. DNA Extraction and Quality Control
Genomic DNA was extracted from peripheral whole-blood samples of patients and both their parents using the QIAsymphony® SP instrument with QIAsymphony® Midi Kit (Qiagen, Venlo, The Netherlands), following the manufacturer’s instructions. The DNA quality was verified through 1% agarose gel electrophoresis and the DNA concentration was measured using the Nanodrop ND 1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA).
2.4. MLPA
MLPA analysis for the identification of deletions and duplications involving the STRC-CATSPER2 and OTOA genes was performed using SALSA® MLPA® probe mixes P461-B1 DIS (MRC-Holland, Amsterdam, the Netherlands. The kit contains different probes spanning the aforementioned genes. In particular, seven probes for the STRC gene (covering exons 19, 20, 23, 24, 25, and 28), five probes for the CATSPER2 gene (covering exons 1, 2, 4, and 7), and ten probes for the OTOA gene (covering exons 2, 5, 7, 8, 11, 12, 16, 17, 18, and 20) are present. Multiple flanking probes are also included in the 15q15.3 and 16q12.2 regions, indicating the extent of possible copy number variations (CNVs). Furthermore, four STRC pseudogene (p-STRC) probes are present to identify possible gene conversions. Samples were performed according to the manufacturer’s instructions. Briefly, 50–100 ng of DNA was denatured and hybridised overnight with the SALSA® probe mix; afterwards, samples were treated with DNA ligase and polymerase chain reaction (PCR) amplification was carried out with specific fluorescent-labelled PCR primers. Amplified product fragment analysis was performed on an ABI 3500dx Genetic Analyzer (Life-Technologies, Carlsbad, CA, USA). Coffalyser.Net software v.220513.1739 (MRC Holland - Amsterdam, the Netherlands) was employed for data analysis in combination with the lot-specific MLPA Coffalyser sheet. The following cut-offs for dosage quotient (DQ) of the probes were applied to interpret the MLPA results: 0.80 < DQ < 1.20 (no deletion/duplication), DQ = 0 (homozygous deletion), 0.40 < DQ < 0.65 (heterozygous deletion), 1.30 > DQ > 1.65 (heterozygous duplication), and 1.75 < DQ < 2.15 (homozygous duplication/heterozygous triplication).
2.5. LR-PCR and STRC Sequencing
LR-PCR was performed on all patients carrying a heterozygous STRC deletion and presenting an audiometric pattern suggestive of Deafness, autosomal recessive 16 (MIM: # 603720), due to biallelic variants in the STRC gene. Considering the presence of a highly homologous p-STRC, in order to avoid p-STRC sequences, two LR-PCR products were generated for subsequent nested PCR using the UltraRun LongRange PCR Kit (Qiagen, Venlo, The Netherlands), as described by Vona et al. [12]. In particular, two LR-PCR fragments, the first from exon 1 to exon 19 and the second from exon 12 to exon 29, were obtained. They both overlap intron 18, which is a non-identity region, where only STRC presents three additional nucleotides. After amplification, a 1% agarose gel was employed to verify the fragments’ length and PCR products were diluted 1:1000 to decrease pseudogene carryover. Intron 18 was therefore amplified and sequenced to confirm pseudogene exclusion. After ensuring the exclusive amplification of STRC, nested PCRs and Sanger sequencing of all exons were performed.
2.6. WES and Data Analysis
WES was performed on an Illumina NextSeq 550 instrument (Illumina Inc., San Diego, CA, USA) using the Twist Human Core Exome and Human RefSeq Panel kit (Twist Bioscience, South San Francisco, CA, USA) according to the manufacturer’s protocol. Briefly, genomic DNA was enzymatically fragmented, ligated to a universal adapter, and amplified using the Unique Dual Index primers (Twist Bioscience, South San Francisco, CA, USA). Samples were therefore hybridised with the Twist Human Core Exome and the Human RefSeq Panel kit, which cover 99% of protein-coding genes. Hybridised fragments were captured, amplified, and sequenced.
The process allows the production of FASTQ files that were analysed through a custom pipeline (Germline-Pipeline) developed by enGenome s.r.l. (https://www.engenome.com/, Pavia, Italy, accessed on 25 January 2023). This pipeline comprises several steps, such as FASTQ trimming, FASTQ quality check, FASTQ mapping, marking of duplicates, base quality score recalibration, and variant calling, thus permitting the identification of germline variants, including single nucleotide variants (SNVs), short insertions/deletions (INDELs), and exon-level CNVs starting from sequence reads. The secondary analysis, therefore, leads to the generation of final VCF files that contain SNVs, INDELs, and CNVs.
VCF files were analysed through the enGenome Expert Variant Interpreter (eVai) software (https://evai.engenome.com/, Pavia, Italy, accessed on 25 January 2023), which allows variant annotation, interpretation, and prioritisation. In particular, eVai completes the prioritisation process by exploiting both artificial intelligence and the American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG/AMP) guidelines to analyse and classify genomic variants [13].
The variant frequency was verified both in NCBI dbSNP build 155 (https://www.ncbi.nlm.nih.gov/snp/, accessed on 25 January 2023) and gnomAD (https://gnomad.broadinstitute.org/, accessed on 25 January 2023) in order to exclude variants previously reported as polymorphisms. The pathogenicity of already-reported variants was assessed through the Human Gene Mutation Database® (HGMD®) (https://my.qiagendigitalinsights.com/bbp/view/hgmd/pro/start.php, accessed on 25 January 2023), Deafness Variation Database (http://deafnessvariationdatabase.org/, accessed on 25 January 2023), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/, accessed on 25 January 2023). All databases were last accessed on 25 January 2023. The effect of all identified variants was evaluated through in silico prediction tools, such as PolyPhen-2 [14], Sorting Intolerant From Tolerant (SIFT) [15], Pseudo Amino Acid Protein Intolerance Variant Predictor (PaPI score) [16], Deep Neural Network Variant Predictor (DANN score) [17], and dbscSNV score [18]. SNVs leading to synonymous amino acid substitutions not predicted as damaging, not affecting splicing, or highly conserved residues were excluded; variants with a quality score (QUAL) < 20 or called in off-target regions were excluded as well.
Variants were discussed within a multidisciplinary team to assess whether they could be possibly matched to each patient’s phenotype; all variants of interest were confirmed with Sanger sequencing. Familial segregation of the identified variants was also performed using Sanger sequencing.
2.7. Prevalence of USH2A and ADGRV1 Pathogenic Variant Carriers in Italian Cohorts
One thousand two hundred eleven individuals from three different Italian cohorts were included in this analysis: (1) the Friuli-Venezia Giulia (FVG) cohort, which includes a collection of samples from six villages (Clauzetto, Erto, Illegio, Resia, San Martino del Carso, and Sauris) located in northeastern Italy [19]; (2) the Val Borbera (VBI) cohort, which consists of samples collected in a geographically isolated valley in the northwest of Italy [19]; (3) the Carlantino (CAR) cohort, which collects samples from a small village located in the Puglia region in southern Italy [19]. Whole genome sequencing (WGS) and detailed phenotypic data of these subjects were already available as an in-house database as a result of previous studies [20]. Only healthy individuals with available WGS data were considered; specifically, 663 individuals were selected from the FVG cohort, 424 from the VBI cohort, and 124 from the CAR cohort. WGS data were generated and analyzed in each cohort separately, as previously described by Cocca et al. [19].
USH2A and ADGRV1 gene variants were extracted considering a minor allele frequency of 5%. Functional annotation was performed using the Variant Effect Predictor tool [21] and frameshift, splice acceptor, splice donor, start lost, stop gained, and stop lost variants were extracted from the generated data using bcftools’ plug-in “Split-VEP” [22]. The pathogenicity of the extracted variants was assessed through HGMD®.
The frequency of the extracted variants was checked in Non-Finnish Europeans in the GnomAD database (https://gnomad.broadinstitute.org/, accessed on 25 January 2023). A sample test of proportions was performed to compare the pathogenic variant carrier frequency of the USH2A and ADGRV1 genes in our cohorts with the prevalence reported in the literature. The statistical significance was set to p-value < 0.05. This analysis was performed with R version 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria).
3. Results
In the last 18 months, 102 patients affected by HHL and negative in GJB2, GJB6 and MT-RNR1 analyses were recruited and analysed through a multistep and integrative approach (Figure 1) that comprises: (1) a detailed clinical characterisation to distinguish NSHL patients from SHL ones; (2) the analysis of STRC-CATSPER2 and OTOA deletions in patients affected by NSHL; (3) STRC sequencing in patients carrying a heterozygous deletion of STRC and an audiometric pattern suggestive of Deafness, autosomal recessive 16; (4) WES analysis in patients negative to steps 2 and 3 and in SHL subjects. The clinical evaluation allowed the identification of 82 patients affected by apparent NSHL and 20 presenting SHL.
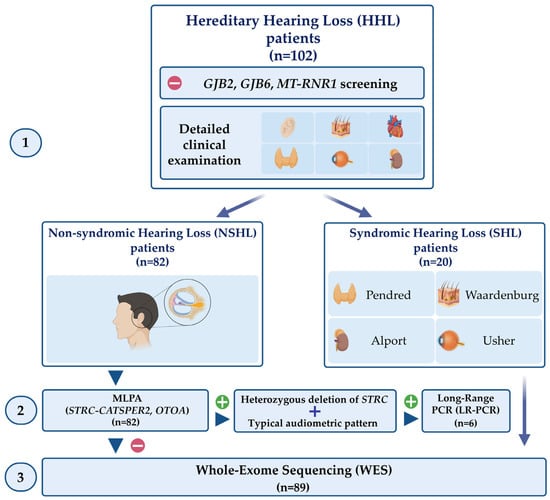
Figure 1.
Schematic representation of the multistep and integrative approach performed in this study. One-hundred and two patients affected by HHL and negative in GJB2, GJB6, and MT-RNR1 screening underwent a detailed clinical examination (1) aimed to differentiate NSHL patients from SHL ones. The patients affected by NHSL were screened for deletions in the STRC-CATSPER2 and OTOA genes (2). For the NSHL patients carrying a heterozygous deletion of STRC with a typical audiometric pattern, STRC sequencing was performed. WES was carried out in all NSHL patients negative for these steps and in SHL patients. Created with BioRender.com (accessed on 25 January 2023).
3.1. STRC Analysis
All NSHL patients first underwent MLPA analysis of STRC-CATSPER2 and OTOA genes in order to identify possible deletions. Six patients (7.3%) carried a homozygous deletion of the entire STRC gene and one patient (Patient 3) presented an entire STRC gene deletion in compound heterozygosity with a deletion involving only exon 19 of the same gene; none of the analysed subjects presented a biallelic deletion involving OTOA (Table 1).

Table 1.
STRC deletions identified through MLPA analysis. Gene: gene carrying the deletion. Type of deletion: entire-gene deletion or specific exon(s) deletion. Genotype: homozygous: the same variant is present on both alleles; compound heterozygous: two different variants are present on each allele. Inheritance: inheritance pattern of the deletion established after parental segregation.
Additionally, six patients (Patients 8, 9, 10, 11, 12, and 13) were carriers of a heterozygous deletion of STRC. Parental segregation of the deletion showed that it had been inherited from the father in four cases and from the mother in the remaining two (Table 2). Five of them (Patients 8, 9, 10, 11, and 12) had previously undergone PTA and their bone conduction audiograms are reported in Figure 2: they all presented an audiometric pattern compatible with Deafness, autosomal recessive 16 (MIM: # 603720), which is characterised by mild-to-moderate bilateral hearing impairment [4]. An ABR test had been performed on one patient (Patient 13) due to her age and revealed a threshold compatible with moderate HL. In these patients, in consideration of their clinical features and the suggestive presence of a heterozygous STRC deletion, LR-PCR analysis of the STRC gene (NM_153700.2) was performed with the final aim of identifying a possible hemizygous in trans variant that could explain their clinical phenotype. Indeed, in all of them, a hemizygous variant was identified and parental segregation showed that it had been maternally inherited in four of them and paternally in the last two cases, thus confirming their presence in trans with the previously identified gene deletion and allowing the achievement of a molecular diagnosis in an additional 7.3% of cases (Table 2).

Table 2.
STRC deletions and SNVs identified in the six patients that underwent both MLPA and LR-PCR analyses. Gene: gene carrying the deletion or the identified variants with NCBI RefSeq accession number of the considered protein-coding transcripts (NM_). cDNA change and Protein change: variant description according to the Human Genome Variation Society (HGVS) nomenclature guidelines. Genotype: compound heterozygous: two different variants are present on each allele. Inheritance: inheritance pattern of every identified variant established after parental segregation. ACMG/AMP classification: variant pathogenicity according to ACMG/AMP guidelines. References: PubMed unique IDentifier (PMID) of publications reporting each variant. *: stop codon. NA: not available.
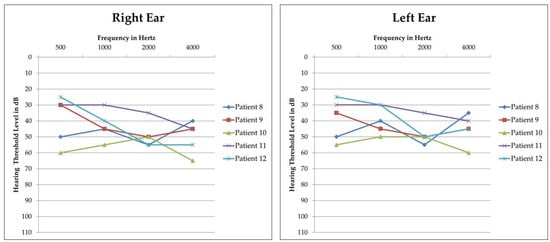
Figure 2.
Audiograms of patients carrying a heterozygous STRC deletion and a hemizygous pathogenic or likely pathogenic in trans variant. Threshold of the right and left ear of each patient is reported separately. All patients display mild-to-moderate bilateral sensorineural HL with a symmetric pattern between the right and left ear. Patient 13 did not undergo PTA but only ABR, due to her age.
3.2. WES Analysis in NSHL Patients
Sixty-nine patients affected by apparent NSHL and negative in steps 2 and 3 of the diagnostic workflow underwent in trio WES analysis. A molecular diagnosis was confirmed in 26 cases (37.9%) (Table 3).

Table 3.
Variants identified through WES analysis in apparent NSHL patients. Gene: list of genes carrying the identified variants with NCBI RefSeq accession number of the considered protein-coding transcripts (NM_). Associated disease: genetic disorder nomenclature according to the Online Mendelian Inheritance in Man® (OMIM®) database. Transmission pattern: AD: autosomal dominant; AR: autosomal recessive. cDNA change and Protein change: variant description according to the Human Genome Variation Society (HGVS) nomenclature guidelines. Genotype: homozygous: the same variant is present on both alleles; heterozygous: the variant affects only one allele; compound heterozygous: two different variants are present on each allele. Inheritance: inheritance pattern of every identified variant established after parental segregation. ACMG/AMP classification: variant pathogenicity according to ACMG/AMP guidelines. References: PubMed unique IDentifier (PMID) of publications reporting each variant. *: stop codon. NA: not available.
Among NSHL patients that received a molecular diagnosis, 10/26 were affected by an isolated AR form of HL, underlining how recessive HL is the most common hereditary deafness, in line with previous studies reported in the literature [23]. These cases are represented by Patients 16, 19, 20, 21, 24, 27, 29, 32, 38, and 39. Alongside the frequent forms of AR NSHL, such as MYO15A- and TMPRSS3-related [24], variants in rarely mutated genes were also identified, as the cases of Patient 21, carrying a homozygous splicing variant in the LRTOMT gene, and Patient 27, presenting two compound heterozygous variants in the CLDN14 gene. Additionally, 2/26 patients were carriers of biallelic variants in OTOF, which is associated with auditory neuropathy, autosomal recessive 1 (MIM: # 601071). Conversely, only 4/26 patients were affected by AD NSHL, due to heterozygous variants in the CECAM16, MYH14, KCNQ4, and PLS1 genes, respectively.
Furthermore, WES analysis allowed the identification of two interesting events: the presence of eight cases of non-syndromic mimics and the occurrence of two patients affected by a dual molecular diagnosis.
Among subjects affected by non-syndromic mimics, Patients 15, 17, and 34 may be included. Patient 15 presents a de novo heterozygous variant in the GATA3 gene, whose pathogenic variants are associated with hypoparathyroidism, sensorineural deafness, and renal dysplasia syndrome (MIM: # 146255). Currently, the five-year-old girl only shows HL; no signs of renal dysplasia are visible upon abdominal ultrasound and calcium and parathormone levels are within the reference range. Patient 17 carries two compound heterozygous variants in the HARS2 gene, which is associated with Perrault syndrome 2 (MIM: # 614926). The patient, a two-year-old girl, currently presents only HL. Additionally, Patient 34, a 17-month-old boy whose HL was the only feature detected during the initial clinical evaluation, presents a known [25] heterozygous de novo pathogenetic variant in the MITF gene, causative of Waardenburg syndrome type 2a (MIM: # 193510). Furthermore, this cohort was strikingly enriched in patients affected by Usher syndrome type 2 that at present only show HL and have not developed retinitis pigmentosa yet. Indeed, they represent 5/26 (19.2%) of the analysed subjects, namely Patients 30 and 33, who are carriers of biallelic variants in the ADGRV1 gene, and Patients 35, 36, and 37, carriers of biallelic variants in the USH2A gene.
Patients 14 and 26 were affected by a dual molecular diagnosis: both of them carry a heterozygous variant in a gene associated with NSHL (respectively, ATP2B2—Deafness, autosomal dominant 82, MIM: # 619804, and COL11A1—Deafness, autosomal dominant 37, MIM: # 618533) and a heterozygous variant in a gene associated with Waardenburg syndrome (respectively, EDN3—Waardenburg syndrome type 4, MIM: # 613265, PAX3—Waardenburg syndrome type 1, MIM: # 193500).
Finally, WES analysis of Patient 39, a 6-year-old boy affected by bilaterally symmetric moderate sensorineural HL, highlighted the presence of two in trans variants in the STRC gene. In consideration of this result, LR-PCR analysis was performed to overcome the p-STRC issue and, indeed, it confirmed the molecular diagnosis, further corroborated by the clinical phenotype of the proband.
3.3. WES Analysis in SHL Patients
Twenty patients affected by SHL underwent WES analysis and a molecular diagnosis was identified in 11 of them (55%) (Table 4).

Table 4.
Variants identified through WES analysis in SHL patients. Gene: list of genes carrying the identified variants with NCBI RefSeq accession number of the considered protein-coding transcripts (NM_). Associated disease: genetic disorder nomenclature according to the Online Mendelian Inheritance in Man® (OMIM®) database. Transmission pattern: AD: autosomal dominant; AR: autosomal recessive; XL: X-linked. cDNA change and Protein change: variant description according to the Human Genome Variation Society (HGVS) nomenclature guidelines. Genotype: homozygous: the same variant is present on both alleles; heterozygous: the variant affects only one allele; compound heterozygous: two different variants are present on each allele. Inheritance: inheritance pattern of every identified variant established after parental segregation. ACMG/AMP classification: variant pathogenicity according to ACMG/AMP guidelines. References: PubMed unique IDentifier (PMID) of publications reporting each variant. *: stop codon. NA: not available.
Five out of 11 patients that were molecularly diagnosed were affected by Usher syndrome type 1 (Patients 44, 48, and 49) or type 2 (Patients 41 and 45); overall, the prevalence of Usher syndrome among the 20 analysed syndromic patients was 25%, confirming the high frequency of this condition in HHL cohorts. Two patients were diagnosed with relatively frequent forms of SHL and both were already suspected upon the initial clinical evaluation: Patient 40 was affected by Charge syndrome (MIM: # 214800) and Patient 43 by branchiootorenal syndrome 1 (MIM: # 602588).
Additionally, two patients were carriers of heterozygous variants in two recently described genes. In particular, Patient 42 presented a heterozygous stop-gain variant in the SF3B2 gene, whose pathogenic variants were reported in 2021 to be causative of craniofacial microsomia (MIM: # 164210) [26]. Indeed, Patient 42 showed facial asymmetry, mandibular hypoplasia, asymmetric ear abnormalities resembling a question mark ear, bilateral preauricular tags, and a pit along the branchial arch, together with bilateral conductive hearing loss, thus matching the phenotype of the other described patients. Furthermore, Patient 47 carried a heterozygous splicing variant in the PPP1R12A gene, which was described in 2020 as mutated in subjects affected by genitourinary and/or/brain malformation syndrome (MIM: # 618820) [27]. This condition is characterised by abnormal internal and external genitalia, structural renal abnormalities, brain malformations, and eye and skeletal anomalies. Patient 47 showed right megaureter and ureterocele, unilateral myopia of the right eye, and symmetric profound sensorineural HL; the latter has not been reported in any of the twelve patients described in the literature.
Finally, among SHL patients, two subjects affected by a dual molecular diagnosis were also identified. Patient 46, affected by congenital bilateral profound sensorineural HL and retinitis pigmentosa, was a carrier both of the most frequent variants associated with NSHL (c.35delG, p.(Gly12Valfs * 2) in the GJB2 gene [28]) and of a novel heterozygous frameshift variant in the RPGR gene, which is causative of X-linked retinitis pigmentosa 3 (MIM: #300029). WES analysis of Patient 50, presenting severe bilateral sensorineural HL and cutaneous signs of pseudoxanthoma elasticum, highlighted the presence of a known heterozygous variant in the WFS1 gene [29], associated with Deafness, autosomal dominant 6/14/38 (MIM: # 600965) and an already-reported homozygous variant in the ABCC6 gene [30], causative of pseudoxanthoma elasticum (MIM: # 264800).
3.4. Prevalence of USH2A and ADGRV1 Pathogenic Variant Carriers in Italian Cohorts
A noteworthy feature of this study is represented by a peculiar enrichment in patients affected by Usher syndrome type 2, carrying biallelic variants in the USH2A (4/7) and ADGRV1 (3/7) genes.
The estimated frequency of pathogenic variant carriers in the USH2A gene is approximately 1:70 in the general population [31] and no data are available regarding ADGRV1 pathogenic variant carriers. Furthermore, the prevalence of USH2A and ADGRV1 pathogenic variant carriers in the Italian population is unknown. In this light, the carrier frequency of pathogenic variants within the USH2A and ADGRV1 genes in the Italian population was evaluated, taking advantage of WGS data of 1211 individuals from three different Italian cohorts, namely FVG, VBI, and CAR. Pathogenic variants within USH2A and ADGRV1 genes were extracted and checked through HGMD®. The complete list of the extracted variants for each cohort is reported in Table S1.
Regarding USH2A, heterozygous pathogenic variants were detected in 22/663 (3.3%) individuals of the FVG cohort, in 3/424 (0.70%) subjects of the VBI cohort, and in 8/124 (6.5%) individuals belonging to the CAR cohort. In these Italian cohorts, the overall carrier frequency of pathogenic variants in USH2A was 33/1211 (2.7%). Concerning the ADGRV1 gene, heterozygous pathogenic variants were identified in 23/663 (3.5%) individuals of the FVG cohort, in 5/424 (1.2%) subjects of the VBI cohort, and in 3/124 (2.4%) individuals belonging to the CAR cohort. The overall prevalence of ADGRV1 pathogenic variant carriers in these Italian cohorts was 31/1211 (2.6%). To compare the frequency of USH2A pathogenic variant carriers in our cohorts with the available literature data, a sample test of proportions was performed. The statistical analysis result revealed that the frequency of pathogenic variant carriers in the USH2A gene in these Italian cohorts was statistically significantly higher (p-value = 0.0001436) with respect to the available literature data [31].
4. Discussion
The multistep and integrated diagnostic workflow described in this study was an effective tool to provide HHL patients with a molecular diagnosis. Indeed, regarding NSHL patients, STRC MLPA and LR-PCR analyses allowed us to solve 13 cases, while WES explained 26 additional ones: overall, the detection rate of this approach was 47.6%. Concerning SHL subjects, WES analysis was fundamental to highlight the molecular cause in 55% of patients. These results are in line with previous studies, from both Italian cohorts and international ones, that verified how the overall diagnostic yield in HHL patients is around 50% [32,33,34,35].
The STRC gene turned out to be a significant contributor to AR NSHL also in this cohort, as previous works already reported [4]; in particular, the incidence of STRC deletions has been estimated at between 1% and 5% in HL patients [36]. Conversely, STRC SNVs have been reported only in a few cohorts [12] due to the complexity of interpretation of STRC sequencing data secondary to the presence of p-STRC. Despite the fact that a detailed genotype–phenotype correlation between STRC loss-of-function and mild-to-moderate HL has been reported in studies involving a limited number of participants, this correspondence seems extremely consistent in all of them [4,37,38,39]. In this light, diagnostic tools able to highlight point mutations in this gene overcoming the pseudogene issue should be implemented in all laboratories specialised in the diagnostic work-up of HHL. Indeed, in our cohort, LR-PCR analysis in six patients with specific audiometric features (i.e., mild-to-moderate HL) and the suggestive presence of a heterozygous STRC deletion led to the identification of an in trans hemizygous variant in all of them. Additionally, this diagnostic technique confirmed the presence of the two SNVs in the STRC gene detected by WES analysis in Patient 39. This finding, in particular, underlines how the possibility to perform STRC sequencing in all patients with mild-to-moderate HL, even without a heterozygous STRC deletion, should be considered to increase the diagnostic yield in this specific subgroup of HHL patients.
Concerning WES analysis in both NSHL and SHL patients, the results of the present study confirm the role of few genes as major players in both categories, such as MYO15A and TMPRSS3 in NSHL patients [24] and CDH23 in SHL ones [40].
Within NSHL subjects, patients affected by non-syndromic mimics represented 8/26 cases (30.8%). Non-syndromic mimics may be divided into two categories: (1) patients that present subtle signs and symptoms attributable to the identified syndrome that were missed during the initial clinical evaluation; (2) patients that currently present only one sign/symptom and may develop other characteristics later in life. Within the first group, Patient 34, a carrier of a known heterozygous variant in the MITF gene, might be included: during the initial clinical evaluation, only HL was detected, but he will undergo further assessment upon readmission to identify possible subtle signs of Waardenburg syndrome, such as mild pigmentary abnormalities of the skin, eyes, and hair. Conversely, in the second group, Patient 15, a carrier of a novel heterozygous variant in the GATA3 gene associated with hypoparathyroidism, sensorineural deafness, and renal dysplasia syndrome, will need to be constantly monitored over time to unveil possible early signs of hypoparathyroidism. Patient 17, presenting two compound heterozygous novel variants in the HARS2 gene causative of Perrault syndrome, also belongs to the second group and clinical features such as primary amenorrhea and infertility would only be evaluated when she reaches the pubertal stage.
Additionally, a peculiar finding of this study is represented by the overall identification of seven patients (7/102—6.9%) affected by Usher syndrome type 2 due to biallelic variants in the USH2A (4/7) and ADGRV1 (3/7) genes. These patients have been identified both among subjects presenting with apparent NSHL (5/7) and within SHL patients (2/7). In the NSHL group, they qualify as non-syndromic mimics [10], as the identified subjects currently present only HL and have not yet developed retinitis pigmentosa, which is an age-dependent feature. In these cases, the identification of the correct molecular diagnosis before the onset of vision loss is fundamental to prompt a specific clinical follow-up. For instance, periodic ophthalmological evaluations should be planned and, whenever patients develop severe progressive HL with poor speech discrimination and communication difficulties even with hearing aids, making these children excellent cochlear implant candidates, surgery should be performed before the onset of ocular problems to maximise communication skills [41]. In the SHL group, Patient 41 came to the geneticist’s attention at 48 years of age and had already developed vision loss together with HL: in this case, the clinical diagnosis was straightforward and thus confirmed by the molecular one. Conversely, Patient 45, a 5-year-old boy, presented HL and mild facial dysmorphisms, such as frontal bossing and dermal translucency of the head: in this subject, the identification of the molecular diagnosis was able to explain his HL and prompt the appropriate ophthalmological follow-up, but was not able to fully explain his facial features.
The prevalence of Usher syndrome type 2 patients in our cohort is slightly higher than expected [31] and, to date, no published study has so far analysed the prevalence of USH2A and ADGRV1 pathogenic variant carriers in the Italian population. Knowing the exact prevalence of Usher syndrome type 2 carriers within a specific ethnic group is fundamental for physicians to plan tailored healthcare intervention in everyday clinical practice. In this light, the frequency of USH2A and ADGRV1 pathogenic variant carriers in the Italian population was evaluated, taking advantage of already available in-house WGS data of 1211 healthy individuals belonging to three different Italian cohorts. Bioinformatic analyses revealed that the frequency of pathogenic variant carriers in the Italian population is around 2:70, for both the USH2A and ADGRV1 genes. Notably, the frequency of USH2A pathogenic variant carriers was statistically significantly higher (p-value < 0.05) compared with previous studies [31]. It has to be considered that the available literature data about USH2A pathogenic variant carrier frequency in the worldwide population is limited and not updated. In this light, further epidemiological data will be needed to accurately evaluate the prevalence of USH2A pathogenic variant carriers across different ethnic populations. Additionally, for the first time, the prevalence of ADGRV1 variant carriers in the Italian population was provided. These data could be considered as a starting point for a better and tailored clinical management: indeed, providing a more accurate picture of Usher syndrome type 2 carrier frequency could contribute, in a long-term perspective, to a more effective implementation of diagnostic and follow-up strategies in the clinical practice.
Finally, in both NSHL and SHL patients, two cases of dual molecular diagnoses have been identified: overall, four out of 89 patients (4.5%) that underwent WES analysis presented a multilocus genomic variation, in line with previous studies [42]. Notably, Patients 14 and 26 both present a variant in a gene associated with NSHL and a variant in a gene associated with Waardenburg syndrome, even if, from a clinical point of view, both subjects were classified as apparently NSHL cases. Waardenburg syndrome is characterised by HL together with pigmentation abnormalities of the skin, hair, and eyes, and other possible features, such as dystopia canthorum, skeletal abnormalities, or Hirschsprung disease [43]. Some of these clinical features might be subtle and, as already discussed, Waardenburg syndrome may also be a non-syndromic mimic. In this light, a thorough clinical evaluation of both patients and their fathers (from whom they inherited the variants) is planned in the following months. Conversely, both patients that were first diagnosed as SHL cases were found to be affected by NSHL together with a second genetic disorder. Indeed, Patient 46 presents GJB2-associated HL and RPGR-associated retinitis pigmentosa while Patient 50 is affected by WFS1-related HL and pseudoxanthoma elasticum due to an ABCC6 mutation. In the first case, the patient was initially suspected to have a single condition that could explain all her clinical features, while the in the second patient the presence of two different genetic disorders was promptly hypothesised upon the clinical evaluation. These two cases underline how sometimes the clinical diagnosis of two genetic disorders is straightforward while in others only a molecular analysis can provide a definitive explanation of patients’ signs and symptoms.
5. Conclusions
In conclusion, this study underlines how the molecular landscape behind HHL is extremely complex, given the high genetic and clinical heterogeneity of this phenotype. Additionally, some peculiar scenarios are emerging, such as the occurrence of non-syndromic mimics and multilocus genomic variation, further challenging the achievement of a correct molecular diagnosis. In this context, a multidisciplinary approach that combines a comprehensive clinical evaluation together with a careful analysis of genomic data is the key to provide patients and their families with precise information about the prognosis, treatment, follow-up, and recurrence risks, thus representing the essential basis of a personalised clinical management.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines11030703/s1, Table S1: Pathogenic variants within USH2A and ADGRV1 genes identified in Italian cohorts.
Author Contributions
Conceptualization, B.S., A.M. and G.G.; methodology, A.S., A.M., G.G.N. and S.L.; validation, S.L. and A.M.; formal analysis, B.S., A.S., A.M., G.G.N. and S.L.; investigation, B.S., C.G., L.G., S.M., S.B. and E.R.; resources, B.S., A.S., A.M. and G.G.; data curation, B.S., A.S., A.M. and G.G.N.; writing—original draft preparation, B.S. and A.S.; writing—review and editing, A.M. and G.G.; visualization, B.S. and A.S.; supervision, G.G.; project administration, G.G.; funding acquisition, G.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by D70-RESRICGIROTTO and BENEFICENTIA Stiftung to G.G., and by the Italian Ministry of Health, through the contribution given to the Institute for Maternal and Child Health I.R.C.C.S. “Burlo Garofolo”, Trieste, Italy (Ricerca Corrente L3_14\22).
Institutional Review Board Statement
The study was conducted in accordance with the tenets of the Helsinki Declaration and was approved by the Ethics Committee of the I.R.C.C.S. “Burlo Garofolo” of Trieste.
Informed Consent Statement
Written informed consent was obtained from all participants involved in the study or their legal guardians.
Data Availability Statement
Data presented in this study are available upon request to the corresponding author. Data are not publicly available due to privacy restrictions.
Acknowledgments
This work has been generated within the European Reference Network on Rare Congenital Malformations and Rare Intellectual Disability (ERN-ITHACA) [EU Framework Partnership Agreement ID: 3HP-HP-FPA ERN-01-2016/739516]. The authors also wish to thank all the patients and their families for their keen participation in the study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Posey, J.E.; O’Donnell-Luria, A.H.; Chong, J.X.; Harel, T.; Jhangiani, S.N.; Coban Akdemir, Z.H.; Buyske, S.; Pehlivan, D.; Carvalho, C.M.B.; Baxter, S.; et al. Insights into Genetics, Human Biology and Disease Gleaned from Family Based Genomic Studies. Genet. Med. 2019, 21, 798–812. [Google Scholar] [CrossRef] [PubMed]
- Korver, A.M.H.; Smith, R.J.H.; Van Camp, G.; Schleiss, M.R.; Bitner-Glindzicz, M.A.K.; Lustig, L.R.; Usami, S.-I.; Boudewyns, A.N. Congenital Hearing Loss. Nat. Rev. Dis. Prim. 2017, 3, 16094. [Google Scholar] [CrossRef] [PubMed]
- Bademci, G.; Cengiz, F.B.; Foster, J.; Duman, D.; Sennaroglu, L.; Diaz-Horta, O.; Atik, T.; Kirazli, T.; Olgun, L.; Alper, H.; et al. Variations in Multiple Syndromic Deafness Genes Mimic Non-Syndromic Hearing Loss. Sci. Rep. 2016, 6, 31622. [Google Scholar] [CrossRef] [PubMed]
- Ito, T.; Kawashima, Y.; Fujikawa, T.; Honda, K.; Makabe, A.; Kitamura, K.; Tsutsumi, T. Rapid Screening of Copy Number Variations in STRC by Droplet Digital PCR in Patients with Mild-to-Moderate Hearing Loss. Hum. Genome Var. 2019, 6, 41. [Google Scholar] [CrossRef]
- Ideura, M.; Nishio, S.; Moteki, H.; Takumi, Y.; Miyagawa, M.; Sato, T.; Kobayashi, Y.; Ohyama, K.; Oda, K.; Matsui, T.; et al. Comprehensive Analysis of Syndromic Hearing Loss Patients in Japan. Sci. Rep. 2019, 9, 11976. [Google Scholar] [CrossRef]
- Gettelfinger, J.; Dahl, J. Syndromic Hearing Loss: A Brief Review of Common Presentations and Genetics. J. Pediatr. Genet. 2018, 07, 001–008. [Google Scholar] [CrossRef]
- Faridi, R.; Rea, A.; Fenollar-Ferrer, C.; O’Keefe, R.T.; Gu, S.; Munir, Z.; Khan, A.A.; Riazuddin, S.; Hoa, M.; Naz, S.; et al. New Insights into Perrault Syndrome, a Clinically and Genetically Heterogeneous Disorder. Hum. Genet. 2022, 141, 805–819. [Google Scholar] [CrossRef]
- Barakat, A.J.; Raygada, M.; Rennert, O.M. Barakat Syndrome Revisited. Am. J. Med. Genet. Part A 2018, 176, 1341–1348. [Google Scholar] [CrossRef]
- Morgan, A.; Faletra, F.; Severi, G.; La Bianca, M.; Licchetta, L.; Gasparini, P.; Graziano, C.; Girotto, G. There Is More Than Meets the Eye: Identification of Dual Molecular Diagnosis in Patients Affected by Hearing Loss. Biomedicines 2022, 10, 12. [Google Scholar] [CrossRef]
- Gooch, C.; Rudy, N.; Smith, R.J.; Robin, N.H. Genetic Testing Hearing Loss: The Challenge of Non Syndromic Mimics. Int. J. Pediatr. Otorhinolaryngol. 2021, 150, 110872. [Google Scholar] [CrossRef]
- Clark, J.G. Uses and Abuses of Hearing Loss Classification. ASHA 1981, 23, 493–500. [Google Scholar] [PubMed]
- Vona, B.; Hofrichter, M.A.H.; Neuner, C.; Schröder, J.; Gehrig, A.; Hennermann, J.B.; Kraus, F.; Shehata-Dieler, W.; Klopocki, E.; Nanda, I.; et al. DFNB16 Is a Frequent Cause of Congenital Hearing Impairment: Implementation of STRC Mutation Analysis in Routine Diagnostics. Clin. Genet. 2015, 87, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.; Jordan, D.M.; Sunyaev, S.R. Predicting Functional Effect of Human Missense Mutations Using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 7, 20. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting Amino Acid Changes That Affect Protein Function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Limongelli, I.; Marini, S.; Bellazzi, R. PaPI: Pseudo Amino Acid Composition to Score Human Protein-Coding Variants. BMC Bioinform. 2015, 16, 123. [Google Scholar] [CrossRef]
- Quang, D.; Chen, Y.; Xie, X. DANN: A Deep Learning Approach for Annotating the Pathogenicity of Genetic Variants. Bioinformatics 2015, 31, 761–763. [Google Scholar] [CrossRef]
- Jian, X.; Boerwinkle, E.; Liu, X. In Silico Prediction of Splice-Altering Single Nucleotide Variants in the Human Genome. Nucleic Acids Res. 2014, 42, 13534–13544. [Google Scholar] [CrossRef]
- Cocca, M.; Barbieri, C.; Concas, M.P.; Robino, A.; Brumat, M.; Gandin, I.; Trudu, M.; Sala, C.F.; Vuckovic, D.; Girotto, G.; et al. A Bird’s-Eye View of Italian Genomic Variation through Whole-Genome Sequencing. Eur. J. Hum. Genet. 2020, 28, 435–444. [Google Scholar] [CrossRef]
- Spedicati, B.; Cocca, M.; Palmisano, R.; Faletra, F.; Barbieri, C.; Francescatto, M.; Mezzavilla, M.; Morgan, A.; Pelliccione, G.; Gasparini, P.; et al. Natural Human Knockouts and Mendelian Disorders: Deep Phenotyping in Italian Isolates. Eur. J. Hum. Genet. 2021, 29, 1272–1281. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Imtiaz, A. ARNSHL Gene Identification: Past, Present and Future. Mol. Genet. Genom. 2022, 297, 1185–1193. [Google Scholar] [CrossRef]
- Duman, D.; Tekin, M. Autosomal Recessive Nonsyndromic Deafness Genes: A Review. Front. Biosci. Landmark Ed. 2012, 17, 2213–2236. [Google Scholar] [CrossRef] [PubMed]
- Pingault, V.; Ente, D.; Dastot-Le Moal, F.; Goossens, M.; Marlin, S.; Bondurand, N. Review and Update of Mutations Causing Waardenburg Syndrome. Hum. Mutat. 2010, 31, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Timberlake, A.T.; Griffin, C.; Heike, C.L.; Hing, A.V.; Cunningham, M.L.; Chitayat, D.; Davis, M.R.; Doust, S.J.; Drake, A.F.; Duenas-Roque, M.M.; et al. Haploinsufficiency of SF3B2 Causes Craniofacial Microsomia. Nat. Commun. 2021, 12, 4680. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.J.; Alkhunaizi, E.; Kruszka, P.; Pyle, L.C.; Grange, D.K.; Berger, S.I.; Payne, K.K.; Masser-Frye, D.; Hu, T.; Christie, M.R.; et al. Loss-of-Function Variants in PPP1R12A: From Isolated Sex Reversal to Holoprosencephaly Spectrum and Urogenital Malformations. Am. J. Hum. Genet. 2020, 106, 121–128. [Google Scholar] [CrossRef]
- Chan, D.K.; Chang, K.W. GJB2-Associated Hearing Loss: Systematic Review of Worldwide Prevalence, Genotype, and Auditory Phenotype. Laryngoscope 2014, 124, E34–E53. [Google Scholar] [CrossRef]
- Marshall, B.A.; Permutt, M.A.; Paciorkowski, A.R.; Hoekel, J.; Karzon, R.; Wasson, J.; Viehover, A.; White, N.H.; Shimony, J.S.; Manwaring, L.; et al. Phenotypic Characteristics of Early Wolfram Syndrome. Orphanet J. Rare Dis. 2013, 8, 64. [Google Scholar] [CrossRef]
- Miksch, S.; Lumsden, A.; Guenther, U.P.; Foernzler, D.; Christen-Zäch, S.; Daugherty, C.; Ramesar, R.K.S.; Lebwohl, M.; Hohl, D.; Neldner, K.H.; et al. Molecular Genetics of Pseudoxanthoma Elasticum: Type and Frequency of Mutations in ABCC6. Hum. Mutat. 2005, 26, 235–248. [Google Scholar] [CrossRef]
- Koenekoop, R.; Arriaga, M.; Trzupek, K.M.; Lentz, J. Usher Syndrome Type II. 1999 Dec 10 [Updated 2020 Oct 22]. In GeneReviews® [Internet]; Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; National Organization for Rare Disorders: Seattle, WA, USA, 1993–2023. Available online: https://www.ncbi.nlm.nih.gov/books/NBK1341/ (accessed on 25 January 2023).
- Morgan, A.; Lenarduzzi, S.; Spedicati, B.; Cattaruzzi, E.; Murru, F.M.; Pelliccione, G.; Mazzà, D.; Zollino, M.; Graziano, C.; Ambrosetti, U.; et al. Lights and Shadows in the Genetics of Syndromic and Non-Syndromic Hearing Loss in the Italian Population. Genes 2020, 11, 1237. [Google Scholar] [CrossRef]
- Sang, S.; Ling, J.; Liu, X.; Mei, L.; Cai, X.; Li, T.; Li, W.; Li, M.; Wen, J.; Liu, X.; et al. Proband Whole-Exome Sequencing Identified Genes Responsible for Autosomal Recessive Non-Syndromic Hearing Loss in 33 Chinese Nuclear Families. Front. Genet. 2019, 10, 639. [Google Scholar] [CrossRef]
- Van Heurck, R.; Carminho-Rodrigues, M.T.; Ranza, E.; Stafuzza, C.; Quteineh, L.; Gehrig, C.; Hammar, E.; Guipponi, M.; Abramowicz, M.; Senn, P.; et al. Benefits of Exome Sequencing in Children with Suspected Isolated Hearing Loss. Genes 2021, 12, 1277. [Google Scholar] [CrossRef]
- Elander, J.; Ullmark, T.; Ehrencrona, H.; Jonson, T.; Piccinelli, P.; Samuelsson, S.; Löwgren, K.; Falkenius-Schmidt, K.; Ehinger, J.; Stenfeldt, K.; et al. Extended Genetic Diagnostics for Children with Profound Sensorineural Hearing Loss by Implementing Massive Parallel Sequencing. Diagnostic Outcome, Family Experience and Clinical Implementation. Int. J. Pediatr. Otorhinolaryngol. 2022, 159, 111218. [Google Scholar] [CrossRef]
- Yokota, Y.; Moteki, H.; Nishio, S.Y.; Yamaguchi, T.; Wakui, K.; Kobayashi, Y.; Ohyama, K.; Miyazaki, H.; Matsuoka, R.; Abe, S.; et al. Frequency and Clinical Features of Hearing Loss Caused by STRC Deletions. Sci. Rep. 2019, 9, 4408. [Google Scholar] [CrossRef] [PubMed]
- Markova, T.G.; Alekseeva, N.N.; Mironovich, O.L.; Galeeva, N.M.; Lalayants, M.R.; Bliznetz, E.A.; Chibisova, S.S.; Polyakov, A.V.; Tavartkiladze, G.A. Clinical Features of Hearing Loss Caused by STRC Gene Deletions/Mutations in Russian Population. Int. J. Pediatr. Otorhinolaryngol. 2020, 138, 110247. [Google Scholar] [CrossRef] [PubMed]
- Simi, A.; Perry, J.; Schindler, E.; Oza, A.; Luo, M.; Hartman, T.; Krantz, I.D.; Germiller, J.A.; Kawai, K.; Kenna, M. Audiologic Phenotype and Progression in Pediatric STRC-Related Autosomal Recessive Hearing Loss. Laryngoscope 2021, 131, E2897–E2903. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.J.; Oh, D.-Y.; Han, J.H.; Oh, J.; Kim, M.Y.; Park, H.-R.; Seok, J.; Cho, S.-D.; Lee, S.-Y.; Kim, Y.; et al. Significant Mendelian Genetic Contribution to Pediatric Mild-to-Moderate Hearing Loss and Its Comprehensive Diagnostic Approach. Genet. Med. Off. J. Am. Coll. Med. Genet. 2020, 22, 1119–1128. [Google Scholar] [CrossRef]
- Usami, S.; Isaka, Y.; Miyagawa, M.; Nishio, S. Variants in CDH23 Cause a Broad Spectrum of Hearing Loss: From Non-Syndromic to Syndromic Hearing Loss as Well as from Congenital to Age-Related Hearing Loss. Hum. Genet. 2022, 141, 903–914. [Google Scholar] [CrossRef]
- Davies, C.; Bergman, J.; Misztal, C.; Ramchandran, R.; Mittal, J.; Bulut, E.; Shah, V.; Mittal, R.; Eshraghi, A.A. The Outcomes of Cochlear Implantation in Usher Syndrome: A Systematic Review. J. Clin. Med. 2021, 10, 2915. [Google Scholar] [CrossRef]
- Posey, J.E.; Harel, T.; Liu, P.; Rosenfeld, J.A.; James, R.A.; Coban Akdemir, Z.H.; Walkiewicz, M.; Bi, W.; Xiao, R.; Ding, Y.; et al. Resolution of Disease Phenotypes Resulting from Multilocus Genomic Variation. N. Engl. J. Med. 2016, 376, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Song, J.; He, C.; Cai, X.; Yuan, K.; Mei, L.; Feng, Y. Genetic Insights, Disease Mechanisms, and Biological Therapeutics for Waardenburg Syndrome. Gene Ther. 2022, 29, 479–497. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).